Abstract
The coordinated induced expression of β-oxidation genes is essential to provide the energy supply for germination and postgerminative development. However, very little is known about other functions of β-oxidation in nonreserve organs. We have identified a gene-specific pattern of induced β-oxidation gene expression in wounded leaves of Arabidopsis. Mechanical damage triggered the local and systemic induction of only ACX1 among acyl-coenzyme A oxidase (ACX) genes, and KAT2/PED1 among 3-ketoacyl-coenzyme A thiolase (KAT) genes in Arabidopsis. In turn, wounding induced KAT5/PKT2 only systemically. Although most of the β-oxidation genes were activated by wound-related factors such as dehydration and abscisic acid, jasmonic acid (JA) induced only ACX1 and KAT5. Reduced expression of ACX1 or KAT2 genes, in transgenic plants expressing their corresponding mRNAs in antisense orientation, correlated with defective wound-activated synthesis of JA and with reduced expression of JA-responsive genes. Induced expression of JA-responsive genes by exogenous application of JA was unaffected in those transgenic plants, suggesting that ACX1 and KAT2 play a major role in driving wound-activated responses by participating in the biosynthesis of JA in wounded Arabidopsis plants.
Plants often undergo the onslaught of chewing insects or larger herbivores that cause damage to the leaves. Preexisting physical barriers may not be enough to prevent injuries and thus, plants require active inducible defense mechanisms. Moreover, once an injury occurs there is no possibility of wound healing by mobilization of specialized cells as it occurs in animals. In plants, every cell has become competent for the activation of wound-triggered defense responses. Wound-activated defense relies on the production or release of signals in the damaged tissues and the local and systemic activation of signaling pathways (León et al., 2001). Wound-activated signaling pathways usually lead to the transcriptional activation of defense-related genes. In addition, damaged areas undergo a severe disorder of tissue and cellular structures that is accompanied by a drastic loss of water (Reymond et al., 2000). Wound-activated gene expression seems to be the result of the combined action of damage and water stress of the wounded leaf (Reymond et al., 2000), processes that require the synthesis, accumulation, and perception of jasmonic acid (JA) and abscisic acid (ABA; Peña-Cortés et al., 1995; Bergey et al., 1996). The signaling function of jasmonates in wound-activated defense has been extensively documented (Turner et al., 2002). Besides, jasmonates are also involved in pathogen-triggered defense in coordination with the function of salicylic acid (SA; Glazebrook et al., 2003).
Plants synthesize jasmonates from linolenic acid through the octadecanoid pathway (Schaller, 2001). This is a complex metabolic pathway involving the participation of different subcellular organelles. The release of linolenic acid from membrane lipids and subsequent redox reactions to 12-oxo-10,15(Z)-octadecatrienoic acid occurs in chloroplasts. By a still unknown mechanism 12-oxo-10,15(Z)-octadecatrienoic acid is transported into peroxisomes where it is first reduced and then the pathway completed by three consecutive steps of β-oxidation to yield JA (Schaller, 2001; Strassner et al., 2002). Although it is well known that the expression of most of the JA biosynthetic genes is induced by different stress factors including pest and pathogen attacks (Schaller, 2001; Turner et al., 2002), nothing is known about the identity and regulation of the β-oxidation genes involved in the biosynthesis of jasmonates and the activation of wound-related defense.
β-Oxidation occurs in glyoxisomes and peroxisomes of plants and requires the consecutive action of acyl-coenzyme A (CoA) oxidases (ACX) and multifunctional proteins (MFP) with, at least, enoyl-CoA hydratase and β-hydroxyacyl-CoA dehydrogenase activities, and 3-ketoacyl-CoA thiolases (KAT), which finally release acetyl-CoA and the substrate with two less carbon units, which may undergo a new β-oxidation cycle (Graham and Eastmond, 2002). The Arabidopis genome (Arabidopsis Genome Initiative [AGI], 2000), includes six genes encoding ACXs, ACX1 and ACX2 (Hooks et al., 1999); ACX3 (Eastmond et al., 2000; Froman et al., 2000); ACX4 (Hayashi et al., 1999); and the still uncharacterized homologs of ACX1 and ACX3, which we have named ACX1.2, and ACX3.2 (ACX5 and ACX6, respectively, in Rylott et al., 2003); at least two MFPs, AIM1 and MFP2 (Richmond and Bleecker, 1999); and three KATs, PED1/KAT2 (Hayashi et al., 1998; Germain et al., 2001), PKT2/KAT5 (Germain et al., 2001), and KAT1, a homolog of PED1/KAT2 in chromosome 1. Although β-oxidation has been traditionally considered as a catabolic machinery devoted to the production of energy through fatty acid degradation, it can be also considered as a processing system to convert complex precursors into simpler molecules. To test the involvement of β-oxidation in wound-activated generation of signaling molecules we have analyzed whether the expression of genes coding for ACXs and KATs in Arabidopsis may be altered in response to mechanical damage, dehydration or treatment with wound- and dehydration-related molecules. We also analyzed whether the induced expression of ACX or KAT genes is affected in Arabidopsis genotypes either insensitive to JA or SA-deficient such as coi1-1 mutant and nahG transgenic plants, respectively (Delaney et al., 1994; Feys et al., 1994), or in the double mutant transgenic coi1-1 nahG that we have generated. After identification of a gene-specific pattern of β-oxidation induction by wounding, we have generated transgenic plants with reduced expression of β-oxidation ACX1 and KAT2 genes regulated by wounding, and we have used them to test the involvement of those gene products in the biosynthesis of JA as well as in the further activation of JA-mediated wound-related defense. Our results provide evidence for the participation of ACX1 and KAT2 in the biosynthesis of JA in wounded Arabidopsis leaves.
RESULTS
Differential Accumulation of ACX and KAT Transcripts in Wounded and Dehydrated Arabidopsis
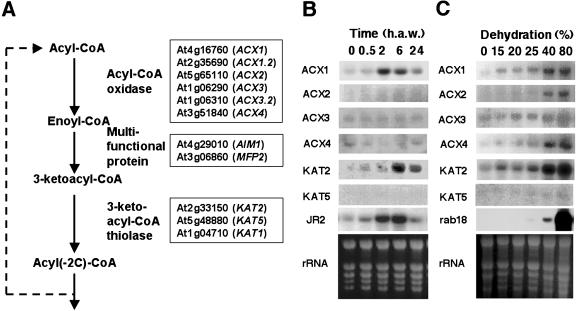
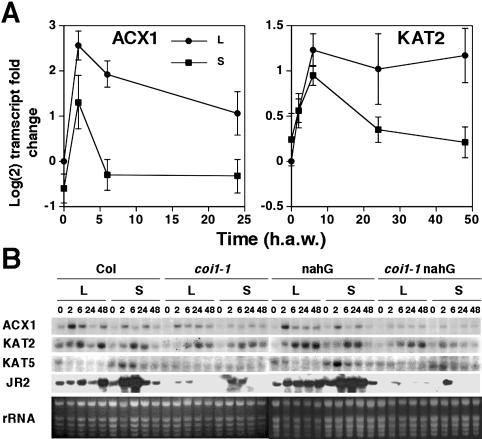
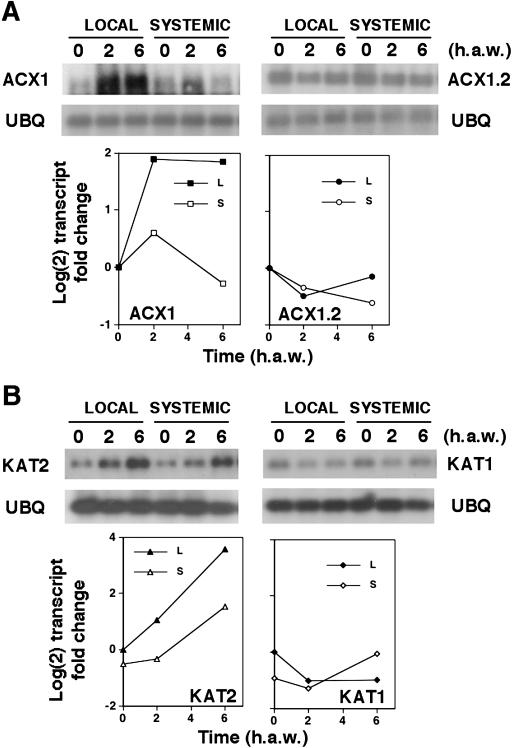
β-Oxidation is a complex biochemical process requiring the function of several enzymes, which are encoded by multiple genes in Arabidopsis. Figure 1A shows a diagram of the β-oxidation pathway including the enzymes and the corresponding AGI loci names and symbols of Arabidopsis β-oxidation genes. We have explored whether the expression of β-oxidation genes might be altered in mechanically wounded plants undergoing damage and water loss. We analyzed the levels of transcripts of ACX and KAT genes in mechanically damaged leaves and also in unwounded dehydrated Arabidopsis plants. Whereas dehydration induced all ACX and KAT genes in Arabidopsis, mechanical damage triggered the activation of just a subset of β-oxidation genes (Fig. 1, B and C). Figure 1C shows that ACX1 and KAT2 transcripts accumulated over basal levels in seedlings undergoing 15% dehydration, and transcript levels increased progressively with the degree of dehydration. Other genes such as ACX2 and ACX4 required water losses of at least 40%, a result similar to that observed for rab18, which is a typical marker of dehydration and cold acclimation responses in Arabidopsis (Lang and Palva, 1992). Genes such as ACX3 and KAT5 were the less sensitive to dehydration, requiring more than 80% of water loss to be induced. In turn, only the ACX1 transcript among those of ACX genes accumulated in wounded leaves (Fig. 1B). Transient induction of ACX1 started as soon as 30 min after wounding (a.w.) and reached a maximum around 2 h a.w. The ACX1 transcript level returned to basal levels detected in nonwounded leaves by 24 h a.w. KAT2 transcript accumulated in wounded leaves starting at 30 min a.w. to peak at around 6 h a.w (Fig. 1B). In contrast to ACX1, KAT2 transcript levels were still significantly elevated by 24 h a.w. (Fig. 1B). The kinetic of wound-induced ACX1 gene expression is similar to that of the jasmonic acid-responsive JR2 gene of Arabidopsis (Fig. 1B), which has been characterized as a marker of the JA-dependent wound-induced signaling pathway in Arabidopsis (Titarenko et al., 1997). We have further analyzed whether wound-induced expression of ACX1 and KAT2 genes occurred not only locally but also systemically in nonwounded leaves of wounded plants. As shown in Figure 2, wounding caused not only local but also systemic accumulation of ACX1 and KAT2 transcripts in wounded plants. Systemic accumulation of ACX1 and KAT2 transcripts peaked at 2 and 6 h a.w., respectively (Fig. 2A). Whereas ACX1 induction was above 6- and 3-fold over basal levels in local wounded and systemic unwounded leaves, respectively, induction of KAT2 was around 2.5-fold both locally and systemically (Fig. 2A). We checked that wounding did not induce both local and systemic accumulation of any of the other ACX or KAT genes (data not shown). However, KAT5, which was not induced locally in wounded leaves (Fig. 1B), was found to be systemically induced with a maximum around 2 h a.w. (Fig. 2B). To gain insight into the mechanism involved in wound-activated expression of β-oxidation genes, we tested local and systemic induction by wounding in the JA-insensitive coi1-1 mutant, in the SA-deficient nahG transgenic Arabidopsis plants, and in the double coi1-1 nahG transgenic mutant plants (generated as described in “Materials and Methods” section). Local and systemic induction of ACX1 was largely reduced in coi1-1 and almost undetectable in coi1-1 nahG compared to that detected in Arabidopsis ecotype Columbia (Col) wild-type plants (Fig. 2B). However, full induction of ACX1 by wounding occurred in nahG plants (Fig. 2B), suggesting that full wound-induced expression of ACX1 requires COI1-mediated JA-dependent signaling but is unaffected by SA deficiency. In contrast, local and systemic induction of KAT2 by wounding was essentially similar in the JA-insensitive coi1-1 background, the SA-deficient nahG transgenics, and the wild-type plants (Fig. 2B), suggesting that induction of KAT2 by wounding requires neither JA perception nor SA accumulation. Accumulation of KAT5 transcript in systemic leaves of wounded wild-type Col plants was abolished in the coi1-1 or coi1-1 nahG mutants (Fig. 2B), suggesting that systemic induction of KAT5 by wounding was fully COI1-dependent, as it has been shown for the wound-responsive JA-dependent JR2 gene.
Figure 1.
Genes coding for enzymes of the β-oxidation pathway and pattern of expression upon wounding and dehydration in Arabidopsis. A, AGI loci name and symbol abbreviation of genes annotated in the Arabidopsis genome coding for ACXs, MFPs, and KATs. Northern-blot analysis of ACX and KAT transcript accumulation in wounded (B) and dehydrated (C) Arabidopsis. Soil- grown plants were wounded by crushing leaves with forceps and leaf samples harvested at the indicated times (h a.w.). Progressive dehydration of 10-d-old seedlings was performed as indicated in “Materials and Methods.” Percentage of dehydration was estimated from the weight ratio of seedlings after dehydration process to zero time at the moment of liquid medium removal. Blots were hybridized with specific probes for different ACX and KAT genes and with probes for JR2 and rab18 as markers of wound- and dehydration-induced gene expression. Ethidium bromide staining of the rRNA is included as loading control.
Figure 2.
Local and systemic accumulation of wound-inducible ACX and KAT genes. Wild-type Col and either coi1-1 mutant, nahG transgenic, or coi1-1 nahG double mutant transgenic Arabidopsis plants, grown for 3 weeks in soil under 16 h light/8 h darkness photoperiod, were used for wounding one-half of the rosette leaves of every plant. At the indicated times (h a.w.), leaf samples were harvested from either wounded (local; L) or nonwounded (systemic; S) leaves and the total RNA isolated. A, Relative transcript levels (-fold) in wild-type Col plants were analyzed by northern and the values displayed in y axis are the mean of Log(2) of the transcript fold change ± se of eight and six independent experiments for ACX1 and KAT2, respectively. B, Levels of ACX1, KAT2, and KAT5 transcripts in wounded leaves and unwounded systemic rosette leaves of mechanically damaged wild-type, JA-insensitive coi1-1 mutants, SA-deficient nahG transgenic plants, and double coi1-1 nahG. The accumulation of the corresponding wound-inducible transcripts is compared to the JA-dependent wound-activated JR2 gene. Ethidium bromide staining of rRNA is included as loading control.
Because of the high sequence homology (more than 85% identity at the nucleotide sequence) between ACX1 (At4g16760) and its homolog ACX1.2 (At2g35690), and between KAT2 (At2g33150) and its homolog KAT1 (At1g04710), we analyzed the expression of these genes by reverse transcription (RT)-PCR. cDNAs obtained by RT of RNAs from local wounded and systemic leaves were amplified by PCR with specific primers for ACX1, ACX1.2, KAT2 and KAT1. Figure 3 shows the corresponding Southern analysis and quantification of transcripts after normalization to the endogenous content of ubiquitin10 (UBQ) transcript. Whereas expression of ACX1 and KAT2 genes was induced in local wounded and in systemic leaves, no induction was detected for ACX1.2 or KAT1 genes (Fig. 3). Quantification of uninduced levels of transcripts indicated that ACX1 and KAT2 levels were between 50- and 100-fold higher than ACX1.2 and KAT1, respectively (data not shown), suggesting that neither ACX1.2 nor KAT1 genes are functionally homologs of ACX1 and KAT2, respectively.
Figure 3.
RT-PCR analysis of wound-induced expression of ACX1and KAT2 genes and their sequence-related ACX1.2 and KAT1 genes. Samples were collected from wounded (local) and unwounded (systemic) leaves, at the indicated times after wounding. Total RNA was isolated, reverse transcribed, amplified by PCR with specific primers, and the resulting DNAs were separated by electrophoresis in 1% agarose gel. DNA was blotted and analyzed by the Southern technique with probes detecting both ACX1 and ACX1.2 (A), and KAT2 and KAT1 cDNAs (B). Amplification of UBQ gene was conducted for normalization of the quantitative transcript analysis. The relative transcript levels were quantified by PhosphorImager analysis, normalized for the UBQ content and were relatives to the levels detected in leaves at zero time after wounding. Values in y axis represent the Log (2) of the transcript fold change.
Differential Induction of ACX and KAT Genes by JA and ABA
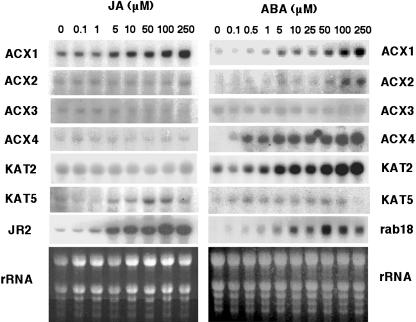
We checked whether JA or ABA induced the expression of ACX and KAT genes in Arabidopsis. Whereas all genes but ACX3 and KAT5 were induced by ABA, JA only up-regulated the expression of ACX1 and KAT5 (Fig. 4). The ACX1 and KAT5 transcripts accumulated in a dose-dependent manner by treatment with JA at concentrations above 5 μm, similarly to the JA-responsive JR2 gene (Fig. 4). The ABA-induced expression of different ACX and KAT genes showed different degrees of sensitivity to ABA. Responsiveness to ABA of ACX1 and KAT2 genes was found to be very similar to that shown by rab18. ACX2 was found to be less sensitive to ABA than rab18 and, by contrast, ACX4 was more sensitive (Fig. 4). Treatment of coi1-1 mutant plants with JA did not lead to induced expression of ACX1, KAT5, and JR2 genes, whereas ABA-induced expression of β-oxidation genes in coi1-1 mutants was similar to that observed in wild-type plants (data not shown).
Figure 4.
Northern-blot analysis of ACX and KAT transcript accumulation in Arabidopsis plantlets treated with JA or ABA. 10-d-old seedlings were treated with JA or ABA at the final concentrations indicated and samples collected at 6 h after application. Ethidium bromide staining of rRNA is included as loading control.
Reduced Wound-Activated Gene Expression and JA Synthesis in Transgenic Antisense Lines of ACX1 and KAT2
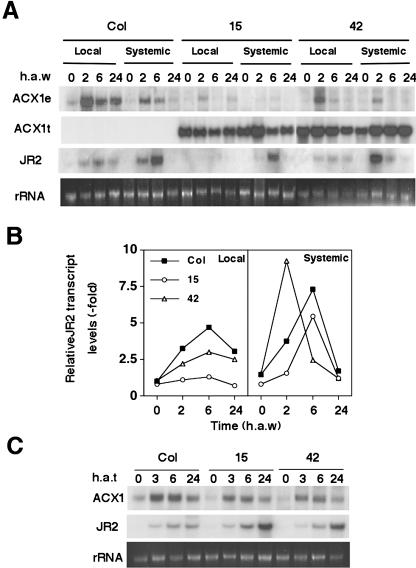
We have generated transgenic Arabidopsis lines expressing antisense mRNAs from ACX1 and KAT2 genes under the control of the 35S cauliflower mosaic virus promoter. From more than 60 independent lines generated for every gene, we selected homozygous lines with a single insertion displaying antisense effect either by reducing basal or wound-induced endogenous transcript levels. Figure 5 shows the northern analysis of basal and systemic endogenous transcript, transgene, and JR2 transcript for wild-type plants and two independent ACX1 antisense transgenic lines at different times after wounding. High levels of expression of the transgene correlated to reductions of both basal (20%–65%) and wound-induced (50%–90%) endogenous transcript levels (Fig. 5A). Concomitantly, wound-induced JR2 transcript accumulation in antisense transgenic lines was lower than in Col plants. ACX1 antisense transgenic line number 15, which showed the best antisense effect, did not induce JR2 gene locally in wounded leaves and exhibited a significant reduction in systemic transcript accumulation (Fig. 5, A and B). We also examined whether antisense lines were affected in the responsiveness to JA. Since accumulation of JR2 transcript in transgenic plants treated with exogenous JA was not reduced when compared to wild-type plants (Fig. 5C), ACX1 antisense lines are not defective in either perception of JA or downstream signaling events.
Figure 5.
Wound-activated gene expression in transgenic lines expressing antisense ACX1 mRNAs. Homozygous transgenic plants with single insertions of antisense ACX1 constructs were wounded by thoroughly crushing one-half of the rosette leaves (A and B) or treated with 250 μm JA (C). Leaf samples from either wounded leaves (local) or nonwounded leaves (systemic) of the rosette were collected at the indicated times (h a.w.). Total RNAs were isolated and analyzed by the northern technique with the following radiolabeled probes: a 557-bp XhoI 5′ fragment of the ACX1 cDNA that hybridizes only to the endogenous transcript; the transgene, corresponding to 3′noncoding sequence of ACX1 cDNA, which hybrizes to both endogenous transcript and transgene; and a fragment of 1.2 kb of JR2 cDNA. When indicated, quantification of endogenous transcripts was performed by PhosphorImager analysis and expressed as relative values compared to transcript levels of nonwounded leaves of wild-type plants. Ethidium bromide staining of rRNA is included as loading control.
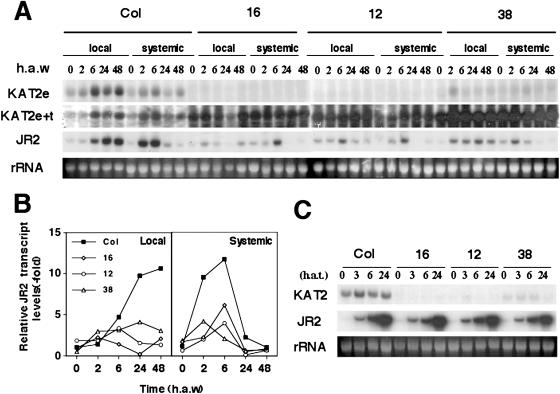
We selected three independent transgenic lines that show strong antisense expression of the KAT2 gene and greater than 90% reduction in the basal and wound-induced endogenous levels of the KAT2 transcript (Fig. 6A). As a result of reduced KAT2 expression, wound-induced expression of the JA-responsive JR2 gene was severely reduced both locally (60%) and systemically (75%) in KAT2 antisense lines (Fig. 6, A and B). However, this effect was not due to a reduced sensitivity or altered JA signaling because all three transgenic lines, similarly to wild-type plants, fully induced JR2 gene expression by exogenous JA application (Fig. 6C).
Figure 6.
Transgenic lines expressing antisense KAT2 mRNAs are defective in JA-mediated wound-triggered expression of JR2 gene. Homozygous transgenic plants with single insertions of antisense KAT2 constructs were wounded by thoroughly crushing one-half of the rosette leaves (A and B) or treated with 250 μm JA (C). Leaf samples from either wounded leaves (local) or nonwounded leaves (systemic) of the rosette were collected at the indicated times (h a.w.). Total RNAs were isolated and analyzed by the northern technique with the following radiolabeled probes: a full-length cDNA of KAT2, which hybridizes to endogenous transcript and transgene; a 241-bp fragment of noncoding 3′end sequence of KAT2 cDNA that is not present in the transgene and, consequently, only hybridized to endogenous transcript; and a fragment of 1.2 kb of JR2 cDNA. When indicated, quantification of endogenous transcripts was performed by PhosphorImager analysis as described in the legend of Figure 5. Ethidium bromide staining of rRNA is included as loading control.
To test whether antisense lines of ACX1 and KAT2 genes are impaired in the synthesis or accumulation of JA, we determined the basal and wound-induced levels of jasmonates in leaves of wild-type and antisense transgenic lines. Table I summarizes the endogenous content of jasmonates in unwounded (at zero time) and wounded leaves at 90 min a.w. from wild-type and transgenic plants. A 40% to 50% reduction in wound-induced accumulation of jasmonates was detected in ACX1 antisense lines and the reduction was between 65% and 80% in KAT2 antisense lines (Table I).
Table I.
Levels of jasmonates in unwounded and wounded leaves of wild-type and antisense ACX1 and KAT2 transgenic lines
| Jasmonates (ng/g FW)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Exp.1
|
Exp.2
|
|||||||||
| Time | Col | asACX1 no. 15 | asACX1 no. 42 | asKAT2 no. 12 | asKAT2 no. 16 | Col | asACX1 no. 15 | asKAT2 no. 12 | asKAT2 no. 16 | |
| min a.w. | ||||||||||
| 0 | 61 ± 21 | 43 ± 5 | 32 ± 1 | 31 ± 10 | 31 ± 4 | 49 ± 4 | 74 ± 8 | 57 ± 17 | 62 ± 11 | |
| 90 | 5361 ± 234 | 2576 ± 246 | 3121 ± 763 | 688 ± 250 | 760 ± 179 | 1745 ± 244 | 1169 ± 84 | 395 ± 31 | 451 ± 46 | |
Mean values ± se of four and six replicate samples for Exp.1 and Exp.2, respectively. Jasmonates were analyzed by gas chromatography mass spectrometry in methanolic extracts from unwounded control plants at zero time (0 min a.w.) and from wounded leaves at 90 min a.w.
Overall, our data suggest that β-oxidation is transcriptionally activated in a gene-specific manner by mechanical wounding. Results presented in this paper point out to ACX1 and KAT2 genes as the major targets for wound-triggered up-regulation, their corresponding products likely being involved in the biosynthesis of jasmonates in response to damage.
DISCUSSION
β-Oxidation plays a critical role in germination and early postgerminative development in higher plants. By degrading fatty acids to acetyl-CoA, which is further metabolized through the glyoxylate cycle, the energy liberated sustains plant growth until cotyledons become green and photosynthesis is functional (Kindl, 1987; Graham and Eastmond, 2002). Besides this essential role, β-oxidation has been suggested to be involved in the generation of stress-related signaling molecules such as JA (Vick and Zimmerman, 1984) and SA (Ribnicky et al., 1998; Hertweck et al., 2001) and also in the regulation of flower development (Richmond and Bleecker, 1999). We have explored the involvement of β-oxidation in wound-activated responses in leaves of Arabidopsis. For this purpose, we have analyzed the expression of the whole set of genes annotated in the Arabidopsis genome that potentially code for ACX and KAT proteins.
Although the expression of β-oxidation genes is coordinately regulated during the mobilization of storage lipids in germinating seeds of Arabidopsis (Rylott et al., 2001), we have found that only a subset of β-oxidation genes is activated in response to stress, and also that this subset is specific for a given kind of stress factor. Whereas dehydration activates the expression of most of the ACX and KAT genes (Fig. 1C), expression in response to wounding is gene-specific (Figs. 1–3). Wounding triggers a cascade of complex signaling events, which include JA-mediated processes that seem to be crucial for systemic wound-activated responses (Stratmann, 2003). Regarding this, the β-oxidation pathway has been proposed to be involved in the biosynthesis and modification of octadecanoid-derived molecules including jasmonates (Schaller, 2001). Although the involvement of β-oxidation gene products in JA biosynthesis has been postulated (Schaller 2001; Strassner et al., 2002), the identity of the related genes has remained unknown to date. In this study, we detected wound-activated expression of only ACX1 (Figs. 1–3), KAT2, and KAT5 genes (Figs. 1 and 2) among the ACX and KAT genes involved in β-oxidation. Of those, KAT5 appears to be induced only systemically and in strict dependence of COI1 and/or perception of JA (Fig. 2B). Moreover, the levels of expression of KAT5 were far below those of KAT2 (e.g. barely detectable by northern analysis; Figs. 1 and 2). It seems that β-oxidation requirements for local and systemic wound-activated responses are mainly driven by the ACX1/KAT2 system. However, the regulation of ACX1 and KAT2 expression, despite both being wound-inducible, differ in terms of JA responsiveness. Wound-activated expression of ACX1 is reduced but still detected in the JA-insensitive coi1-1 mutant (Fig. 2). However, wound-activated expression of KAT2 is unaffected in the coi1-1 mutant (Fig. 2). Moreover, whereas JA induces ACX1, KAT2 is not responsive to that phytohormone. These data suggest that wound-activated β-oxidation may be, at least in part, driven by the induced expression of ACX1 and KAT2 genes through the previously described JA-independent pathway in Arabidopsis (Titarenko et al., 1997; Rojo et al., 1999; León et al., 2001). However, induction of KAT5, as well as ACX1 by exogenous JA (Fig. 4), and systemic induction of KAT5 by wounding (Fig. 2), seem to point to a role of KAT5 as partner of ACX1 in the wound-activated JA-dependent systemic responses.
We detected a generalized β-oxidation gene induction upon dehydration (Fig. 1C), which contrasts to the gene-specific wound-activated expression of β-oxidation genes in Arabidopsis (Fig. 1B). Dehydration has been proposed as a component of both mechanical wounding and senescence, most likely mediated by the action of jasmonates (Reymond et al., 2000, He et al., 2002). We have shown that the expression of a subset of β-oxidation genes, which are induced by wounding, dehydration, and treatment with JA or ABA do not overlap (Figs. 1, 2, and 4), suggesting the existence of distinct signaling pathways, triggered by different stimuli, that can activate the expression of β-oxidation genes. Our data also indicate the uncoupling of dehydration- and damage-related components in the wound-induced expression of β-oxidation genes. Analysis of 150 Arabidopsis genes by cDNA microarrays revealed that a large fraction, but not all of the wound-inducible genes, required damage-related water losses to be induced by wounding (Reymond et al., 2000). ACX1 is included among those genes, in agreement with our results showing wound- and dehydration-induced accumulation of ACX1 transcript (Fig. 1). However, although microarray analysis revealed a lack of significant induction of PED1/KAT2 by wounding and dehydration (Reymond et al., 2000), we have reproducibly detected PED1/KAT2 transcript accumulation both in wounded and dehydrated Arabidopsis leaves (Figs. 1–3). Moreover, Reymond and colleagues proposed a strictly COI1-dependent mechanism for wound-induced expression of ACX1. However, our results indicate that although mainly induced through a COI1-dependent pathway, ACX1 induction by wounding is still detectable in the coi1-1 mutant (Fig. 2B), suggesting that COI1-independent mechanisms may also be involved in wound-activated expression of this gene. Systemic wound-induced β-oxidation system involving ACX1 and KAT2 may be operating through a COI1-independent mechanism, which is not likely to be JA-responsive since KAT2 was found to be insensitive to JA (Fig. 4). Whether the ACX1-KAT2 pathway might be activated by a mobile wound-generated signal other than JA or by similar signals through different mechanisms will require further analysis. Remarkably, ACX1 and KAT2 are strongly induced by application of ABA, whereas KAT5 does not seem to be responsive to this phytohormone (Fig. 4). Activation of wound responses in solanaceous plants is tightly linked to local and systemic accumulation of both JA and ABA (Peña-Cortés et al., 1995). However, in Arabidopsis and other plants, the coordinated actions of JA and ABA activating the expression of wound-inducible genes have been uncoupled (Lee et al., 1996, Dammann et al., 1997). Nevertheless, the involvement of ABA in wound-activated gene expression remains controversial. It has been reported that in tomato (Lycopersicon esculentum) perception of ABA is required for wound activation of proteinase inhibitor genes (Carrera and Prat, 1998), but that ABA does not function as a primary wound signal (Birkenmeier and Ryan, 1998). It is worth noting that studies with tomato and potato (Solanum tuberosum) were often made using proteinase inhibitors as a representative of wound-inducible genes although they may not be good markers of JA-independent wound-signaling pathways. This could be the case of wound-activated expression of β-oxidation genes reported in this study in Arabidopsis. Our results support the existence of both JA-dependent and JA-independent, maybe ABA-dependent, systemic wound activation of β-oxidation genes. These data support also the involvement of de novo JA biosynthesis in systemic responses to wounding in Arabidopsis.
In addition to the ACX and KAT genes previously reported we have characterized ACX1.2 and KAT1, which code for proteins very similar in amino acid sequence to ACX1 and KAT2, respectively. RT-PCR analysis showed that both genes are expressed, although their transcript levels were far below those detected for ACX1 and KAT2. Moreover, neither ACX1.2 nor KAT1 were responsive to wounding (Fig. 3). We also checked that neither JA nor ABA induced ACX1.2 or KAT1 genes (data not shown). These data suggest that ACX1.2 and KAT1 are not functional homologs of ACX1 and KAT2 in wound-related defense.
We explored whether ACX1 and KAT2 genes may be involved in the wound-induced synthesis of JA and JA-mediated expression of defense-related genes. For that, we generated transgenic lines of Arabidopsis expressing ACX1 and KAT2 mRNAs in antisense orientation. Further analysis of transcript levels of JA-responsive genes in response to wounding showed a good correlation between ACX1 and KAT2 endogenous transcript reduction with decreased wound-activated expression of JR2 in antisense lines when compared to wild-type plants (Figs. 5 and 6). Despite the reduced activation of JR2 by wounding in transgenic lines, these plants are fully responsive to exogenous application of JA (Figs. 5 and 6), suggesting that transgenic plants are affected in the wound-triggered synthesis of JA and not in its perception or downstream signaling of this molecule. We have confirmed that ACX1 and KAT2 antisense transgenic plants have reduced ability to accumulate JA in response to mechanical damage, and also that JA deficiency parallels the antisense effect on the accumulation of the corresponding endogenous transcripts (Figs. 5 and 6; Table I). However, the fact that ACX1 antisense plants, despite displaying reduced expression of endogenous ACX1 gene, only show reduced JA accumulation and JR2 induction in wounded leaves but not systemically, likely means that the levels of ACX1 expression remaining in antisense plants may be enough to guarantee systemic accumulation of JA and JR2 expression in response to wounding. Alternatively, wound-induced systemic induction of JR2 gene may require the function of other signaling molecule upstream JA in the pathway. Nevertheless, our data regarding KAT2 antisense expression seem to point to KAT2 as an essential component for local and systemic JA accumulation, whereas ACX1 would not be a rate-limiting step, at least for the systemic synthesis.
In Arabidopsis, wound-triggered synthesis of JA reaches a maximum around 90 min a.w. to decrease thereafter (Stenzel et al., 2003). Although ACX1 and KAT2 transcripts peaked at 2 and 6 h a.w., we found that both genes start to be induced already by 0.5 h a.w. (Fig. 1B and data not shown). Wound-induced accumulation of JA biosynthetic gene transcripts, such as allene oxide synthase and allene oxide cyclase, also peak after maximum wound-induced accumulation of JA (Ziegler et al., 2001; Stenzel et al., 2003). However, we cannot exclude that basal levels of expression of most JA biosynthetic genes, including β-oxidation genes, may be enough to ensure the initial wound-triggered synthesis of JA in damaged leaves. A subsequent positive feedback in JA biosynthesis may then occur through JA-mediated activation of JA biosynthetic genes as proposed by different groups (Stenzel et al., 2003, and references therein). In contrast to other JA biosynthetic genes that are JA-responsive, KAT2 was not activated by exogenous application of JA to unwounded plants (Fig. 4). The lack of JA-responsiveness of KAT2 may represent in fact a limiting step for the JA-mediated positive regulatory loop in wound-activated responses in Arabidopsis. As a consequence, KAT2 function may exert an overall regulation on the net rate of JA biosynthesis and on the prevalence of JA-dependent or JA-independent wound-activated responses in Arabidopsis.
MATERIALS AND METHODS
Plant Material
Seeds of Arabidopsis Col (Lehle Seed, Tucson, AZ) and the transgenic nahG (kindly donated by Dr. John Ryals) were either sown in moistened soil and grown under photoperiod cycles of 16 h day and 8 h night (20°C) under 150 μE m−2 s−1 cool-white fluorescent lamps and 60% relative humidity, or surface sterilized and germinated in sterile liquid or agar-supplemented Murashige and Skoog medium (Duchefa, Haarlem, The Netherlands). For microplate liquid culture, 8 to 10 seeds per well were transferred to 24-well tissue culture clusters (Costar, Cambridge, MA) containing 1 mL/well of sterile Murashige and Skoog medium supplemented with 0.5% Suc, and further grown with continuous shaking for 10 d under photoperiod cycles as described above. Fresh medium (500 μL) was added to every well 8 d after sowing and experiments were conducted 2 d later. Seeds from the JA insensitive Arabidopsis coi1-1 mutant (Feys et al., 1994), kindly supplied by John Turner, were sown on Murashige and Skoog-agar plates supplemented with 2% Suc and 20 μm JA (Duchefa), and selected 8 d post-germination as those showing normal root growth. An F2 segregating population of seeds, obtained by selfing the F1 obtained by crossing male-sterile coi1-1 mutants with pollen from nahG homozygous transgenic plants, were germinated in Murashige and Skoog-agar plates supplemented with 20 μm JA and 50 mg/L kanamycin to select double coi1-1 nahG transgenic mutants plants resistant to antibiotic and with normal root growth. They were subsequently transferred to microplates and grown for an additional 2 d in liquid medium as described above or transferred to moistened soil.
Constructs for Antisense Expression
A fragment of 559 bp (from nucleotide 1,797–2,356) of the 3′end of ACX1 cDNA obtained by RT-PCR with oligonucleotides OP-DDRT9 (5′-TCGGTCATAG-3′) and oligo(dT)(11) MN (Operon Technologies, Alameda, CA) was originally cloned in pUC18 (Titarenko et al., 1997). After subcloning in pBluescript-SK, an XbaI/SalI fragment was directionally cloned in antisense orientation in a BinA7 binary vector under the control of 35S promoter and the octopine synthase 3′termination sequences (Höfgen and Willmitzer, 1990). Plant transformants were selected for resistance to kanamycin. Homozygous transgenic lines with a single insertion were selected by screening kanamycin resistance in the progeny of original transformants. Similarly, a cDNA containing the complete coding sequence of KAT2 was amplified by RT-PCR with oligonucleotides KAT-1F (5′-CCG GAA AAA ATG GAG AAA GCG ATC GAG A-3′) and KAT-1R (5′-CGG TTT TGG TGC ATG GTC CTC TCT AGC G-3′) and cloned in antisense orientation in the SmaI site of the previously-mentioned BinA7 vector under the control of 35S promoter and the octopine synthase 3′termination sequences. Selection of transformants was performed as mentioned for ACX1 antisense lines. Further selection of the transgenic lines with the best antisense effects was achieved by northern analysis of the basal and wound-induced levels of the corresponding endogenous transcripts.
Dehydration, Wounding, and Phytohormone Treatments
Culture medium was removed from the wells where plantlets were grown and replaced by 1 mL of fresh medium. Plantlets were treated with JA (Duchefa) or ABA (Sigma-Aldrich Quimica, Madrid) at the indicated concentrations. Seedlings were dehydrated by removing liquid medium from the wells, by blotting plantlets softly on Whatman 3MM paper, and by incubation in the covered microplate well without liquid medium. Samples were harvested at the indicated times after treatment with JA or ABA, or after removal of liquid medium from wells, then frozen in liquid nitrogen and used for total RNA isolation. Percent of dehydration was calculated as the ratio of fresh weight of plantlets at the time they were harvested to their fresh weight at zero time after removing liquid culture medium. Wounding of soil-grown plants was performed by thoroughly crushing with forceps one-half of the rosette leaves. Wounded and nonwounded leaves were harvested at the indicated times and frozen in liquid nitrogen for further total RNA isolation and analysis of local and systemic transcript accumulation, respectively.
RNA Isolation and Northern-Blot Analysis
Total RNA was isolated, separated, and analyzed by northern techniques following standard procedures (Sambrook et al., 1989). The inserts of clones used as probes were labeled with [32P]dCTP Redivue and the Rediprime labeling kit from Amersham Pharmacia Biotech (Uppsala). rab 18 probe was a PCR product obtained by amplification from Arabidopsis genomic DNA with primers RAB18A 5′-CCC CTG CAG TCC ATA TCC GAA ACC GGA CT-3′and RAB18B 5′-GGG GAA TTC ACG TAC CGA GCT AGA GCT GG-3′. After hybridization, filters were washed in 3× SSC, 0.5% SDS at 55°C to 65°C (1× SSC buffer is 150 mm NaCl and 15 mm C6H5Na3O7) and exposed for autoradiography. Prior to any subsequent hybridization, filters were stripped with hot water containing 0.1% SDS as recommended by the manufacturers. Equal RNA loading was confirmed by ethidium bromide staining of ribosomal RNAs. Hybridized filters were quantified by Phosphorimager analysis using ImageReader and ImageGauge software (Fuji, Tokyo).
RT-PCR Analysis
First strand cDNA synthesis, generated from 1 μg total RNA with oligo(dT) and Revertaid H minus MMuLV Reverse Transcriptase (MBI Fermentas, Vilnius, Lithuania), was used as template for PCR with the following primers: 5′-GGG GCA GGG TAC AGA GGA GCA GCA GAA G-3′ and 5′-TAA AAT TCC GCC ATA TGA CGA TCG TAC A-3′ for ACX1; 5′-CGG AAT GAA GTT TGG AAA CGG GG-3′and 5′-CCA AGA TAC TGG TCC GTG TAG TC-3′for ACX1.2; 5′-CCG GAA AAA ATG GAG AAA GCG ATC GAG A-3′ and 5′-CGG TTT TGG TGC ATG GTC CTC TCT AGC G-3′ for KAT2; 5′-CCT GGA TCT CAG AGA GC-3′ and 5′-GGC CTT ATT GTC ATT AGA C-3′ for KAT1; and 5′-GAT CTT TGC CGG AAA ACA ATT GGA GGA TGG T-3′ and 5′-CGA CTT GTC ATT AGA AAG AAA GAG ATA ACA GG-3′ for UBQ. PCR products were separated by electrophoresis in 1% agarose gels and blotted to Hybond-N membranes (Sambrook et al., 1989). Probes for Southern hybridization were obtained from PCR amplification products with specific primers described above or by digestion of the corresponding plasmids with the appropriate inserts. Bands were excised from the gel and purified by the QIAEXII kit (Qiagen, Valencia, CA). DNA was 32P-labeled with the Rediprime system (Amersham) and hybridized filters quantified by Phosphorimager (Fuji) as described above. Quantification of radioactively labeled bands was normalized with the hybridization of the corresponding UBQ band.
Determination of Jasmonates
At 0 and 90 min a.w., mechanically damaged leaves of the rosettes were harvested (to a final fresh weight between 0.5 and 1.0 g), frozen in liquid nitrogen, and stored at −80°C until JA extraction. A modified protocol proposed by Gundlach et al. (1992) was used for extraction and gas chromatography mass spectrometry analysis as previously reported (Heck et al., 2003) with 9,10-dihydro-jasmonate spikes as standards for calculating recovery. Three to six replicates for every measurement were used. Values are the mean ± se.
Acknowledgments
We thank John Ryals and John Turner for kind provision of nahG transgenics and coi1-1 Arabidopsis mutant seeds, respectively. We are grateful to Dr. Paloma Pérez for critical reading of the manuscript.
This work was supported by the Spanish Comisión Interministerial de Ciencia y Tecnología as part of the Programa Nacional de Biotecnología (BIO99–1129 and BIO2002–03533) and by Consellería de Agricultura, Pesca y Alimentación de la Generalitat Valenciana (GV–CAPA–0013–CO2–02). M.C.C. and C.M. were recipients of fellowships from Generalitat Valenciana (Spain) and CSIC (Programa I3P, Spain), respectively, and J.P.M. received a grant from the Swiss National Science Foundation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.039925.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bergey DR, Howe GA, Ryan CA (1996) Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc Natl Acad Sci USA 93: 12053–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier GF, Ryan CA (1998) Wound signaling in tomato plants. Evidence that ABA is not a primary signal for defense gene activation. Plant Physiol 117: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Prat S (1998) Expression of the Arabidopsis abi1-1 mutant allele inhibits proteinase inhibitor wound-induction in tomato. Plant J 15: 765–771 [DOI] [PubMed] [Google Scholar]
- Dammann C, Rojo E, Sánchez-Serrano JJ (1997) Abscisic acid and JA activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J 11: 773–782 [DOI] [PubMed] [Google Scholar]
- Delaney T, Ukness S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Hooks MA, Williams D, Lange P, Bechtold N, Sarrobert C, Nussaume L, Graham IA (2000) Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J Biol Chem 275: 34375–34381 [DOI] [PubMed] [Google Scholar]
- Feys BSF, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froman BE, Edwards PC, Bursch AG, Dehesh K (2000) ACX3, a novel medium-chain acyl-CoA oxidase from Arabidopsis. Plant Physiol 123: 733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde J-P, Bryce JH, Graham IA, Smith SM (2001) Requirement of 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28: 1–12 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Métraux JP, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Graham I, Eastmond PJ (2002) Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog Lipid Res 41: 156–181 [DOI] [PubMed] [Google Scholar]
- Gundlach H, Muller MJ, Kutchan TM, Zenk MH (1992) Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc Natl Acad Sci USA 89: 2389–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, De Bellis L, Ciuri A, Kondo M, Hayashi M, Nishimura M (1999) A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plants. J Biol Chem 274: 12715–12721 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M (1998) 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in Glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck S, Grau T, Buchala A, Métraux J-P, Nawrath C (2003) Genetic evidence that expression of NahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis- Pseudomonas syringae pv. tomato interaction. Plant J 36: 342–352 [DOI] [PubMed] [Google Scholar]
- Hertweck C, Jarvis AP, Xiang L, Moore BS, Oldham NJ (2001) A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid β-oxidation. Chembiochem 10: 784–786 [DOI] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (1990) Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato (Solanum tuberosum). Plant Sci 66: 221–230 [Google Scholar]
- Hooks MA, Kellas F, Graham IA (1999) Long-chain acyl-CoA oxidases of Arabidopsis. Plant J 20: 1–13 [DOI] [PubMed] [Google Scholar]
- Kindl H (1987) β-Oxidation of fatty acids by specific organelles. In PK Stumpf, ed, The Biochemistry of Plants, Vol 9. Academic Press, New York, pp 31-50
- Lang V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Lee J, Parthier B, Löbler M (1996) Jasmonate signalling can be uncoupled from abscisic acid signalling in barley: identification of jasmonate-regulated transcripts which are not induced by abscisic acid. Planta 199: 625–632 [DOI] [PubMed] [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Fisahn J, Willmitzer L (1995) Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc Natl Acad Sci USA 92: 4106–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribnicky DM, Shulaev V, Raskin I (1998) Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol 118: 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB (1999) A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11: 1911–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, León J, Sánchez-Serrano JJ (1999) Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J 20: 135–142 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Hooks MA, Graham IA (2001) Co-ordinate regulation of genes involved in storage lipid metabolism in Arabidopsis thaliana. Biochem Soc Trans 29: 283–287 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larsson TR, Graham IA (2003) Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. J Biol Chem 278: 21370–21377 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaller F (2001) Enzymes of the synthesis of octadecanoid-derived signalling molecules. J Exp Bot 52: 11–23 [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C (2003) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51: 895–911 [DOI] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick UB, Howe GA, Weiler EM, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601 [DOI] [PubMed] [Google Scholar]
- Stratmann JW (2003) Long distance run in the wound response: Jasmonic acid is pulling ahead. Trends Plant Sci 8: 247–250 [DOI] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, León J, Sánchez-Serrano JJ (1997) Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell Suppl 14: S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC (1984) Biosynthesis of jasmonic acid by several plant species. Plant Physiol 75: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Keinänen M, Baldwin IT (2001) Herbivore-induced of allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry 58: 729–738 [DOI] [PubMed] [Google Scholar]