Abstract
The polycomb group (PcG) genes encode a family of proteins that methylate and ubiquitinate histones to close chromatin and suppress gene expression. PcG proteins are present at elevated levels in cancer cells, and this is associated with reduced tumor suppressor protein level and enhanced cell survival. Agents that reduce PcG protein level are regarded as potentially cancer-preventative agents. Sulforaphane (SFN) is a biologically important isothiocyanate found in cruciferous vegetables that is an important candidate chemopreventive agent. However, the impact of SFN on the level and function of PcG proteins in skin cancer cells has not been assessed. We show that SFN treatment causes a concentration-dependent reduction in PcG protein (Bmi-1, Ezh2) expression in SCC-13 skin cancer cells and also reduces trimethylation of lysine 27 of histone H3. This is associated with accumulation of cells in G2/M phase; reduced levels of cyclin B1, cyclin A, cyclin dependent kinases 1 and 2; and increased p21Cip1 expression. Sulforaphane treatment also increases cleavage of procaspase 3, 8, and 9 and enhances PARP cleavage and apoptosis. Similar results are observed in other skin-derived cell immortalized and transformed cell lines. Forced expression of the Bmi-1 polycomb protein in SCC-13 cells reverses these effects. The SFN-dependent loss of Bmi-1 and Ezh2 is due to proteasome-associated degradation. These results suggest that dietary isothiocyanates may suppress cancer progression by reducing PcG protein level via a proteasome-dependent mechanism, thereby inhibiting PcG-dependent pro-survival epigenetic events.
Introduction
Polycomb group (PcG) proteins are an important group of epigenetic regulators that repress gene expression by modifying chromatin structure (Simon and Kingston, 2009; Mills, 2010). PcG proteins function as two complexes. The polycomb repressive complex (PRC) 2 includes four core proteins—Ezh2, Suz12, eed (embryonic ectoderm development), and retinoblastoma-binding protein p48. The catalytic subunit is Ezh2, a methyltransferase that trimethylates lysine 27 of histone H3 (H3K27) (Simon and Kingston, 2009). The complex contains three noncatalytic subunits, including Suz12, eed and retinoblastoma-binding protein p48. Suz12 and eed are required for optimal Ezh2 histone methyltransferase activity. Interaction of Suz12 and eed with Ezh2 results in a 1000-fold increase in Ezh2 catalytic activity, showing that the full complex is required for optimal H3K27me3 formation (Pasini et al., 2004). The core of the PRC1 protein complex includes Ring1B, Bmi-1, polyhomeotic homolog 1, and chromobox homolog (Simon and Kingston, 2009). The catalytic subunit Ring1B ubiquitinylates H2A-K119 and is optimally active in association with Bmi-1 (Li et al., 2006). An important role of the CBX protein is interaction with H3K27me3 to anchor the PRC1 complex to chromatin (Eckert et al., 2011). PRC1 ubiquitinylates H2A-K119 as part of the process leading to a closed chromatin state.
PcG protein expression is altered in cancer cells and tumors, and this is linked to increased cancer cell proliferation and survival (Mills, 2010; Eckert et al., 2011). For example, Bmi-1 level is increased in a host of cancer cell types (Mills, 2010) and can cooperate with myc to promote B- and T-cell lymphoma formation (Jacobs et al., 1999). Elevated Bmi-1 expression is associated with repression of the cyclin dependent kinase inhibitors p16ink4a and p19arf in several systems (Jacobs et al., 1999). Elevated Ezh2 expression is associated with poor prognosis and aggressive tumor metastasis (Kleer et al., 2003). Interfering with Ezh2 expression retards cell proliferation and induces cell cycle arrest at the G2/M transition in the primary human fibroblasts (Tang et al., 2004). Conversely, overexpression of Ezh2 shortens G1 and causes cell accumulation in S phase in cultured mouse embryonic fibroblasts (Bracken et al., 2003).
Epidemiological studies indicate that consumption of cruciferous vegetables may reduce epithelial cancer (van Poppel et al., 1999). Sulforaphane [SFN; 1-isothiocyanato-4-(methylsulfinyl) butane] is an abundant bioactive isothiocyanate found in broccoli and broccoli sprouts and is an important candidate chemopreventive agent (Clarke et al., 2008). It activates phase II detoxification enzymes such as glutathione transferase and suppresses phase I carcinogen activation enzymes (Brooks et al., 2001). SFN suppresses cancer cell proliferation and inhibits cancer formation in carcinogen-induced, genetic, and xenograft animal cancer models (Myzak and Dashwood, 2006). SFN suppresses cancer cell survival via a number of mechanisms (Clarke et al., 2008). Of particular interest in the present work are findings that SFN modulates epigenetic phenomena (Myzak and Dashwood, 2006). For example, Myzak et al. (2004) reported that SFN inhibits histone deacetylase activity and enhances p21Cip1 promoter-associated acetylated histone and p21Cip1 expression in human HCT116 colon cancer cells. Sulforaphane-dependent suppression of histone deacetylase activity has also been reported in BPH-1, PC-3, and LNCaP prostate cancer cells (Myzak and Dashwood, 2006).
SFN has been shown to be effective in preventing skin cancer. For example, topical application of sulforaphane-containing broccoli sprout extract protects against UVB-induced skin carcinogenesis in SKH-1 mice (Dinkova-Kostova et al., 2006). SFN is also inhibits 7,12-dimethylbenz(a)anthracene/12-O-tetradecanoylphorbol-13-acetate-induced cancer in C57BL/6 mice (Xu et al., 2006). However, knowledge is limited regarding the molecular mechanisms of SFN action in inhibiting skin cancer formation. In the present study, we examine the role of PcG proteins as mediators of SFN regulation of skin cancer cell survival.
Materials and Methods
Chemicals and Reagents.
Sulforaphane was obtained from LKT Laboratories (St. Paul, MN). Trypsin and Dulbecco's modified Eagle's medium were purchased from Invitrogen (Carlsbad, CA). Lactacystin and mouse anti-β-actin were obtained from Sigma (St. Louis, MO). Mouse monoclonal anti-Bmi-1, rabbit anti-histone H3, and rabbit anti-UTX were obtained from Abcam (Cambridge, MA). Rabbit anti-procaspase 3, anti-procaspase 9, and mouse anti-procaspase 8 were from Cell Signaling Technology (Danvers, MA). Mouse monoclonal anti-Suz12 and rabbit polyclonal anti-H3K27me3 were purchased from Millipore (Billerica, MA). Mouse monoclonal antibody specific for poly(ADP-ribose) polymerase (PARP) was from BD Pharmingen (San Diego, CA). Rabbit polyclonal antibodies specific for cdc25c, cdk2, cdk4, and ubiquitin, and mouse monoclonal antibodies were specific for p21Cip1, cyclin A, cdk1, and cyclin B1 were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal antibody specific for Ezh2 was obtained from BD Transduction Labs (San Jose, CA). Rabbit anti-JMJD3 was purchased from Abgent, Inc. (San Diego, CA). Horseradish peroxidase-conjugated sheep anti-mouse IgG and donkey anti-rabbit IgG were obtained from GE Healthcare (Piscataway, NJ).
Cell Culture and Cell Cycle Analysis.
A431 and SCC-13 squamous cell carcinoma and HaCaT immortal but not transformed cell lines were obtained from American Type Culture Collection (Manassas, VA). The cells are maintained in a Dulbecco's modified Eagle's medium containing 5% fetal calf serum, antibiotics, d-glucose, l-glutamine, and sodium pyruvate (Balasubramanian et al., 2010). For cell proliferation assays, A431, HaCaT, and SCC-13 cells were plated at low density in 9.5-cm2 dishes and allowed to attach for 24 h. Cells were then treated with fresh medium containing 0.1% DMSO or indicated concentration of SFN. After 48 h, the cells were harvested with 0.025% trypsin containing 1 mM EDTA, fixed in isotonic buffer and counted using Coulter counter (Beckman Coulter, Fullerton, CA). For cell cycle analysis, subconfluent A431, HaCaT, and SCC-13 cells were treated with SFN (0–20 μM) or vehicle for 24 h. The cells were then trypsinized, washed twice in ice-cold phosphate-buffered saline (PBS) and centrifuged (4°C). The cell pellets were resuspended in PBS and fixed in ice-cold methanol. The cells were then washed and suspended in PBS, incubated with RNase and stained with propidium iodide before flow cytometric analysis.
Bmi-1 Adenovirus Infection.
Construction of human Bmi-1 encoding adenovirus (tAd5-hBmi-1), empty virus (tAd5-EV), and the helper TA virus (Ad5-TA) are described elsewhere (Lee et al., 2008). SCC-13 cells were plated into 35-mm dishes at 20% confluence and allowed to attach. After 24 h, the cells were infected with 5 MOI of multiplicity of hBmi-1 encoding or empty virus along with 5 MOI of Ad5-TA helper virus in the presence of 2.5 μg/ml hexadimethrine (Polybrene).
Immunological Methods.
Subconfluent cells were treated in the presence of 20 μM SFN or 0.1% DMSO for 24 h. The cells were then washed in phosphate-buffered saline and suspended in lysis buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate, 1 mM sodium vanadate, 1 μg of leupeptin, and 1 mM phenylmethylsulfonyl fluoride. Equal amounts of protein were electrophoresed on denaturing and reducing polyacrylamide gels (8–12%) and transferred to nitrocellulose membrane for immunoblot. The membrane was incubated in 10 mM Tris-HCl, pH 7.2, with 100 mM NaCl, 0.1% Tween 20, and 5% nonfat dry milk to block nonspecific binding and then with the primary antibody. Binding of the primary antibody was detected using a corresponding horseradish peroxidase-conjugated secondary antibody and chemiluminescent detection methods (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK).
Quantitative Real-Time RT-PCR.
Subconfluent cultures of SCC-13 cells were treated with 0, 15, or 20 μM SFN. After 24 h, total RNA was extracted using the RNeasy RNA isolation kit (QIAGEN, Valencia, CA). Superscript reverse transcriptase and oligo (dT) primers (Invitrogen) were used for reverse transcription from total RNA. The cDNA samples were amplified using primers specific for Ezh2 (forward, 5′-GCA TCT ATT GCT GGC ACC ATC TGA; reverse, 5′-TTG TTA CCC TTG CGG GTT GCA T), Bmi-1 (forward, 5′-TGT TCG TTA CCT GGA GAC CAG CAA; reverse, 5′-ATT GGC AGC ATC AGC AGA AGG ATG), and GAPDH (forward, 5′-TCC ACT GGC GTC TTC ACC, reverse, 5′-GGC AGA GAT GAT GAC CCT TT).
Detection of Ubiquitinated PcG Proteins.
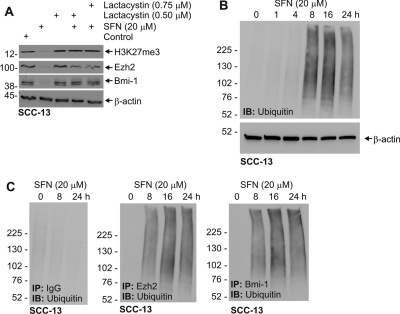
Subconfluent SCC-13 cells were treated with 0 or 20 μM SFN for 0, 1, 4, 8, 16, and 24 h and total cells extract was prepared in lysis buffer. Total extract (30 μg) was electrophoresed for detection of total ubiquitin level. To detect specific ubiquitination of Bmi-1 and Ezh2, 75 μg of total extract was immunoprecipitated with anti-IgG, anti-Bmi-1, or anti-Ezh2, and the precipitate was electrophoresed for immunoblot with anti-ubiquitin.
Results
SFN Suppresses SCC-13 Cancer Cell Proliferation, PcG Protein Level, and Histone H3K27me3 Formation.
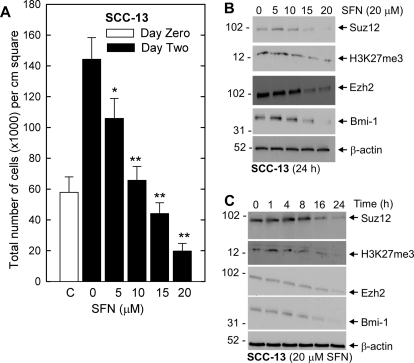
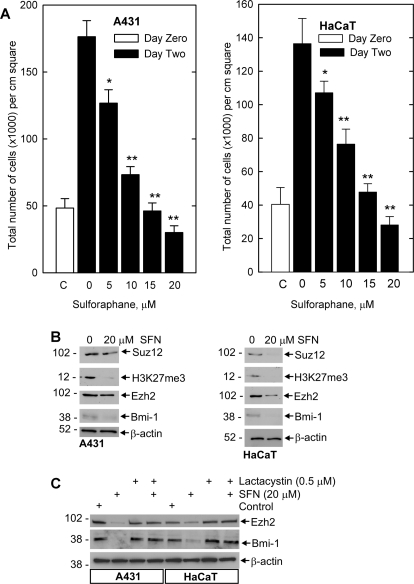
SFN has been reported to suppress cancer cell survival in a number of systems (Singh et al., 2004a,b; Myzak and Dashwood, 2006). We began by examining the impact of SFN treatment on SCC-13 cell proliferation. SFN treatment suppresses SCC-13 cell number in a concentration-dependent manner and treatment with intermediate levels of SFN (10 μM) maintains cell number at the initial plating density (Fig. 1A). Because increased PcG protein expression is known to be associated with increased skin cancer cell survival (Balasubramanian et al., 2010), we determined whether SFN treatment is associated with reduced PcG expression. Figure 1B shows that the reduction in cell number is associated with a parallel reduction in Suz12, Ezh2, and Bmi-1 level. The end product of Ezh2 action is H3K27me3 formation (Pasini et al., 2004), and our studies confirm a reduction H3K27me3 level with the reduction in Ezh2 expression (Fig. 1B). Time course studies indicate that the suppression is complete within 24 h after additional of 20 μM SFN (Fig. 1C).
Fig. 1.
SFN impact on skin cancer cell proliferation and PcG protein level. A, SCC-13 cells growing at low density in 35-mm dishes were treated with indicated concentrations of SFN or 0.1% DMSO for 48 h before harvest and counting. Error bars represent the mean ± S.E.M. for three separate experiments and the open bar indicates cell number at the initiation of treatment. Cell number is significantly reduced by SFN treatment compared with the 2-day treatment with 0 mM SFN (Student's t test, P < 0.05*, P < 0.001**). B and C, SFN modulates SCC-13 cell PcG protein level. Subconfluent cultures of SCC-13 were incubated with 0 - 20 μM SFN for 24 h or with 20 μM SFN for 0 to 24 h. The cells were harvested for immunoblot detection of Bmi-1, Ezh2, and H3K27me3. β-Actin level was monitored to assure equal protein loading. Similar results were observed in each of three experiments.
SFN Modulates Cell Cycle.
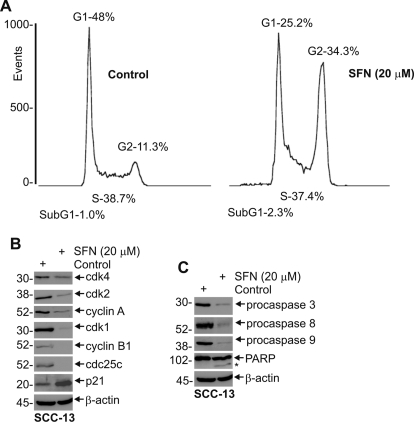
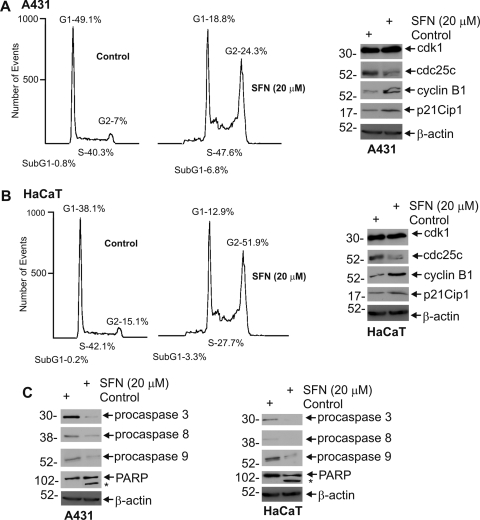
We next assessed the impact of SFN on cell cycle progression and apoptosis. SFN treatment causes PC-3 prostate cancer cells to arrest in G2/M (Singh et al., 2004b), whereas G1/S arrest is observed in DU145 and LNCaP cells (Wang et al., 2004). SFN-induced G2/M cell cycle arrest has been reported to involve increased expression of p21Cip1 and altered expression of cyclin B1, cdc25b, and cdc25c in different cancer cells (Gamet-Payrastre et al., 2000; Singh et al., 2004b; Herman-Antosiewicz et al., 2007). Figure 2A shows that SFN treatment of SCC-13 cells increases the number of G2/M events from 11 to 34% and is associated with a concomitant decrease in G1 events from 48 to 25%. In contrast, the number of S-phase cells is not altered. Figure 2B shows that this arrest is associated with a reduction in cdk1, cdk2, cyclin A, and cyclin B1. The level of cdc25c, the phosphatase that dephosphorylates and activates cdk1 (Donzelli and Draetta, 2003), is also reduced. In contrast, p21Cip1 level is increased (Fig. 2B). These findings are consistent with cessation of cell proliferation.
Fig. 2.
Impact of SFN on cell cycle. A, subconfluent SCC-13 cells were treated without or with 20 μM SFN for 24 h. The cells were then trypsinized, washed, fixed in methanol, and stained with propidium iodide for cell cycle analysis. The experiment is representative of three separate experiments. The propidium iodide level (DNA content) is represented on the x-axis and number of events on the y-axis. B and C, effect of SFN on cell cycle and apoptosis regulatory proteins. SCC-13 cells were treated as above and harvested. Total cell lysate was prepared and 30 to 75 μg of protein was electrophoresed for immunoblot. β-Actin level was measured to normalize loading. These results are representative of three separate experiments. The asterisk indicates cleaved activated PARP.
SFN Affects Apoptosis.
Figure 2C shows a SFN-dependent decrease in the level of procaspase 3, 8, and 9 and an increase in PARP cleavage. The SFN impact is similar to that observed for the bioactive green tea polyphenol, (−)-epigallocatechin-3-gallate, in that it alters cell cycle regulatory protein expression to reduce cell proliferation and increases apoptosis (Balasubramanian et al., 2010). However, SFN alters the relative distribution of cells within the cell cycle, whereas (−)-epigallocatechin-3-gallate (EGCG) does not (Balasubramanian et al., 2010). Considering the strong caspase and PARP cleavage observed in 20 μM SFN treated cells, cell cycle analysis shows only a modest increase in sub-G1 events (Fig. 2A). However, the sub-G1 fraction is markedly increased when cells are treated for longer periods or with increased levels of SFN. Treatment with 40 μM SFN for 24 h, for example, increases G1 content to 55%, reduces G2 content to 28%, and increases sub-G1 events to 11% (not shown).
Impact of Bmi-1 on SFN-Dependent Events.
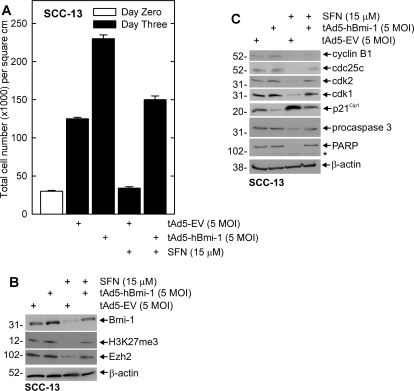
The experiments in Fig. 1 demonstrate that SFN treatment reduces PcG protein level. Because PcG proteins generally promote cell survival, we examined whether maintaining PcG protein level during SFN challenge reverses the SFN-associated response. Cells were infected with tAd5-hBmi-1 or tAd5-EV for 24 h and then treated with DMSO or 15 μM SFN for 48 h. Figure 3A shows that total cell number increases during the 3-day growth period and that this is further increased by Bmi-1. Addition of 15 μM SFN to tAd5-EV-infected cells reduces cell number. It is interesting that this suppression is reversed by Bmi-1 expression. Figure 3B shows that SFN reduces Bmi-1, Ezh2, and H3K27me3 level and that these changes are reversed by maintaining Bmi-1. These studies indicate that suppression of PcG protein level and cell number is associated with SFN treatment, and these responses can be reversed by Bmi-1.
Fig. 3.
Bmi-1 reverses SFN-dependent events in SCC-13 cells. A, SCC-13 cells were infected with 5 MOI of empty (tAd5-EV) or Bmi-1 encoding adenovirus (tAd5-hBmi-1) in the presence of 5 MOI of Ad5-TA helper virus. At 24 h after infection, the cells were treated with fresh virus-free medium with or without 15 μM SFN. After an additional 48 h, the cells were harvested for counting. Cell number on day 0 of virus infection is indicated by the open bar, whereas cell counts on day 3 are indicated by shaded bars. The values mean ± S.E.M., n = 3. B, impact of Bmi-1 overexpression on SFN suppression of Ezh2 level and H3K27me3 formation. Cells were infected and treated as in A before harvest and preparation of extracts for electrophoresis and detection of the indicated proteins by immunoblot. β-Actin level was used to normalize protein loading. Similar results were observed in each of three experiments. C, SCC-13 cells were infected with tAd5-EV or tAd5-hBmi-1 as in A, and after 24 h, the cells were treated with 20 μM SFN or 0.1% DMSO for 48 h. Total extracts were prepared for electrophoresis and detection of the indicated proteins. β-Actin was monitored as a loading control. Similar results were observed in each of three individual experiments.
To understand the mechanism of Bmi-1 reversal of the SFN-dependent responses, we monitored the impact of Bmi-1 overexpression on key cell cycle and apoptotic endpoints. Figure 3C shows that SFN treatment increased p21Cip1 and suppresses cdk1, cdk2, cyclin B1, and cdc25C levels, changes that are consistent with reduced proliferation. SFN also activates procaspase 3 and PARP cleavage. However, these changes are partially reversed in the presence of Bmi-1 (Fig. 3C).
SFN Increases Proteasome-Dependent PcG Protein Degradation.
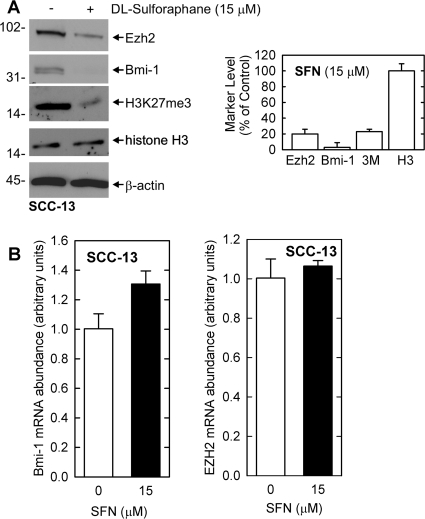
A key consideration is the mechanism whereby PcG protein level is reduced by SFN treatment. The mechanism responsible for SFN-dependent loss of PcG protein expression in skin cancer cells is not known. The reduced level could be due to a direct impact on the PcG protein level or it could be associated with a reduction in mRNA level. To assess this, SCC-13 cells were treated with SFN and PcG protein and mRNA levels were monitored. As shown in Fig. 4A, SFN reduces Bmi-1 and Ezh2 protein level and reduces H3K27me3 formation. The graph compares the average protein level in control and SFN-treated cultures based on three independent experiments. Ezh2 and Bmi-1 level and H3K27me3 formation are significantly reduced in SFN treated cells. We next used quantitative PCR to compared Bmi-1 and Ezh2 mRNA level in control and SFN-treated cells. Figure 4B shows that Bmi-1 and Ezh2 mRNA levels are not altered by SFN treatment.
Fig. 4.
SFN regulation of PcG protein mRNA level. A, SCC-13 cells were treated with 15 μM SFN for 24 h and total protein extracts were prepared for detection of Bmi-1, Ezh2, and H3K27me3. The graphical analysis presents the mean ± S.E.M. for three separate experiments. Student's t test analysis reveals that Ezh2, Bmi-1, and H3K27me3 (3M) levels are significantly reduced in SFN-treated cells compared with control (Student's t test, p < 0.001). B, SFN does not alter PcG protein encoding mRNA. For detection of RNA, 1 μg of total RNA was reverse-transcribed followed by quantitative PCR. Bmi-1 and Ezh2 mRNA level was normalized to GAPDH. Similar results were observed in each of three separate experiments.
These findings suggest that the SFN-dependent protein reduction is due to direct effects on protein level and not mRNA level. The proteasome has been reported to be involved in PcG turnover in selected cell types (Tan et al., 2007; Dimri et al., 2010). To assess this possibility, we treated SCC-13 cells with SFN in the presence or absence of lactacystin and monitored the impact on Bmi-1, Ezh2, and H3K27me3 formation. Figure 5A shows that lactacystin inhibits the SFN-dependent loss of these markers. To provide additional evidence for proteasome involvement we treated cells with SFN for 0–24 h and monitored for the presence of ubiquitinated Ezh2 and Bmi-1. Figure 5B shows that SFN stimulates a general increase in ubiquitination. To determine whether Bmi-1 and Ezh2 are among the ubiquitinated proteins, we treated cells for 0–24 h with 20 μM SFN and prepared extract for immunoprecipitation with anti-IgG, anti-Bmi-1, and anti-Ezh2. Immunoblot of the immunoprecipitates with anti-ubiquitin reveals evidence for specific ubiquitination of Bmi-1 and Ezh2 (Fig. 5C). No ubiquitination was observed in the IgG immunoprecipitated group. The onset of increased ubiquitination was at 8 h after application of SFN.
Fig. 5.
Impact of SFN on proteasome-dependent loss of Ezh2 and Bmi-1. A, SCC-13 cells were treated with 20 μM SFN in the presence or absence of 0.5 μM lactacystin for 24 h. Total cell extracts were then prepared for immunodetection of Ezh2, Bmi-1, H3K27me3, and β-actin. B, SFN treatment increases general ubiquitination. Subconfluent SCC-13 cells were treated in the presence of 20 μM SFN for indicated times, and total cell extract was prepared and electrophoresed for detection of ubiquitin. β-Actin served as a loading control. C, SCC-13 cells were treated as above, and total extracts were immunoprecipitated with the indicated antibodies. The precipitates were then electrophoresed and immunoblotted to detect ubiquitin. Similar results were observed in each of three independent experiments.
SFN Regulation of PcG Protein Level in A431 and HaCaT Cells.
To assess whether proteasome regulation of PcG proteins is generally observed, we examined the impact of sulforaphane on Ezh2, Bmi-1, and H3K27me3 formation in A431 and HaCaT cells. We first examined the impact of SFN on cell survival to determine whether SFN reduces cell number in these cells. Figure 6A shows that A431 and HaCaT cells display dose-dependent decrease in cell survival and that this reduction is associated with decreased expression of Bmi-1 and Ezh2 and reduced H3K27me3 formation (Fig. 6B). We anticipated that these changes may also be due to proteasome-dependent degradation. We therefore treated cells with SFN in the presence or absence of lactacystin. As shown in Fig. 6C, lactacystin inhibits the SFN-dependent reduction in Bmi-1 and Ezh2 level. These results confirm that SFN-dependent suppression of PcG level is proteasome-dependent in these cell lines.
Fig. 6.
Impact of SFN on PcG protein expression in A431 and HaCaT cells. A, A431 and HaCaT cells were incubated with 0 to 20 μM SFN and after 24 h the cells were harvested and counted. The graph depicts the mean and S.E. (n = 3). Cell number is significantly reduced by SFN treatment compared with 0 SFN (2 day) (Student's t test; *, P < 0.05; **, P < 0.001). B, A431 and HaCaT cells were treated as described above and extracts were prepared for immunoblot detection of the indicated proteins. C, SFN suppression of PcG protein level involves proteasome-dependent degradation. A431 and HaCaT cells were treated for 24 h in the presence of 20 μM SFN and/or 0.5 μM lactacystin. Cell extracts were then prepared and electrophoresed for immunoblot detection of the Bmi-1 and Ezh2 protein. Similar findings were observed in each of three separate experiments.
SFN Regulation of Cell Cycle and Apoptosis in A431 and HaCaT Cells.
A431 cells are tumor-forming skin cancer cells and HaCaT cells are immortalized nontumorigenic keratinocytes that have p53 mutations. SFN treatment of A431 cells increases G2/M events from 7 to 24% with a parallel reduction in G1 events (Fig. 7A). In HaCaT cells, SFN increases G2/M events from 15 to 51% with a concomitant decrease in G1 and S phase (Fig. 7A). These changes are associated with increased p21Cip1 and a marked reduction in cdc25c (Fig. 7B). In contrast, cdk1 level is not regulated and cyclin B1 level is increased. We also examined the impact of SFN on apoptosis indicators, because sub-G1 events are increased in SFN-treated A431 and HaCaT cells (Fig. 7, A and B). As shown in Fig. 7C, we observe reduced levels of procaspase-3, -8, and -9 and also increased PARP cleavage in SFN-treated A431 and HaCaT cells.
Fig. 7.
SFN modulates cell cycle and apoptosis in A431 and HaCaT cells. A and B, A431 and HaCaT cells growing at lower confluence were treated in the absence or presence of 20 μM SFN for 24 h. The cells were then trypsinized and fixed in methanol, and cell cycle phase was determined by flow cytometry of propidium iodide-stained cells. These findings are representative of three different experiments. Cell cycle regulatory proteins were detected by immunoblot of extracts prepared from these same cells. β-Actin was used as a gel loading normalization control. B, SFN impact on apoptotic markers. A431 and HaCaT cells treated as above and harvested for cell lysate preparation. Protein extract (25–50 μg) was used for immunoblot detection of indicated proteins. β-Actin was used to confirm equal protein loading. The asterisks indicate the PARP cleavage product.
SFN Impact on H3K27me3 Demethylases.
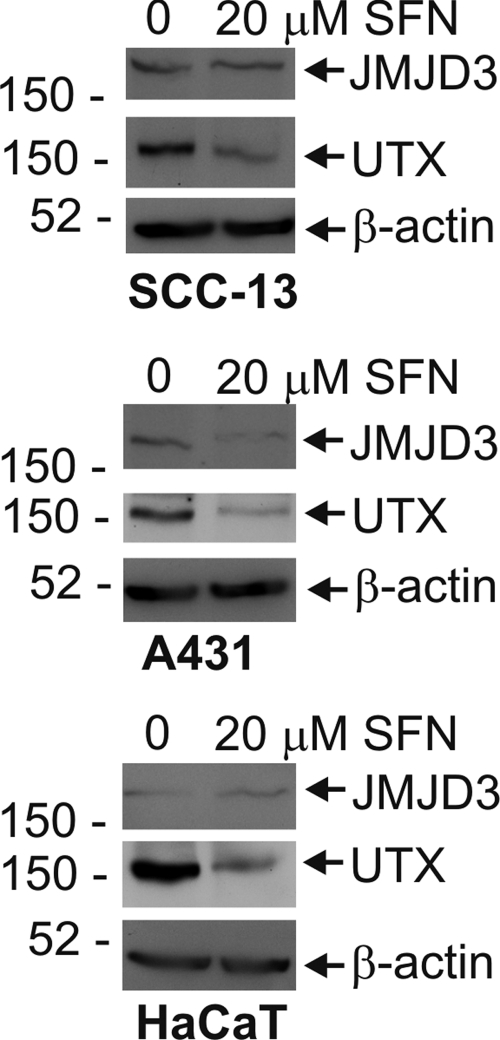
Jumonji domain protein 3 (JMJD3) and ubiquitously transcribed tetratricopeptide repeat, X protein (UTX) are histone demethylases that oppose PcG protein action and demethylate H3K27me3 (Agger et al., 2007, 2009; Barradas et al., 2009). We therefore examined the possibility that the change in H3K27me3 level may result, in part, from a change in expression level of these demethylases. SCC-13, A431, and HaCaT cells were treated for 24 h with 20 μM SFN before assessment of JMJD3 and UTX level. Figure 8 shows that SFN treatment reduces UTX level in all three cells lines and JMJD3 level in A431 cells. The loss of JMJD3 and UTX should lead to an increase in H3K27me3. Because the reverse is observed, it seems likely that the decrease we observe is due to loss of Ezh2.
Fig. 8.
Impact of SFN on UTX and JMJD3 levels. SCC-13, A431, and HaCaT cells growing at low density were treated with 20 μM SFN or vehicle (0.1% DMSO) for 24 h. Total cell extracts were prepared and equivalent amounts of protein were electrophoresed on a 6–10% denaturing polyacrylamide gel and transferred onto a nitrocellulose membrane. JMJD3, UTX, and β-actin levels were monitored using specific antibodies. Similar results were observed in each of three experiments.
Discussion
Sulforaphane is an important candidate dietary cancer preventive agent (Clarke et al., 2008). However, the mechanism of action is not fully elucidated, and the impact of SFN treatment on PcG protein function in skin cancer has not been studied. Our data indicate that SFN suppresses proliferation of epidermis-derived skin cancer cell lines, including SCC-13 and A431. In addition, SFN treatment suppresses proliferation of HaCaT cells, an immortalized but nontumorigenic epidermis-derived cell line. To understand the mechanism by which SFN suppresses proliferation, we examined the impact of SFN on cell-cycle control proteins. Flow cytometry analysis reveals cell-cycle arrest in G2/M and an associated reduction in cyclin B1, cdc25c, and cdk1 level. Based on the reduction in these cell cycle regulatory proteins, we anticipate that SFN treatment causes reduced cdk1/cyclin B1 and cdk2/cyclin A complex formation. This, in conjunction with accumulation of p21Cip1, the cell-cycle kinase inhibitor, would be expected to induce cell-cycle arrest. These results are consistent with previous reports in prostate cancer. For example, Singh et al. (2004b) described SFN induced G2/M arrest in PC-3 cells as associated with a marked decrease in cyclin B1, cdc25B, and cdc25c level and accumulation of Tyr-15-phosphorylated (inactive) cdk1. We also noticed a marked decrease in the level of cdk2 and cyclin A, although we did not observe a change in the percentage of cells at S phase of the cell cycle.
Additional studies reveal that SFN-treatment induces apoptosis as evidenced by increased sub-G1 DNA content and the loss of procaspase-3, -8, and -9 and increased PARP cleavage in SCC-13 cells. Our finding of SFN-dependent G2/M cell accumulation and activation of apoptosis in SCC-13 cells is in agreement with previous studies in prostate cancer cells (Singh et al., 2004b; Myzak et al., 2006). The general pattern of response was similar in the other epidermis-derived cell lines, A431 cells, and HaCaT cells. One difference, however, was an increase in cyclin B1 in A431 and HaCaT but not in SCC-13 cells. SFN-dependent induction of cyclin B1 has been reported in the context of G2/M cell cycle arrest (Gamet-Payrastre et al., 2000; Herman-Antosiewicz et al., 2007). Although an increase in cyclin B1 is a pro-proliferation response, it did not seem to counter the ability of SFN to suppress proliferation of the skin cell lines.
Recent studies suggest that PcG proteins play an important role in enhancing epithelial cancer cell proliferation and survival (Lee et al., 2008; Balasubramanian et al., 2010). Elevated Bmi-1 expression is observed in prostate, lung, breast, ovarian, and skin cancer (Glinsky et al., 2005; Lee et al., 2008), and Ezh2 is amplified and overexpressed in prostate, pancreas, breast, and ovarian cancer (Liu et al., 1992; Kleer et al., 2003). Overexpression of Ezh2 is linked with the invasion and progression of malignant tumors, especially in prostate cancer (Liu et al., 1992), and knockdown of Ezh2 or Bmi-1 inhibits cancer cell growth, motility, invasion, and tumor-forming ability (Chen et al., 2007; Xu et al., 2011). We have demonstrated that Bmi-1, Ezh2, and H3K27me3 are elevated in skin cancer cells and tumors (Lee et al., 2008).
In the present study, we show that SFN treatment suppresses Bmi-1 and Ezh2 levels and that the loss of Ezh2 does not influence histone H3 level but is associated with reduced H3K27me3 formation. These changes are observed as early as 8 to 16 h after initiation of treatment with 15 to 20 μM SFN. Moreover, SFN treatment reduces JMJD3 and UTX level, which would lead to increased H3K27me3 formation. JMJD3 and UTX are demethylases that remove the trimethyl mark from H3K27 (Agger et al., 2007, 2009; Barradas et al., 2009). Taken together, these findings suggest that the major reason for reduced H3K27 level is loss of Ezh2. Basal PcG expression in tumors is controlled by a host of mechanisms including regulation of transcription and control of mRNA level and mRNA translation rate. Of particular interest are recent findings suggesting microRNA control of Ezh2 expression (Alajez et al., 2010; Smits et al., 2010). miR-101 level is reduced in glioblastoma, and this is associated with increased Ezh2 expression and increased cancer cell survival and migration (Smits et al., 2010). Several microRNAs (including miR-26a, miR-101, and miR-98) suppress Ezh2 level, and loss of miR-98 expression is associated with increased Ezh2 expression in recurring tumors of nasopharyngeal carcinoma (Alajez et al., 2010). miR-138 has also suppresses Ezh2 level (Kisliouk et al., 2011). In most cases, it is not known whether these miRNA species control Ezh2 level by reducing mRNA stability or inhibiting protein translation (Cannell et al., 2008). Because of the possibility that miRNA may be important in mediating SFN action, we examined the impact of SFN treatment on Ezh2 and Bmi-1 mRNA level and found that the level of mRNA encoding these proteins is not reduced by SFN treatment. This suggests that reduced mRNA level, whether associated with miRNA-mediated or transcriptional mechanism, is not the basis for the SFN-dependent reduction in Ezh2 or Bmi-1 level.
Given that the loss of PcG protein expression does not involve reduced mRNA level, we looked to mechanisms that control protein stability and in particular the potential role of the proteasome. Recent studies highlight a role for the proteasome in regulating PcG protein level (Tan et al., 2007; Dimri et al., 2010). 3-Deazaneplanocin A, an s-adenosylhomocysteine hydrolase inhibitor, reduces Ezh2 and Suz12 level in MCF-7 cells, and this response is partially inhibited by proteasome inhibitors (Tan et al., 2007). In addition, omega-3 polyunsaturated fatty acids increase proteasome-dependent degradation of Ezh2 in MDA-MB-231 breast cancer cells (Dimri et al., 2010). Our studies are consistent with a role for the proteasome in SFN-dependent reduction of PcG protein level in skin cancer cells. First, the loss of PcG protein in SFN-treated cells is reversed by treatment with lactacystin, a proteasome inhibitor. Second, we show that Bmi-1 and Ezh2 are ubiquitinated in SFN-treated cells but not in control cells. Thus, ubiquitination is not a major mechanism controlling Bmi-1 and Ezh2 expression in resting cells but is specifically activated by SFN treatment. Third, the time course of Bmi-1 and Ezh2 loss from cells matches the time course of appearance of ubiquitinated forms of these proteins. Bmi-1 and Ezh2 are ubiquitinated as early as 8 h after SFN treatment and that loss of these proteins is evident by 8 to 16 h after treatment. Fourth, lactacystin treatment inhibits Bmi-1 and Ezh2 loss in several epidermis-derived cell lines including SCC-13, A431, and HaCaT, suggesting that proteasome degradation is a general mechanism of SFN-catalyzed loss of PcG proteins in epidermis-derived cells.
It is worth noting that the action of SFN may involve more than simply targeting ubiquitinated PcG proteins to the proteasome, because there is evidence that SFN directly increases the level of proteasome protein components. Kwak et al. (2003) showed that SFN treatment increases PSMB5 level in murine embryonic fibroblasts. PSMB5 is a key protein component of the 20S proteasome, and in this system, SFN triggers Nrf2 transcription factor interaction with the PSMB5 gene promoter to drive increased PSMB5 expression (Kwak et al., 2003). This is consistent with a report showing that SFN prevention of skin cancer is in part mediated by Nrf2 (Xu et al., 2006), because Nrf2 knockout mice are more susceptible to DMBA/TPA skin tumor development. Thus, the increase in proteasome activity in SCC-13 cells may be due, in part, to a SFN-induced increase in proteasome subunit gene expression.
A recent study documents increased PcG protein expression in SCC-13 cells and shows that EGCG, a bioactive green tea polyphenol, reduces this expression (Balasubramanian et al., 2010). Moreover, vector-mediated maintenance of Bmi-1 expression in EGCG-treated cells restores cell number and expression of pro-proliferation cell-cycle markers, and suppresses EGCG-stimulated proapoptotic events (Balasubramanian et al., 2010). Our present studies describe a similar response in that the SFN-dependent reduction in cell number, PcG protein expression, H3K27me3 formation, and apoptotic marker activation is partially reversed by forced maintenance of Bmi-1 expression. Such an impact of Bmi-1 has been reported in other systems. For example, ectopic expression of Bmi-1 renders glioma cells resistant to doxorubicin- and UV irradiation induced cell death, whereas knockdown of endogenous Bmi-1 accelerates cell death (Li et al., 2010). Although Bmi-1 antagonism of cell death is an emerging theme, the downstream targets of Bmi-1 that drive enhanced survival vary among cell types. For example, Bmi-1 suppresses Ink4a/Arf expression leading to enhanced cell survival in lymphoid cells (Jacobs et al., 1999). Although a host of cell cycle proteins are PcG targets in skin cancer cells (Balasubramanian et al., 2010), signaling cascades are also important targets (Li et al., 2010).
In summary, our studies examine the impact of SFN on PcG protein expression in a skin cancer system. We show that the SFN-dependent reduction in PcG protein level is mediated via the proteasome. We also show that the impact of SFN treatment can be attenuated when Bmi-1 levels are maintained. Our findings suggest that sulforaphane may reduce skin cancer cell survival reducing PcG protein expression via a mechanism that requires the proteasome.
Acknowledgments
We acknowledge the technical assistance of Wen Xu.
This work was supported by the National Institutes of Health National Cancer Institute [Grant R01-CA131074]; and the National Institutes of Health National Institute of Arthritis, Musculoskeletal and Skin Diseases [Grant R01-AR053851].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.111.072363.
- PcG
- Polycomb group
- H3K27
- lysine 27 of histone H3
- eed
- embryonic ectoderm development
- Ezh2
- enhancer of zeste homolog 2
- Suz12
- suppressor of zeste 12 protein homolog
- H3K27me3
- histone H3 lysine 27 trimethylation
- PRC
- polycomb repressive complex
- Bmi-1
- B-cell-specific Moloney murine leukemia virus integration site 1
- CBX
- chromobox homolog
- cdk
- cyclin-dependent kinase
- cdki
- cyclin-dependent kinase inhibitor
- SFN
- sulforaphane
- PARP
- poly(ADP-ribose) polymerase
- DMSO
- dimethyl sulfoxide
- PBS
- phosphate-buffered saline
- MOI
- multiplicity of infection
- EGCG
- (−)-epigallocatechin-3-gallate
- JMJD3
- Jumonji domain protein 3
- UTX
- ubiquitously transcribed tetratricopeptide repeat, X protein.
Authorship Contributions
Participated in research design: Balasubramanian and Eckert.
Conducted experiments: Balasubramanian and Chew.
Contributed new reagents or analytic tools: Chew.
Performed data analysis: Balasubramanian and Eckert.
Wrote or contributed to the writing of the manuscript: Balasubramanian and Eckert.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449:731–734 [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. (2009) The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev 23:1171–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alajez NM, Shi W, Hui AB, Bruce J, Lenarduzzi M, Ito E, Yue S, O'Sullivan B, Liu FF. (2010) Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis 1:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Adhikary G, Eckert RL. (2010) The Bmi-1 polycomb protein antagonizes the (−)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis 31:496–503 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Barradas M, Anderton E, Acosta JC, Li S, Banito A, Rodriguez-Niedenführ M, Maertens G, Banck M, Zhou MM, Walsh MJ, et al. (2009) Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev 23:1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. (2003) EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22:5323–5335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks JD, Paton VG, Vidanes G. (2001) Potent induction of phase 2 enzymes in human prostate cells by sulforaphane. Cancer Epidemiol Biomarkers Prev 10:949–954 [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Bushell M. (2008) How do microRNAs regulate gene expression? Biochem Soc Trans 36:1224–1231 [DOI] [PubMed] [Google Scholar]
- Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, Hui CK, Lau GK, He ML, Sung J, et al. (2007) Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology 46:200–208 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Dashwood RH, Ho E. (2008) Multi-targeted prevention of cancer by sulforaphane. Cancer Lett 269:291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri M, Bommi PV, Sahasrabuddhe AA, Khandekar JD, Dimri GP. (2010) Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis 31:489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Jenkins SN, Fahey JW, Ye L, Wehage SL, Liby KT, Stephenson KK, Wade KL, Talalay P. (2006) Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett 240:243–252 [DOI] [PubMed] [Google Scholar]
- Donzelli M, Draetta GF. (2003) Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep 4:671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Adhikary G, Rorke EA, Chew YC, Balasubramanian S. (2011) Polycomb group proteins are key regulators of keratinocyte function. J Invest Dermatol 131:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamet-Payrastre L, Li P, Lumeau S, Cassar G, Dupont MA, Chevolleau S, Gasc N, Tulliez J, Tercé F. (2000) Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res 60:1426–1433 [PubMed] [Google Scholar]
- Glinsky GV, Berezovska O, Glinskii AB. (2005) Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 115:1503–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman-Antosiewicz A, Xiao H, Lew KL, Singh SV. (2007) Induction of p21 protein protects against sulforaphane-induced mitotic arrest in LNCaP human prostate cancer cell line. Mol Cancer Ther 6:1673–1681 [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. (1999) Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13:2678–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisliouk T, Yosefi S, Meiri N. (2011) MiR-138 inhibits EZH2 methyltransferase expression and methylation of histone H3 at lysine 27, and affects thermotolerance acquisition. Eur J Neurosci 33:224–235 [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. (2003) EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 100:11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. (2003) Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol 23:8786–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Adhikary G, Balasubramanian S, Gopalakrishnan R, McCormick T, Dimri GP, Eckert RL, Rorke EA. (2008) Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol 128:9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gong LY, Song LB, Jiang LL, Liu LP, Wu J, Yuan J, Cai JC, He M, Wang L, et al. (2010) Oncoprotein Bmi-1 renders apoptotic resistance to glioma cells through activation of the IKK-nuclear factor-kappaB pathway. Am J Pathol 176:699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM. (2006) Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem 281:20643–20649 [DOI] [PubMed] [Google Scholar]
- Liu S, Wolfe MS, Borchardt RT. (1992) Rational approaches to the design of antiviral agents based on S-adenosyl-l-homocysteine hydrolase as a molecular target. Antiviral Res 19:247–265 [DOI] [PubMed] [Google Scholar]
- Mills AA. (2010) Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer 10:669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myzak MC, Dashwood RH. (2006) Histone deacetylases as targets for dietary cancer preventive agents: lessons learned with butyrate, diallyl disulfide, and sulforaphane. Curr Drug Targets 7:443–452 [DOI] [PubMed] [Google Scholar]
- Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. (2006) Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis 27:811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myzak MC, Karplus PA, Chung FL, Dashwood RH. (2004) A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res 64:5767–5774 [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23:4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10:697–708 [DOI] [PubMed] [Google Scholar]
- Singh AV, Xiao D, Lew KL, Dhir R, Singh SV. (2004a) Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis 25:83–90 [DOI] [PubMed] [Google Scholar]
- Singh SV, Herman-Antosiewicz A, Singh AV, Lew KL, Srivastava SK, Kamath R, Brown KD, Zhang L, Baskaran R. (2004b) Sulforaphane-induced G2/M phase cell cycle arrest involves checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C. J Biol Chem 279:25813–25822 [DOI] [PubMed] [Google Scholar]
- Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J, Krichevsky AM, et al. (2010) miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget 1:710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. (2007) Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 21:1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. (2004) Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 23:5759–5769 [DOI] [PubMed] [Google Scholar]
- van Poppel G, Verhoeven DT, Verhagen H, Goldbohm RA. (1999) Brassica vegetables and cancer prevention. Epidemiology and mechanisms. Adv Exp Med Biol 472:159–168 [DOI] [PubMed] [Google Scholar]
- Wang L, Liu D, Ahmed T, Chung FL, Conaway C, Chiao JW. (2004) Targeting cell cycle machinery as a molecular mechanism of sulforaphane in prostate cancer prevention. Int J Oncol 24:187–192 [PubMed] [Google Scholar]
- Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN. (2006) Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res 66:8293–8296 [DOI] [PubMed] [Google Scholar]
- Xu Z, Liu H, Lv X, Liu Y, Li S, Li H. (2011) Knockdown of the Bmi-1 oncogene inhibits cell proliferation and induces cell apoptosis and is involved in the decrease of Akt phosphorylation in the human breast carcinoma cell line MCF-7. Oncol Rep 25:409–418 [DOI] [PubMed] [Google Scholar]