Abstract
To identify new risk variants for cutaneous basal cell carcinoma, we performed a genome-wide association study of 16 million SNPs identified through whole-genome sequencing of 457 Icelanders. We imputed genotypes for 41,675 Illumina SNP chip-typed Icelanders and their relatives. In the discovery phase, the strongest signal came from rs78378222[C] (odds ratio (OR) = 2.36, P = 5.2 × 10−17), which has a frequency of 0.0192 in the Icelandic population. We then confirmed this association in non-Icelandic samples (OR = 1.75, P = 0.0060; overall OR = 2.16, P = 2.2 × 10−20). rs78378222 is in the 3′ untranslated region of TP53 and changes the AATAAA polyadenylation signal to AATACA, resulting in impaired 3′-end processing of TP53 mRNA. Investigation of other tumor types identified associations of this SNP with prostate cancer (OR = 1.44, P = 2.4 × 10−6), glioma (OR = 2.35, P = 1.0 × 10−5) and colorectal adenoma (OR = 1.39, P = 1.6 × 10−4). However, we observed no effect for breast cancer, a common Li-Fraumeni syndrome tumor (OR = 1.06, P = 0.57, 95% confidence interval 0.88–1.27).
Basal cell carcinoma (BCC) is the most common cancer in people of European ancestry. Sun exposure is the primary risk factor for BCC, but genetic predisposition also plays a substantial role1,2. High penetrance mutations in Hedgehog pathway genes (PTCH1, PTCH2 and SUFU) cause Gorlin syndrome, also known as basal cell nevus syndrome1. Candidate gene and genome-wide association studies have shown that common, low-penetrance susceptibility alleles exist at MC1R and several other loci3–7.
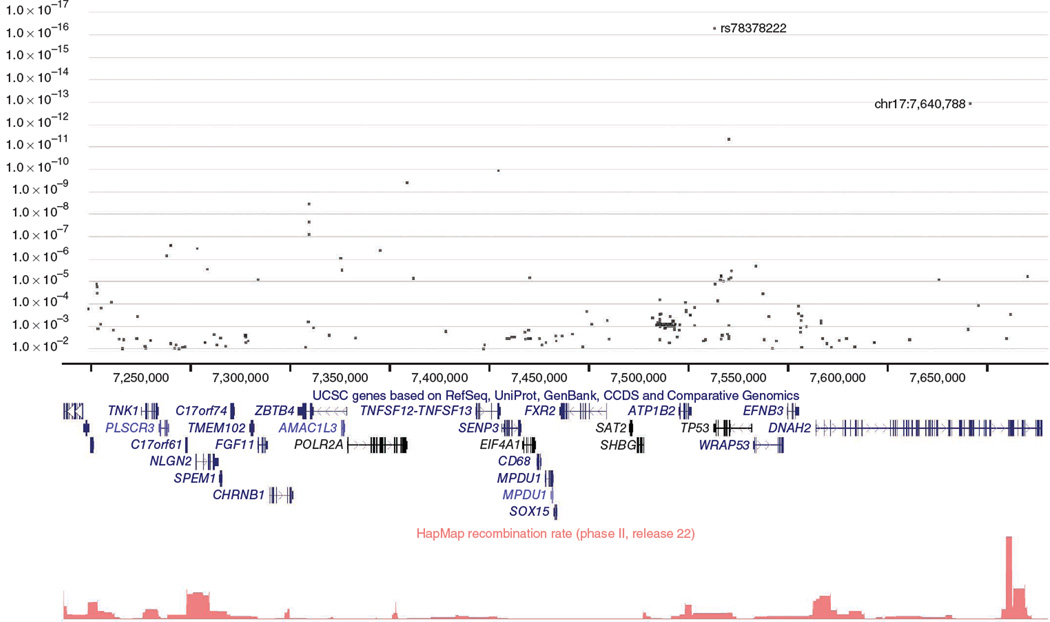
Previously, we described a large genome-wide association study of the Icelandic population using common SNPs and showed how genotypes can be phased over long distances8,9. For BCC, we initially generated Illumina SNP chip data for 1,366 affected individuals (cases) and 40,309 controls. Haplotype association analysis based on long-range phasing showed that several 0.3-cM haplotypes at 17p13 were strongly associated with BCC. The most significant signals were produced by haplotype A6 (OR = 2.04, P = 2.0 × 10−10), spanning the region chr17: 7,186,095–7,425,536 and by a highly correlated haplotype, A8 (OR = 2.00, P = 3.0 × 10−10), spanning an adjacent region, chr17:7,431,901–7,680,389. The region covered by these haplotypes is illustrated in Figure 1.
Figure 1.
Overview of single-point SNP association data obtained from genomic sequencing in the 17p13 region covered by haplotypes A6 and A8. The region shown is chr17:7,186,095–7,680,389 (HG18 Build 36). The upper panel shows BCC association P values for SNPs in the region identified by whole-genome sequencing of 457 individuals. We determined association by two-way imputation (see the text for details); only SNPs with P < 0.01 are plotted. The positions of the TP53 SNP rs78378222 and the newly discovered SNP giving the second-highest signal in the region (chr17:7,640,788) are indicated. The locations of UCSC genes in the region are shown in the middle panel. The lower panel shows recombination rates calculated as described previously23 from HapMap2 release 22 data.
To search for variants that might not be covered well by the chips, we used high-capacity DNA sequencing techniques to sequence the entire genomes of 457 Icelanders to an average depth of over 10× (Online Methods), which identified approximately 16 million SNPs. To ensure that all the rare risk alleles that might be carried on the A6 or A8 backgrounds would be sequenced, we included ten individuals who carried these haplotypes among the 457 individuals selected for sequencing. Using imputation assisted by long-range haplotype phasing, we used the sequence data to determine the genotypes of the 16 million SNPs in the 41,675 Icelanders who had been genotyped on the SNP chips. Moreover, knowledge of Icelandic genealogy allowed us to propagate genotypic information into individuals for whom we have neither SNP chip nor sequence data, a process we refer to as ‘genealogy-based in silico genotyping’. We refer to the combined method of imputing sequence-derived data into phased chromosomes from chip-typed individuals and using genealogy-based in silico genotyping to infer the sequence of ungenotyped individuals as ‘two-way imputation’ (Supplementary Note).
We conducted a two-way-imputation–based genome-wide BCC association analysis of the 16 million SNPs, which we designated the ‘discovery phase’. This analysis identified a number of SNPs with strong associations in the region covered by the two haplotypes. The strongest signal (OR = 2.36, P = 5.2 × 10−17) came from rs78378222, located in the 3′ untranslated region of TP53 (Fig. 1 and Table 1). This signal was not only the strongest in the region covered by the two candidate haplotypes, but it was also the strongest signal of the 16 million SNPs observed genome wide. The minor (C, at risk) allele is present in the Icelandic population at a frequency of 0.0192. There was no deviation from Hardy-Weinberg equilibrium, and we observed C/C homozygotes (although they are rare), so the variant is not recessive lethal. The SNP was not on the Illumina chips and is not in HapMap2 or HapMap3. The best on-chip tag SNP was rs4796305, which produced only a modest signal (OR = 1.18, P = 0.0024, minor allele frequency = 0.119, r2 = 0.15). A quantile-quantile plot is shown in Supplementary Figure 1. Aside from the 17p13 locus, all other SNPs that surpassed the P = 3.0 × 10−9 Bonferroni-adjusted threshold for genome-wide significance were in previously published loci5,6.
Table 1.
Association between BCC and rs78378222[C]
| Sample set | P | OR | 95% CI | Ncases | Frequency in cases |
Ncontrols | Frequency in controls |
Pahet |
|---|---|---|---|---|---|---|---|---|
| Iceland discovery phase two-way imputation | 5.2 × 10−17 | 2.36 | 1.93–2.89 | 2,121b | 0.0442 | >39,614b | 0.0192c | |
| Iceland follow-up phase two-way imputationd | 3.1 × 10−13 | 2.28 | 1.83–2.85 | 1,813b | 0.0428 | >36,709b | 0.0192c | |
| Iceland follow-up phase single-track genotypedd | 3.2 × 10−7 | 2.19 | 1.62–2.96 | 1,044 | 0.0398 | 7,200 | 0.0185 | |
| Iceland follow-up phase combined | 5.4 × 10−19 | 2.25 | 1.88–2.69 | 2,857b | NA | >43,909b | NA | 0.83 |
| Denmark | 0.0057 | 2.06 | 1.23–3.44 | 308 | 0.0341 | 3,441 | 0.0168 | |
| Eastern Europee | 0.12 | 2.04 | 0.84–4.93 | 526 | 0.0133 | 532 | 0.0066 | |
| Spain | 0.77 | 0.86 | 0.34–2.21 | 628 | 0.0040 | 3,928 | 0.0046 | 0.79 |
| Combined non-Icelandic replication | 0.0060 | 1.75 | 1.17–2.62 | 1,462 | NA | 7,901 | NA | 0.27 |
| Combined Iceland follow up and replication | 2.2 × 10−20 | 2.16 | 1.83–2.54 | 4,319b | NA | >51,810b | NA | 0.27 |
NA, not applicable; OR, odds ratio; CI, confidence interval.
The P value for heterogeneity based on combining groups using the Mantel-Haenszel model.
Effective sample size estimate taking into account the efficiency of the Icelandic genealogy-based in silico genotyping.
The control frequency given is derived from 40,309 directly chip-typed control individuals without BCC.
The Iceland follow-up phase two-way imputation set consists of individuals who were chip-typed and individuals whose genotypes were determined by genealogy-based in silico genotyping. This set differs from the Iceland discovery phase two-way imputation set in that it lacks those individuals whose genotypes were determined by genealogy-based in silico genotyping in the discovery phase but who were subsequently directly single-track genotyped. Such individuals form part of the Iceland follow-up phase single-track genotyped set, which consists of individuals whose genotypes were determined solely by a Centaurus single-track assay. The follow-up phase two-way imputation and single-track genotyped sets are non-overlapping (Supplementary Fig. 2).
Samples from Hungary, Slovakia and Romania.
To further investigate the rs78378222 association, we conducted a follow-up phase. The design of the discovery and follow-up phases is detailed in Supplementary Figure 2. We devised a Centaurus10 single-track assay for rs78378222 and used it to genotype all available samples from Icelandic cases with BCC (n = 2,322) and 7,200 controls. Among these, 1,044 out of the 2,322 single-track genotyped cases had not previously been genotyped by the Illumina chip. An association analysis with these 1,044 cases and 7,200 controls yielded OR = 2.19 and P = 3.2 × 10−7 (Table 1, see the Iceland follow-up single-track genotyped category). Note that this is not a fully independent replication of the discovery phase result, as some of the individuals who had been assigned genealogy-based in silico genotypes in the discovery phase were single-track genotyped in the follow-up phase (Supplementary Fig. 2). We then repeated the two-way-imputation–based analysis using a non-overlapping sample that excluded the 1,044 single-tracked cases (Table 1, see the Iceland follow-up two-way–imputation category). Combining the results from the two follow-up groups provided strong evidence for the association of rs78378222 with BCC (OR = 2.25, P = 5.4 × 10−19; Table 1, see the Iceland follow-up phase combined category). Association based on all 2,322 cases who had been single-track genotyped (irrespective of whether or not they had been chip typed) produced comparable results (Supplementary Table 1).
We then typed rs78378222 in replication samples from Denmark, eastern Europe and Spain (the replication sets are described in Supplementary Table 2). We found the risk allele in all the populations tested, with frequencies that seemed to decline with each population’s distance from Iceland (Table 1). Combined, the evidence for replication of the BCC association in non-Icelandic samples was significant and showed no evidence of heterogeneity (OR = 1.75, P = 0.0060, P of heterogeneity (Phet) = 0.27; Table 1). Combined with the Icelandic follow-up phase data, the overall association was highly significant (OR = 2.16, P = 2.2 × 10−20).
The second strongest signal in the genome originated from a previously unreported SNP (designated chr17:7,640,788; OR = 2.41, P = 1.1 × 10−13) which was also in the region covered by the two candidate haplotypes (Fig. 1). Based on single-track genotyping of 2,281 cases and 6,858 controls for both SNPs, chr17:7,640,788 was correlated (r2 = 0.61) with rs78378222 and so may capture the same signal. When adjusted for the effect of rs78378222, there was no residual signal from chr17:7,640,788 (ORadj = 1.07, Padj = 0.72), whereas rs78378222 remained significant after adjustment for chr17:7,640,788 (ORadj = 2.00, Padj = 3.0 × 10−5). Therefore, we did not investigate chr17:7,640,788 further. A common germline variant in TP53 (rs1042522, p.Pro72Arg) has been studied extensively for cancer susceptibility, generally with equivocal results11. We saw no evidence that rs1042522 was associated with BCC (OR = 1.00, P = 0.98). We confirmed this by single-track genotyping. Thus, it appears that p.Pro72Arg does not confer risk of BCC and is unrelated to the rs78378222 signal.
p53 is induced by ultraviolet irradiation of skin and is a primary mediator of the tanning response12,13. We looked for associations between rs78378222 and sensitivity of skin to sun in 11,131 samples from Iceland and The Netherlands14,15. There was no significant association between rs78378222[C] and self-reported sun sensitivity (Fitzpatrick score I and II compared to III and IV; OR = 1.14, P = 0.23, 95% confidence interval (CI) 0.92–1.41). We also noted that rs78378222[C] was somewhat more frequent in individuals with tumors at sun-exposed sites (0.0412) than in those with tumors at non–sun-exposed sites (0.0302); however, this difference was not significant (P = 0.18).
Inspection of the sequence surrounding rs78378222 indicated that it occurs in the sole polyadenylation signal of TP53, with the risk-associated variant changing the sequence AATAAA to AATACA, thus disrupting the signal sequence. This class of mutations was first observed in the polyadenylation signal of HBA2 (encoding alpha 2 globin), leading to alpha-thalassemia16. We obtained RNA from blood and adipose tissue from rs78378222[A/C] heterozygotes and rs78378222[A/A] homozygotes. Using RT-PCR with primers internal to the TP53 mRNA, we observed that rs78378222[A/C] heterozygotes expressed somewhat less TP53 transcript than wild-type homozygotes (P = 0.041; Supplementary Fig. 3a). To investigate polyadenylation site usage of wild-type and variant TP53, we selected total RNA samples from rs78378222[A/C] heterozygotes and carried out 3′ rapid amplification of complementary DNA ends (RACE; Supplementary Fig. 3b). Amplification using a TP53 gene-specific forward primer produced a band of 1.3 kb, which is the expected length of correctly terminated mRNA (Supplementary Fig. 3c). However, sequencing of this band indicated that correctly terminated polyA(+) mRNAs were produced predominantly from the wild-type allele, with 73% of mRNAs containing the wild-type A allele and 27% containing the variant C allele (P = 1.6 × 10−6; Supplementary Fig. 3c). We then carried out RT-PCR using a ‘run-on’ reverse primer, located in the genomic sequence approximately 320 bp beyond the normal 3′ end of TP53 (Supplementary Fig. 3b). Sequencing of RT-PCR products from heterozygotes showed that this RNA species was comprised almost entirely of variant C allele transcripts (Supplementary Fig. 3d). Taken together, these data suggest that the rs78378222[C] variant impairs proper termination and polyadenylation of the TP53 transcript.
Next, we searched for associations between rs78378222 and 20 major tumor types by cross-referencing genotypes to the Icelandic Cancer Registry and national pathology records. After correcting for multiple phenotype testing, we observed significant associations for prostate cancer, brain cancers and colorectal adenoma (but not colorectal cancer) (Supplementary Table 3). We conducted a follow-up phase in an attempt to get further evidence of these associations. The follow-up phase was analogous to that described in Supplementary Figure 2, and the sample numbers are detailed in Supplementary Table 4. We directly genotyped rs78378222 in all available Icelandic cases with prostate cancer, colorectal adenoma or brain cancer and determined follow-up phase two-way–imputation-based and single-track genotype-based association values (Table 2). For prostate cancer, we further genotyped replication samples from five countries. The association with prostate cancer outside Iceland was significant (OR = 1.63, P = 1.1 × 10−4), as was the combined Iceland follow-up and replication sample result (OR = 1.44, P = 2.4 × 10−6; Table 2). For the colorectal adenoma follow-up phase, single-track genotyping gave a comparable result to two-way imputation, and the combined analysis yielded a significant association result (OR = 1.39, P = 1.6 × 10−4; Table 2 and Supplementary Table 1). However, we still observed no significant association with colorectal cancer after genotyping replication samples from four countries (combined OR = 1.06, P = 0.51, 95% CI 0.89–1.27; Supplementary Table 5). This raises the possibility that rs78378222[C] might predispose to colorectal adenomas with a low propensity for progression to invasive cancer.
Table 2.
Association between rs78378222[C] and prostate cancer, glioma and colorectal adenoma
| Sample set | Tumor type | P | OR | 95% CI | Ncases | Frequency in cases |
Ncontrols | Frequency in controls |
Pahet |
|---|---|---|---|---|---|---|---|---|---|
| Iceland follow-up phase two-way imputationb | Prostate cancer | 0.0058 | 1.35 | 1.09–1.67 | 2,671c | 0.0258 | >36,331c | 0.0192d | |
| Iceland follow-up phase single-track genotypedb | Prostate cancer | 0.27 | 1.28 | 0.82–1.99 | 635 | 0.0236 | 7,200 | 0.0185 | |
| Iceland follow-up phase combined | Prostate cancer | 0.0030 | 1.34 | 1.10–1.62 | 3,306c | NA | >43,531c | NA | 0.83 |
| The Netherlands | Prostate cancer | 0.093 | 1.41 | 0.94–2.10 | 1,085 | 0.0207 | 1,796 | 0.0147 | |
| UK | Prostate cancer | 0.24 | 1.39 | 0.80–2.42 | 521 | 0.0192 | 1,407 | 0.0139 | |
| Romania | Prostate cancer | 0.41 | 1.38 | 0.64–2.95 | 639 | 0.0110 | 815 | 0.0080 | |
| United States | Prostate cancer | 0.0026 | 2.24 | 1.33–3.79 | 1,454 | 0.0155 | 1,293 | 0.0070 | |
| Spain | Prostate cancer | 0.037 | 2.52 | 1.06–5.99 | 785 | 0.0070 | 1,787 | 0.0028 | |
| Combined non-Icelandic replication | Prostate cancer | 1.1 × 10−4 | 1.63 | 1.27–2.09 | 4,484 | NA | 7,098 | NA | 0.5 |
| Combined Iceland follow up and replication | Prostate cancer | 2.4 × 10−6 | 1.44 | 1.24–1.68 | 7,790c | NA | >50,629c | NA | 0.43 |
| Iceland follow-up phase two-way imputationb | Glioma | 0.0074 | 2.61 | 1.29–5.27 | 135c | 0.0486 | >37,881c | 0.0192d | |
| Iceland follow-up phase single-track genotypedb | Glioma | 0.22 | 1.90 | 0.69–5.27 | 72 | 0.0347 | 7,200 | 0.0185 | |
| Iceland follow-up phase combined | Glioma | 0.0036 | 2.36 | 1.32–4.20 | 207c | NA | >45,081c | NA | 0.62 |
| USA UCSF | Glioma | 0.0063 | 2.65 | 1.32–5.32 | 658 | 0.0228 | 573 | 0.0087 | |
| USA Mayo Clinic | Glioma | 0.053 | 2.05 | 0.99–4.25 | 530 | 0.0321 | 283 | 0.0159 | |
| Combined non-Icelandic replication | Glioma | 9.2 × 10−4 | 2.34 | 1.42–3.88 | 1,188 | NA | 856 | NA | 0.62 |
| Combined Iceland follow up and replication | Glioma | 1.0 × 10−5 | 2.35 | 1.61–3.44 | 1,395c | NA | >45,937c | NA | 0.88 |
| Iceland follow-up phase two-way imputationb | Colorectal adenoma | 0.0064 | 1.32 | 1.08–1.61 | 3,057c | 0.0252 | >36,022c | 0.0192d | |
| Iceland follow-up phase single-track genotypedb | Colorectal adenoma | 0.0052 | 1.60 | 1.15–2.23 | 1,038 | 0.0294 | 7,200 | 0.0185 | |
| Iceland follow-up phase combined | Colorectal adenoma | 1.6 × 10−4 | 1.39 | 1.17–1.65 | 4,095c | NA | >43,022c | NA | 0.33 |
NA, not applicable; OR, odds ratio; CI, confidence interval.
The P value for heterogeneity based on combining groups using the Mantel-Haenszel model.
The Iceland follow-up phase two-way imputation set consists of individuals who were chip-typed and individuals whose genotypes were determined by genealogy-based in silico genotyping. This set differs from the Iceland discovery phase two-way imputation set shown in Supplementary Table 2 in that it lacks those individuals whose genotypes were determined by genealogy-based in silico genotyping in the discovery phase but who were subsequently directly single-track genotyped. Such individuals form part of the Iceland follow-up phase single-track genotyped set, which consists of individuals whose genotypes were determined solely by Centaurus single-track assay. The follow-up phase two-way imputation and single-track genotyped sets are non-overlapping. See Supplementary Figure 2 and Supplementary Table 3 for further details.
The effective sample size estimate taking into account efficiency of Icelandic genealogy-based in silico genotyping.
The control frequency given is derived from 40,309 directly chip-typed control individuals without BCC.
The discovery two-way–imputation category of ‘all brain cancers’ contained meningiomas (International Classification of Diseases (ICD) 10 code C70) and gliomas (C71–C72). When glioma and meningioma were considered separately, the association appeared stronger for glioma (OR = 2.50, P = 0.0055) than for meningioma (OR = 1.48, P = 0.36). Although this difference was not significant (P = 0.88), we focused on glioma in the follow-up phase (Table 2). In Iceland, the follow-up phase yielded a suggestive association with glioma (OR = 2.36, P = 0.0036). This was confirmed in two case-control samples of adult glioma from the United States (OR = 2.34, P = 9.2 × 10−4). Combined with the Icelandic data, we obtained firm evidence that rs78378222[C] was associated with glioma (OR = 2.35, P = 1.0 × 10−5; Table 2).
We also investigated potential associations with melanoma (because of the link between rs78378222 and skin cancer risk) and breast cancer (because of the involvement of TP53 mutations in Li-Fraumeni and Li-Fraumeni–like syndromes). We did not see any convincing evidence that rs78378222 confers risk of either of these tumors (OR = 1.07, P = 0.64, 95% CI 0.81–1.42 for melanoma and OR = 1.06, P = 0.57, 95% CI 0.88–1.27 for breast cancer; Supplementary Table 5). There was no evidence of specific associations with estrogen-receptor–negative breast cancer (OR = 1.15, P = 0.61) or high-risk breast cancer (defined as an age at diagnosis under 50 or a history of multiple primary breast cancers; OR = 0.87, P = 0.36).
Given its central role in tumor biology and the high frequency of somatic mutation17, intensive efforts have been devoted to searching for germline cancer susceptibility variants in TP53. Aside from gain-of-function mutations typically found in Li-Fraumeni and Li-Fraumeni–like syndromes, no germline variants of TP53 have so far been implicated reproducibly in cancer predisposition11,18. Cross-referencing the Icelandic genealogical database with cancer registry records did not reveal any rs78378222[C] carriers with family histories fitting the criteria of Li-Fraumeni or Li-Fraumeni–like syndrome19,20. TP53 mutations leading to Li-Fraumeni and Li-Fraumeni–like syndromes are rare, occurring in 1 in 5,000 to 1 in 20,000 births21,22. Although the penetrance of rs78378222[C] for the cancers it affects is much less, carriers are expected in frequencies of up to 4% in some populations. The effect of rs78378222[C] on 3′-end processing suggests a new mechanism by which TP53 may promote oncogenesis, with a distinctive spectrum of tumors being affected.
URLs. Picard version 1.17, http://picard.sourceforge.net/.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
Acknowledgments
The work at deCODE genetics was funded in part by contract number 202059 (PROMARK) from the 7th Framework Program of the European Union. The Danish study ‘Diet, Cancer and Health’ was supported by grants from the Danish Cancer Society and ‘Europe against cancer’: European Prospective Investigation into Cancer and Nutrition (EPIC). The Inter99 study ‘A population-based primary prevention study on cardiovascular disease and type 2 diabetes’ was initiated by T. Jørgensen (principal investigator), K. Borch-Johnsen (co-principal investigator), H. Ibsen and T.F. Thomsen. The steering committee comprises the former two individuals and C. Pisinger. The establishment of the cohort was financially supported by research grants from the Danish Research Council, The Danish Centre for Health Technology Assessment, Novo Nordisk Inc., Research Foundation of Copenhagen County, the Ministry of Internal Affairs and Health, The Danish Heart Foundation, The Danish Pharmaceutical Association, The Augustinus Foundation, The Ib Henriksen Foundation and the Becket Foundation. The University of California, San Francisco (UCSF) Adult Glioma Study also acknowledges the people who have made substantial contributions to subject recruitment, specimen processing, pathology review and data analysis, including L.S. McCoy, I. Smirnov, J.S. Patoka, M.D. Prados, S.M. Chang and M.S. Berger (Department of Neurological Surgery, UCSF), J.L. Wiemels (Department of Epidemiology and Biostatistics, UCSF) and T. Tihan (Department of Pathology, UCSF). Work at the University of California, San Francisco has been supported by US National Institutes of Health (NIH) grants R01CA52689 and UCSF Brain Tumor SPORE, P50CA097257, as well as by grants from the National Brain Tumor Foundation, the UCSF Lewis Chair in Brain Tumor Research and by donations from families and friends of J. Berardi, H. Glaser, E. Olsen, R.E. Cooper and W. Martinusen. Work at the Mayo Clinic has been supported by the Mayo Clinic Brain Tumor SPORE (NIH P50 CA108961), the Mayo Clinic Comprehensive Cancer Center (NIH P30 CA15083) and an American Recovery and Reinvestment Act (ARRA) Recovery grant (NIH NS068222). Members of the Swedish Low-risk Colorectal Cancer Study Group are: D. Edler, Karolinska Universitetssjukhuset, Solna, Stockholm, Sweden; C. Lenander, Mag-tarm-centrum, Ersta sjukhus, Stockholm, Sweden; J. Dalén, St Görans sjukhus, Stockholm, Sweden; F. Hjern, Danderyds sjukhus, Danderyd, Sweden; N. Lundqvist, Norrtälje sjukhus, Norrtälje, Sweden; U. Lindforss, Södertälje sjukhus, Södertälje, Sweden; L. Påhlman, Akademiska sjukhuset, Uppsala, Sweden; K. Smedh, Centrallasarettet, Västerås, Sweden; A. Törnqvist, Centralsjukhuset, Karlstad, Sweden; J. Holm, Länssjukhuset Gävle-Sandviken, Gävle. Sweden; M. Janson, Karolinska Universitetssjukhuset, Huddinge, Huddinge, Sweden; M. Andersson, Universitetssjukhuset, Örebro, Sweden; S. Ekelund, Södersjukhuset, Stockholm, Sweden; and E. Olsson, Mälarsjukhuset, Eskilstuna, Sweden. Work on the US Prostate Cancer sample set was supported in part by the Urological Research Foundation, Prostate SPORE grant (P50 CA90386-05S2) and the Robert H. Lurie Comprehensive Cancer Center grant (P30 CA60553). For the UK Prostate Cancer sample set, the UK Department of Health funded the ProtecT study through the National Institute for Health Research (NIHR) Health Technology Assessment programme (projects 96/20/06, 96/20/99). We acknowledge the contribution of all members of the ProtecT study research group. We acknowledge the support of the NIHR Cambridge Biomedical Research Centre and the National Cancer Research Institute (ProMPT) Prostate Cancer Collaborative. The views and opinions expressed herein are those of the authors and do not necessarily reflect those of the UK Department of Health. Sample collection in Romania was supported in part by the Romanian National Council For Scientific Research (CNCSIS-UEFISCSU), grant PNII-IDEI 1184/2008. In Spain, J.I.M. is funded by Red Tematica de Investigacion Cooperative en Cancer RD06/0020/1054.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
The study was designed and the results were interpreted by S.N.S., P.S., G.M., D.F.G., O.T.M., J.H.O., A.K., U.T., T. Rafnar and K.S. Subject ascertainment and recruitment was carried out by S.N.S., J.G., B.S., K.T., R.R., K.R.B., B.A.N., A.T., K.O., P.R., E.G., K.K., K.H., C.C., V.F., P.G., S.N., F.F., M.D.G.-P., E.S., A.P., A.D.J., A.G., F.R., D.P., V. Soriano, C.R., K.K.A., M.M.v.R., R.G.H.M.C., I.M.v.O., D.-J.v.S., J.A.S., W.H.M.P., B.T.H., J.L.D., F.C.H., D.B., O.C., M.J., I.E.C., V.C., P.B., I.N.M., D.E.D., A.C., D.M., S.K., B.A.A., E.J., R.B.B., G.V.E., F.S., P.H.M., T.S., T.V., O.T.J., H.S., T. Jonsson, J.G.J., L.T., T. Rice, H.M.H., Y.X., D.H.L., B.P.O., M.L.K., P.A.D., V. Steinthorsdottir, A.L., R.S.S., T.O.K., K.B., T. Jørgensen, D.R.W., T.H., O.P., V.J., D.E.N., W.J.C., M.W., J.W., R.B.J., E.N., U.V., L.A.K., R.K., J.I.M., J.H.O., U.T. and T. Rafnar. The sequencing, genotyping and expression analysis was carried out by S.N.S., A.J., J.G., O.T.M., H.J., H.T.H., A.S. and U.T. The statistical and bioinformatics analysis was carried out by S.N.S., P.S., G.M., D.F.G., S.A.G., G.T. and A.K. S.N.S., P.S., D.F.G., T. Rafnar and K.S. drafted the manuscript. All authors contributed to the final version of the paper. Principal collaborators for the case-control population samples were: S.K. (Colorectal Adenoma), G.V.E. (Iceland Prostate), V.J. (Romania Prostate), D.E.N. (UK Prostate), W.J.C. (US Prostate), M.W. (US UCSF Glioma), R.B.J. (US Mayo Clinic Glioma), E.J. and J.I.M. (Spain BCC and Prostate), U.V. and O.P. (Denmark BCC), L.A.K. (Netherlands Prostate), R.K. (Eastern Europe BCC) and J.H.O. (Iceland BCC).
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturegenetics/.
References
- 1.de Zwaan SE, Haass NK. Genetics of basal cell carcinoma. Australas. J. Dermatol. 2010;51:81–92. doi: 10.1111/j.1440-0960.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- 2.Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat. Rev. Cancer. 2008;8:743–754. doi: 10.1038/nrc2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Box NF, et al. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. J. Invest. Dermatol. 2001;116:224–229. doi: 10.1046/j.1523-1747.2001.01224.x. [DOI] [PubMed] [Google Scholar]
- 4.Gudbjartsson DF, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 5.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stacey SN, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat. Genet. 2008;40:1313–1318. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 7.Stacey SN, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong A, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutyavin IV, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat. Rev. Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 12.Cui R, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128:853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 13.Miller AJ, Tsao H. New insights into pigmentary pathways and skin cancer. Br. J. Dermatol. 2010;162:22–28. doi: 10.1111/j.1365-2133.2009.09565.x. [DOI] [PubMed] [Google Scholar]
- 14.Sulem P, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 15.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 16.Higgs DR, et al. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- 17.Junttila MR, Evan GI. p53—a Jack of all trades but master of none. Nat. Rev. Cancer. 2009;9:821–829. doi: 10.1038/nrc2728. [DOI] [PubMed] [Google Scholar]
- 18.Palmero EI, Achatz MI, Ashton-Prolla P, Olivier M, Hainaut P. Tumor protein-53 mutations and inherited cancer: beyond Li-Fraumeni syndrome. Curr. Opin. Oncol. 2010;22:64–69. doi: 10.1097/CCO.0b013e328333bf00. [DOI] [PubMed] [Google Scholar]
- 19.Li FP, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48:5358–5362. [PubMed] [Google Scholar]
- 20.Birch JM, et al. Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res. 1994;54:1298–1304. [PubMed] [Google Scholar]
- 21.Gonzalez KD, et al. Beyond Li Fraumeni syndrome: clinical characteristics of families with p53 germline mutations. J. Clin. Oncol. 2009;27:1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 22.Lalloo F, et al. Prediction of pathogenic mutations in patients with early-onset breast cancer by family history. Lancet. 2003;361:1101–1102. doi: 10.1016/S0140-6736(03)12856-5. [DOI] [PubMed] [Google Scholar]
- 23.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.