Significance
Desorption electrospray ionization mass spectrometry imaging (DESI-MSI) is a label-free molecular imaging technique that provides a window into the biochemical processes present in benign and malignant prostate tissue. This is important both in improving the understanding of tissue biology and in achieving rapid cancer diagnosis. We applied DESI-MSI to record lipid, carbohydrate, and most importantly, small metabolite images from 54 normal and malignant prostate tissue specimens. Several Krebs cycle intermediates were present at different concentrations in prostate cancer compared with normal tissue. Statistical calculations identified panels of metabolites that could readily distinguish prostate cancer from normal tissue with nearly 90% accuracy in a validation set. The results also indicated that the ratio of glucose to citrate ion signals could be used to accurately identify prostate cancer.
Keywords: prostate cancer, Krebs cycle, metabolism, desorption electrospray ionization, mass spectrometry
Abstract
Accurate identification of prostate cancer in frozen sections at the time of surgery can be challenging, limiting the surgeon’s ability to best determine resection margins during prostatectomy. We performed desorption electrospray ionization mass spectrometry imaging (DESI-MSI) on 54 banked human cancerous and normal prostate tissue specimens to investigate the spatial distribution of a wide variety of small metabolites, carbohydrates, and lipids. In contrast to several previous studies, our method included Krebs cycle intermediates (m/z <200), which we found to be highly informative in distinguishing cancer from benign tissue. Malignant prostate cells showed marked metabolic derangements compared with their benign counterparts. Using the “Least absolute shrinkage and selection operator” (Lasso), we analyzed all metabolites from the DESI-MS data and identified parsimonious sets of metabolic profiles for distinguishing between cancer and normal tissue. In an independent set of samples, we could use these models to classify prostate cancer from benign specimens with nearly 90% accuracy per patient. Based on previous work in prostate cancer showing that glucose levels are high while citrate is low, we found that measurement of the glucose/citrate ion signal ratio accurately predicted cancer when this ratio exceeds 1.0 and normal prostate when the ratio is less than 0.5. After brief tissue preparation, the glucose/citrate ratio can be recorded on a tissue sample in 1 min or less, which is in sharp contrast to the 20 min or more required by histopathological examination of frozen tissue specimens.
Prostate cancer (PCa) is the most commonly diagnosed solid-organ cancer and the second leading cause of cancer death in men in the United States (1). Because of prostate-specific antigen (PSA) screening in the United States, most PCas are discovered when they are confined to the prostate (2). Many of these localized PCas are treated by surgical removal of the entire prostate (radical prostatectomy). The presence of cancer cells at the edge of the surgical resection, or positive surgical margins, is associated with higher rates of recurrence and death from PCa (3, 4). Therefore, an important clinical challenge in PCa management is to devise a rapid and highly accurate method to detect cancerous cells in real time to allow resection of additional periprostatic tissues and reduce cancer recurrence after surgery. Over the last decade, several innovative analytical techniques (5–12) have been developed to distinguish cancer from benign tissue in various organs. However, none has achieved wide clinical adoption for various reasons including inconvenience, narrow information content, unavailability, poor sensitivity, slowness of adoption, and operating room workflow incompatibility. In PCa, intraoperative frozen sections have been used to attempt to identify PCa at the margin based on analysis of histology. However, frozen sections have been shown to have poor sensitivity and specificity for the detection of PCa and currently are not recommended (13, 14).
Recently, a label-free molecular imaging method called desorption electrospray ionization mass spectrometric imaging (DESI-MSI) has been developed (15–17). DESI-MSI can rapidly evaluate the tissue metabolome in situ by simultaneously characterizing hundreds of lipids and metabolites. In the last 5 y, reports from our group (16–19) and others (15, 20–22) have demonstrated the usefulness of DESI-MSI in viewing the metabolite and lipid distribution in tissue samples from cancers and other diseases. Global changes in metabolism are a cardinal feature of neoplasia (23, 24), and DESI-MSI can determine global metabolite levels and their spatial distributions across a tissue sample on a microscopic slide. DESI-MSI allows tissue analysis with little sample preparation, and provides quantitative multiplex molecular information within minutes. In patients undergoing surgery for gastric cancer, DESI-MSI helped in assessing the surgical margin status for the presence of cancer (16). This rapid assessment could be used as a tool to guide surgeons in removing more tissue to achieve negative margins and improve surgical outcomes (19).
Given the known alterations in metabolic pathways in PCa, we tested whether DESI-MSI of metabolites and lipids could have utility in discriminating cancer from normal tissue obtained from radical prostatectomy. Using tandem and high-resolution mass spectrometry we have characterized the distinct metabolite and lipid profiles of normal and malignant prostate. Although many earlier DESI-MSI studies considered only lipid profiles in identification of cancer, here we report imaging small metabolite distribution to detect PCa. The biochemical basis of this diagnostic accuracy lies in apparent differences in the Krebs cycle between cancerous and benign tissue. Finally, because DESI-MSI provides information on hundreds of molecules (small metabolites and lipids), we used a statistical method called least absolute shrinkage and selection operator (Lasso) (25, 26) to build and validate a classifier that distinguishes normal from malignant prostate tissue.
Results
Metabolite/Lipid Imaging of Prostate Tissue Specimens.
To investigate the lipidomic and metabolic profiles in normal and malignant prostate tissues, we selected 54 fresh frozen prostate tissue samples harvested at the time of radical prostatectomy and performed negative ion mode DESI-MSI. For each case, a 5-μm frozen section was taken for histopathological evaluation. After hematoxylin and eosin (H&E) staining, slides were annotated by a genitourinary pathologist to delineate areas of cancerous and normal tissue. A 15-μm section taken immediately adjacent to the H&E section was used for DESI-MSI. The details of DESI-MSI are given in Materials and Methods and depicted in SI Appendix, Fig. S1.
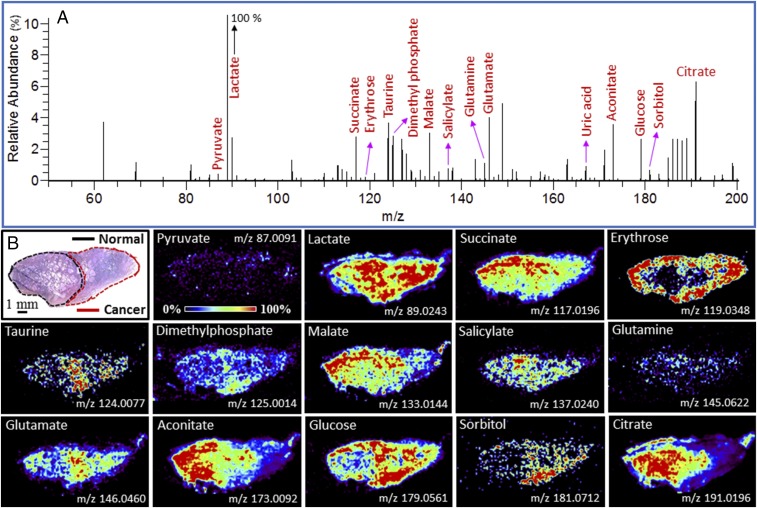
Metabolite and lipid ions were detected in the m/z range 50–1000. Fig. 1A shows a representative negative ion mode DESI mass spectrum of a typical prostate tissue specimen in the m/z range 50–200, where most of the small metabolites and glucose were detected. SI Appendix, Fig. S2 shows a representative image in the m/z range 200–1,000 where most lipids were detected. We identified and characterized several ion signals from the data (SI Appendix, Table S1) by using high mass accuracy, isotopic distribution, and tandem mass spectrometry. Typical molecular characterization data are presented in SI Appendix, Figs. S3 and S4 A–P. The detected species (SI Appendix, Table S1) were mostly deprotonated small metabolites related to energy production in the Krebs cycle, and deprotonated lipids including free fatty acids (FAs), FA dimers, phosphatidic acids, and glycerophospholipids. Using pixel-to-pixel mass spectral data, detailed 2D molecular images of the tissue were constructed to visualize the spatial distribution of selected individual metabolites and lipids. The spatial resolution of the image is ∼200 μm, which compares well with the thickness of a surgical knife. For example, Fig. 1B and SI Appendix, Fig. S2C show ion images of typical small metabolites and lipids, respectively, some of which appear to discriminate cancer from normal based on their relative abundance. The spatial distribution of many additional intense ion signals (m/z 89.0244, 303.2316, 423.2496, 585.4859, etc.) did not appear to be useful in differentiating normal from cancerous tissue (Fig. 1 and SI Appendix, Fig. S2).
Fig. 1.
(A) Negative ion mode DESI mass spectrum in the m/z range 50–200 from a typical prostate tissue specimen showing ion signals of various small metabolites. (B) Spatial distribution of 14 different small metabolites in a prostate tissue specimen (Upper Left, the corresponding H&E staining of the tissue sample) that contains both normal (black outline) and cancer (red outline). In B some individual small metabolite distributions are mapped throughout the tissue, whereas in A the abundance of all small metabolites is displayed by averaging all pixels over a typical line scan. See SI Appendix, Tables S1 and S2 for identification of species with different m/z values. The abundance of the given ion in the corresponding ion image is normalized to 100%.
Selection of Lipids for PCa Diagnosis.
Previous DESI-MSI studies have identified lipidomic profiles that distinguish normal from malignant tissues (16, 20, 21). We also attempted to distinguish cancerous from normal prostate tissues based on the abundance of specific lipid species. Given the large number of lipid ions (m/z 200–1,000) interrogated, we first tried an unbiased statistical approach, Lasso (26), to select a parsimonious set of lipids that could classify normal from cancerous tissues (SI Appendix, Fig. S5) (27). Using a training set of 45 specimens (17 normal and 28 cancerous), the Lasso considered m/z values that best characterized the two classes (cancer vs. normal) and yielded a cross-validation error of nearly 29% on pixel-based predictions (SI Appendix, Fig. S5C) and 27% on patient-based predictions. The lipid-based Lasso model did not work well when applied to the validation set, possibly because of the large heterogeneity in lipidomic profiles of PCa patients. Consistent with this finding, we were unable to validate previous work (28) identifying cholesterol sulfate as a candidate lipid biomarker for PCa (SI Appendix, Fig. S6).
Selection of Small Metabolites with Diagnostic Features.
We unambiguously identified and characterized a large number of small metabolites in the m/z window 50–200 that were differentially present in cancerous vs. normal tissue (Fig. 1A and SI Appendix, Table S1). For example, Fig. 1B shows higher abundance of erythrose (m/z 119.0348), glutamate (m/z 146.0464), glucose (m/z 179.0561), and sorbitol (m/z 181.0712), and lower abundance of succinate (m/z 117.0196), malate (m/z 133.0144), salicylate (m/z 137.0240), aconitate (m/z 173.0092), and citrate (m/z 191.0196) in cancerous tissues compared with normal. Most of these metabolites are important components of the Krebs cycle (Fig. 2). It should be noted that we could not construct DESI images of ionic species with very low abundances, even though they are important metabolites of the Krebs cycle and other metabolic pathways (see SI Appendix, Fig. S7 as typical example). Although imaging the spatial distribution and characterization of individual ions (metabolites) is not necessary for tissue diagnosis by DESI-MSI/Lasso, doing so can pinpoint important metabolites and their relation to PCa biochemistry (vide infra).
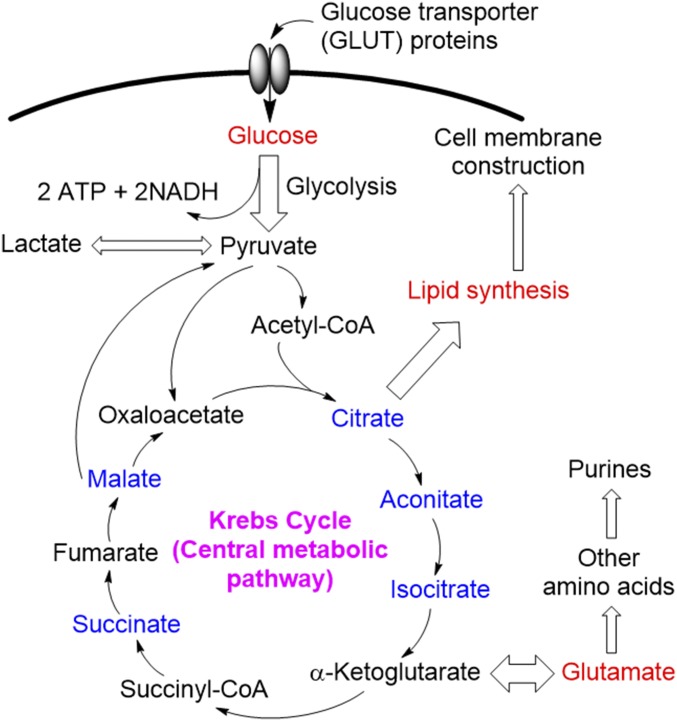
Fig. 2.
Schematic overview of the metabolic flux in the Krebs cycle. DESI-MSI study (e.g., Fig. 1B) shows that the abundances of metabolites labeled in red are up-regulated and blue are down-regulated in cancer compared with normal tissue.
Lasso Analysis for the Training and Validation Sets.
We used Lasso to develop a classifier that estimates the probability that a pixel in each DESI-MS image is either cancer or normal. DESI-MS was performed on a training set of 36 tissue samples with histologically demarcated areas of normal and malignant tissue (18 pure normal and 18 pure cancer tissues from 36 patients). We selected the top 54 peaks of metabolites, which had been mostly characterized (SI Appendix, Tables S1 and S2), to perform the Lasso analysis. Two independent Lasso classifiers were built, one based on individual ion signals, and another based on ratios of different ion signals. The first classifier performed poorly in the validation set, perhaps caused by normalization issues. However, the second approach showed promising results in classification as detailed next. We divided the data in two m/z sets for Lasso: one with m/z range 50–1,000, which considers all detected metabolites and lipids, and the other with the m/z range 50–200, which considers only the small metabolites (SI Appendix, Table S3). Table 1 shows the patient-based prediction results from these calculations (see Statistical Analysis in Materials and Methods for details). The cross-validation analysis of all samples from the training set achieved nearly 92% overall agreement in the m/z range 50–1,000, and nearly 89% overall agreement in the m/z range 50–200, compared with standard histopathologic evaluation (H&E).
Table 1.
Lasso prediction results for 36 prostate tissue specimens in the training set and 18 specimens in the validation set, in comparison with histopathological analyses
| Set | Small metabolites and lipids-based prediction (m/z 50–1,000) | Small metabolites-based prediction (m/z 50–200) | ||||||
| Pathology* | Normal | Cancer | % Agreement | Pathology* | Normal | Cancer | % Agreement | |
| Training set | Normal | 17 | 1 | 94.4 | Normal | 16 | 2 | 88.9 |
| Cancer | 2 | 16 | 88.9 | Cancer | 2 | 16 | 88.9 | |
| Overall agreement: 91.7% | Overall agreement: 88.9% | |||||||
| Validation set | Normal | 8 | 2 | 80 | Normal | 9 | 1 | 90 |
| Cancer | 0 | 8 | 100 | Cancer | 0 | 8 | 100 | |
| Overall agreement: 88.9% | Overall agreement: 94.4% | |||||||
Pathologic evaluation was performed by two pathologists on the same frozen tissue section used for DESI-MSI study. The tissues were H&E-stained after recording DESI-MSI.
We also analyzed 18 independent validation specimens (10 normal and 8 cancer tissues) to test the performance of our Lasso classifier. Table 1 also shows the Lasso prediction obtained from the validation set in both m/z domains, per patients. Compared with histopathology (H&E), nearly 89% and 94% overall agreements were achieved in the m/z ranges 50–1,000 and 50–200, respectively. All these data collectively suggest that analysis of small metabolites can exhibit high accuracy in cancer diagnosis by DESI-MSI.
Glucose/Citrate Ratio as a Biomarker.
We also investigated whether we could preserve the good accuracy of the above analysis using fewer metabolites to make the testing more rapid. As discussed above, DESI-MSI reveals in situ metabolic flux by detecting the relative abundance of numerous metabolites. For example, Fig. 1B shows high levels of glucose (m/z 179.0561; see SI Appendix, Fig. S4I for identification) and low levels of citrate (m/z 191.0196; see SI Appendix, Fig. S4J for identification) in cancer (29). Decreased levels of citrate in PCa are well described and can also be detected using magnetic resonance (MR) spectroscopic imaging (30). Based on these findings, we attempted to use imaging of glucose and citrate (Fig. 1B) to characterize whether a prostate tissue specimen was cancerous or normal. Interestingly, when we calculated the ratio of these two ion signals (glucose/citrate) from the tissue specimen, and constructed the image of that ratio, we accurately differentiated PCa from benign prostate tissue as shown in Fig. 3. Furthermore, this specific ratio can also be chosen from the Lasso model (SI Appendix, Table S3), which is a linear function of log ratios and there are different but nearly equivalent ways of expressing the same fitted model.
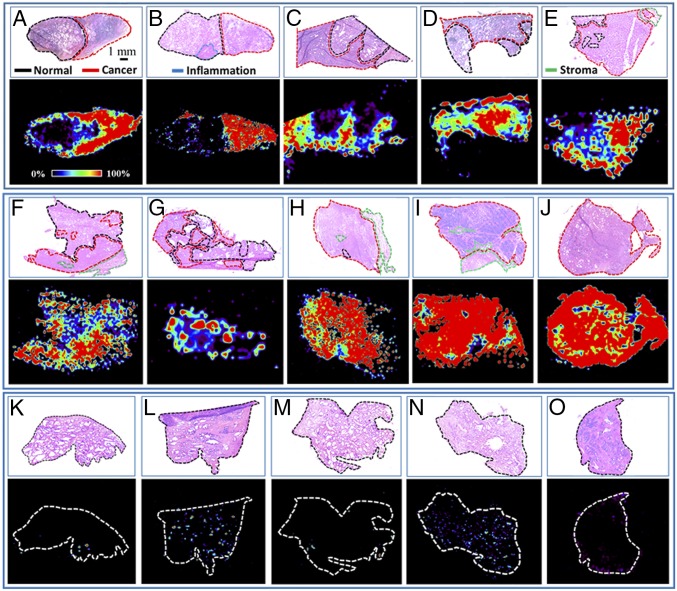
Fig. 3.
Distribution of glucose/citrate ratio of some representative prostate tissue specimens showing significant elevation of the glucose/citrate ratio in cancer. The Top of each panel (A–O) shows the histopathological evaluation (H&E) of the corresponding tissue, where cancer areas have been demarcated by red, benign areas by black, stroma areas by green, and inflammation areas by blue.
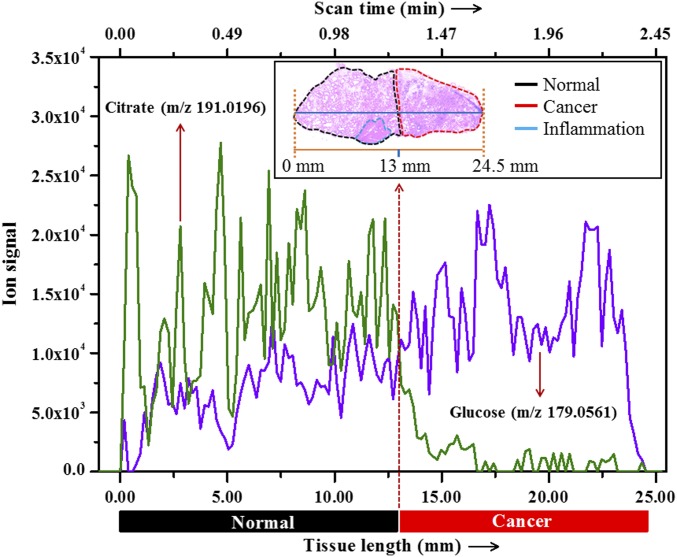
In several malignancies including PCa, molecular changes have been reported in the normal tissues immediately adjacent to the cancer (field defects) and this could adversely impact detection of cancer by blurring the border between malignant and normal tissues (31). To determine whether there was a distinct border of the relative quantities of glucose and citrate between normal and malignant prostate tissues, we set our DESI probe to scan along a single line (32) over a prostate tissue specimen (Fig. 4). Among hundreds of ions recorded on the line scan, we extracted the signal intensities of glucose and citrate to plot them over the scan time (tissue length). Fig. 4 shows that there is a distinct cutoff at the border between normal and malignant prostate tissues as determined by the glucose/citrate ratio. This difference appears to be determined largely by citrate levels as its signal intensity drastically decreases in cancer, whereas the increase of glucose signal intensity was less marked. This difference could be caused by the fact that it is more difficult to ionize glucose than citrate in the gas phase by electrospray (SI Appendix, Fig. S8). Ion images of glucose or citrate alone did not work so well as the ratio between the two metabolites (Fig. 3) in identifying PCa. It should be noted that inflammation is often observed in a PCa specimen (Fig. 3B) and this glucose/citrate ratio fails to distinguish it. A follow-up study is required in the future for finding biomarkers that can distinguish inflammation from malignant or normal tissue.
Fig. 4.
Extracted ion chronogram of glucose and citrate over a line scan of a typical prostate tissue specimen that contains both benign (black outline) and cancer (red outline) areas. (Inset) H&E staining of the tissue and the position of the line scan (blue).
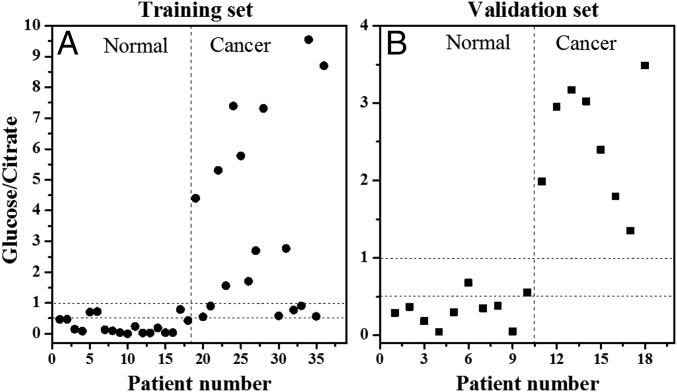
To test the ability of the glucose/citrate ratio to diagnose PCa, we used the same training set that was used above for the Lasso, comprising 36 specimens. We evaluated the average glucose/citrate signal ratio from each individual specimen for the cancerous and normal areas based on histology on the adjacent H&E slide. There was a significant difference in the distribution of the glucose/citrate ratios between PCa and normal tissues, and specimens could be accurately classified as cancer if the glucose/citrate signal ratio was >1 and normal if the ratio was <0.5 (Fig. 5A). In an independent set of 18 specimens (8 cancer and 10 normal tissues) we were able to validate that all specimens with glucose/citrate signal ratio >1 were malignant and all with ratios <0.5 were normal (Fig. 5B). There were nine specimens in the training set (Fig. 5A), and two specimens in the validation set (Fig. 5B) with a ratio between 0.5 and 1; the significance of this ratio is unknown, although some of those were classified as normal and some as cancer by H&E.
Fig. 5.
Negative ion mode DESI-MS ion signal intensity ratios for glucose/citrate are plotted for (A) the training set (18 benign and 18 cancer specimens), and (B) the validation set (10 benign and 8 cancer specimens) by averaging the ion signals of glucose and citrate from all pixels acquired from the individual tissue sample. From these plots, a tissue can be classified as cancer when glucose/citrate signal ratio is >1, and benign when the ratio is <0.5.
Discussion
DESI-MSI is a novel method that enables rapid multiplex mapping of metabolites and lipids in fresh tissue specimens. Using DESI-MSI, we identified key differences in the distribution of various metabolites and lipids between cancerous and normal prostate tissue. Prior studies of DESI-MSI for cancer diagnosis focused primarily on lipids as biomarkers. We began by evaluating lipids as well but the Lasso model relying on lipids only performed poorly in differentiating normal prostate from cancer. Incorporation of small metabolites into the Lasso model improved the accuracy of PCa identification (Table 1). Our findings add to the understanding of the metabolism of PCa and may have applications in the clinical care of men with PCa.
The metabolic profile of cancer cells is strikingly different from normal cells. Metabolism in the prostate gland shows distinctly different kinetics (metabolic flux) of the Krebs cycle compared with other organs (Fig. 2) (29). DESI-MSI is an ideal method to exploit these differences in cancer biochemistry in vivo because it is tissue based, does not require tissue fixation, and provides reasonably good spatial resolution (∼200 µm).
In the cancerous specimens evaluated using DESI-MSI (Fig. 1B) we detected significantly lower levels of several Krebs cycle intermediates (m/z <200) including citrate/isocitrate, aconitate, succinate, and malate. Very likely, these decreased levels are the product of faster transformation of intermediates. Indeed, previous work has suggested that the Krebs cycle in malignant prostate is bioenergetically more efficient (gaining more ATP per glucose metabolized) compared with normal prostate (29). Further, a number of anaplerotic and cataplerotic reactions, involved in biosynthetic pathways, likely contribute to the reduced levels of the Krebs cycle intermediates in PCa. One noteworthy example is the higher levels of glutamate we detected in PCa (Fig. 1B), which are known to be formed from α-ketoglutarate (Fig. 2). Glutamate has been implicated as a biomarker of PCa aggressiveness, suggesting that modulation of the pathways and the metabolome could modulate tumor behavior (33, 34).
A second interesting cataplerotic reaction in the Krebs cycle involves shunting citrate into lipogenesis in PCa, presumably to construct cell membranes for proliferating cancer cells (Fig. 2) (35). Indeed, we detected higher levels of lipids (m/z 200–1,000) in PCa compared with normal prostate tissues (SI Appendix, Fig. S5B), a finding consistent with metabolic reprogramming in PCa associated with increased expression and activity of several lipogenic enzymes including FA synthase (FASN) (35). Furthermore, unlike many other cancers, FA oxidation is the main source of energy required for cellular proliferation and progression of PCa (35).
Development of PCa is associated with the metabolic switch from a high level of citrate secretion to citrate oxidation (29). Costello and Franklin (36–38) and Costello et al. (39) proposed that high uptake of zinc by normal prostate mitochondria inhibits the activity of m-aconitase, thereby slowing the citrate oxidation in the Krebs cycle (Fig. 2) that results in a high level of citrate in normal prostate tissue (Figs. 1 and 4) (39). The gene encoding the zinc uptake transporters (ZIP1) is down-regulated in PCa, leading to a faster oxidation of citrate driven by low level of zinc (38). In this study, we were able to image the known changes in citrate levels in PCa and normal tissues (Fig. 4) using DESI-MSI to distinguish normal from malignant tissues with high accuracy and excellent spatial resolution.
MR spectroscopy imaging (MRSI) uses the choline/citrate ratio to diagnose and prognosticate PCa (30). However, we could not use this ratio because in a DESI-MSI run (which has much higher spatial resolution than MRSI) all species need to have the same charge to be detected together. Choline appears protonated and has a positive charge, whereas citrate is deprotonated and has a negative charge. We found glucose to be a reliable partner with citrate in the negative ion mode DESI-MSI and we have used that ratio to distinguish PCa from normal in a single scan.
With further development, DESI-MSI/Lasso could find several potential clinical applications in managing PCa. Two important strengths of DESI-MSI—its speed and the need for little sample preparation—make it particularly promising as a rapid, point-of-care clinical test. DESI-MSI could be used to complement frozen-section analysis by histopathology, particularly when the H&E-based frozen-section analysis is challenging because of morphological mimics, artifacts, and heterogeneity. The method described in this study could be adapted to rapidly detect cancer cells at the margin of resection during prostatectomy––effectively providing a real-time evaluation of surgical margins to guide the surgeons on the extent of their resection. We are keen to work further in this aspect. Positive surgical margins are a common problem, occurring in 11–48% of cases (40, 41). Avoiding positive surgical margins would reduce the incidence of cancer recurrence and need for secondary treatments. Indeed, we have shown before that DESI-MSI can have similar application in gastric and pancreatic cancer margin analyses (16, 19). In addition, our method could be used on fresh prostate biopsy tissues to provide real-time diagnosis for men undergoing prostate biopsy. It should be noted, however, that this preliminary study is based on a limited number of samples. Therefore, future study will include a larger number of samples to further validate the current results and construct a more powerful Lasso classifier that could improve the overall accuracy.
Materials and Methods
Preparation of Prostate Tissue Specimens.
Following Institutional Review Board approval, we accessed fresh-frozen prostate tissue obtained from prostatectomy specimens and stored in the Stanford University Department of Urology tissue bank. The samples included 64 prostate tissue specimens (28 benign only, 36 cancer only, and 10 cancer mixed with benign tissue) that had been obtained from different patients and stored at −80 °C until sectioning. A 5-μm section of each tissue sample was mounted on a glass slide and stained with H&E. A genitourinary pathologist (C.A.K.) reviewed and annotated normal and cancerous areas on the H&E-stained 5-µm adjacent tissue sections. Benign tissue included areas of normal prostate and areas of inflammation. 15-µm-thick adjacent sections were obtained using a Leica CM1950 cryostat (Leica Biosystems). These tissue sections were mounted on glass microscope slides and stored at −80 °C before DESI-MS analysis. The 54 specimens were used to construct a training set (n = 36) composed of 18 normal and 18 cancer (Gleason pattern 3 and/or Gleason pattern 4) specimens, and a validation set (n = 18) composed of 10 normal and 8 cancerous specimens for DESI-MS evaluation. The remaining 10 specimens of mixed grade were evaluated by the distribution of the glucose/citrate ion signal ratio to determine cancer margins.
DESI-MSI Study.
The detailed method of tissue imaging by DESI has been described elsewhere (21, 42). Briefly, a laboratory-built DESI source coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) was used for tissue imaging. DESI-MSI was performed in the negative ion mode (−5 kV) from m/z 50–1,000 with a spatial resolution of 200 µm (spray spot diameter) using a histologically compatible solvent system 1:1 (vol/vol) dimethylformamide/acetonitrile (DMF/ACN) (43) at a flow rate of 0.7 µL/min. The HPLC-grade solvents (DMF, ACN, etc.) were purchased from Sigma-Aldrich. Nitrogen, at a pressure of 170 psi, was used as the sheath gas for electrospray nebulization. The prostatic tissues were scanned under impinging charged droplets using a 2D moving stage in horizontal rows separated by a 200-µm (spatial resolution) vertical step. All imaging experiments were carried out under identical experimental conditions including geometrical parameters, e.g., spray tip-to-surface distance ∼2 mm, spray incident angle of 55°, and spray-to-inlet distance ∼5 mm. Data acquisition was performed using XCalibur 2.2 software (Thermo Fisher Scientific Inc.). An in-house program allowed the conversion of the XCalibur 2.2 mass spectral files (.raw) into the image file, which could be read by a biomedical image analysis software called Biomap (freeware, https://ms-imaging.org/wp/. The distributions of different metabolites, lipids, and the glucose/citrate ratio were plotted (Figs. 1B and 3, and SI Appendix, Fig. S2C) using the Biomap software. In ion images, we have used rainbow color order to represent the highest concentration by red and the lowest concentration by violet.
Metabolite/Lipid Identification.
The ions observed in the DESI-MS study were identified by searching the MassBank (www.massbank.jp) and the LIPID Metabolites and Pathways Strategy (www.lipidmaps.org/) databases based on high mass accuracy and isotopic distribution. When the database listed multiple isobaric/isomeric metabolites or lipids, we performed collision-induced dissociation (CID) and compared the corresponding fragmentation profile with that of the standard from the above database to characterize the species (44) wherever applicable (see SI Appendix, Fig. S6 A–P as typical examples). The CID spectra of the mass selected ions from the PCa tissue specimen are sometimes complex, although the majority of fragment ions matches with that of standards. This complexity can be interpreted by the interference of isomeric/isobaric ions derived from the biological matrix (tissue). As the position and stereochemistry of the double bond in an FA complicates the structural elucidation, they are often tentatively assigned in FAs and glycerophospholipids. However, some common and well-known metabolites and lipids that are commercially available were identified from the tissue by comparing their MS/MS data with standards.
Post-DESI-MSI Histopathological Reevaluation.
As the DESI solvent DMF/ACN (1:1; vol/vol) is histologically compatible (43), the same tissue samples, after analyzing in DESI-MS, were subjected to H&E staining followed by histopathological reevaluation. The dyes and solvents used in H&E staining were purchased from Fisher Scientific. High-resolution (0.43-µm) optical images of H&E-stained tissues were recorded using a Hamamatsu NanoZoomer 2.0-RS slide scanner. The pathology examination was made without the knowledge of DESI-MS evaluation.
Statistical Analysis.
XCalibur raw data files were converted to .txt files for statistical analysis. The raw data in .txt file format were imported to the R language for statistical analysis. Although hundreds of metabolites and lipids were detected by DESI-MS, we selected the top 54 peaks (SI Appendix, Tables S1 and S2), whose abundances were significant, and most of them (SI Appendix, Table S1) were characterized by tandem mass spectrometry. We performed statistical analysis both by using the individual peaks and by using all possible ratios of two peaks in this list (SI Appendix, Tables S1 and S2). Within the training set (18 benign, and 18 cancer), we applied the Lasso method (multiclass-logistic regression with L1 penalty) using the glmnet package in the CRAN R language library (45).
The Lasso yields sparse models, that is, models that involve only a subset of the variables/predictors (27). Therefore, models generated using the Lasso are simpler and easier to interpret than those from other linear regression methods. In our application, the Lasso method yields a model with parsimonious sets of features for discriminating cancer and normal prostate tissue. A mathematical weight for each statistically informative feature is calculated by the Lasso depending on the importance that the mass-spectral feature has in characterizing a certain class. Because the features selected by the Lasso can occur at a valley or a shoulder of an actual mass-spectra peak, identification of the selected features was performed by characterizing the nearest mass-spectra peak to the statistically selected feature. Classification was done on a pixel-by-pixel basis into one of two classes: cancer or normal, and then results were converted to patientwise predictions using a majority rule: if the majority of a tissue (patient) pixels are predicted to be cancer, the tissue is predicted as cancerous. We used cross-validation, leaving out one patient at a time, to select the Lasso tuning parameter and to assess the predictive accuracy within the training set. Then, the chosen model was applied to the test set of 18 patients.
Additional Information.
Supporting information accompanies this paper in the SI Appendix.
Supplementary Material
Acknowledgments
S.B. and R.N.Z. thank Livia S. Eberlin for her help. G.A.S. thanks the Urology Care Foundation for a Research Scholar Award and the National Comprehensive Cancer Network for a Young Investigator Award. J.D.B. was supported by the National Institutes of Health (Grant NIH 1U01CA196387). This work was supported by the Air Force Office of Scientific Research through Basic Research Initiative Grant AFOSR FA9550-12-1-0400 and by Grant NIH 1R21DA039578-01.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700677114/-/DCSupplemental.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Brooks JD. Managing localized prostate cancer in the era of prostate-specific antigen screening. Cancer. 2013;119(22):3906–3909. doi: 10.1002/cncr.28301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yossepowitch O, et al. Positive surgical margins in radical prostatectomy: Outlining the problem and its long-term consequences. Eur Urol. 2009;55(1):87–99. doi: 10.1016/j.eururo.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 4.Brooks JD, et al. The impact of tumor volume on outcomes after radical prostatectomy: Implications for prostate cancer screening. Open Prostate Cancer J. 2008;1:1–8. [Google Scholar]
- 5.Erguvan-Dogan B, et al. Specimen radiography in confirmation of MRI-guided needle localization and surgical excision of breast lesions. AJR Am J Roentgenol. 2006;187(2):339–344. doi: 10.2214/AJR.05.0422. [DOI] [PubMed] [Google Scholar]
- 6.Haka AS, et al. In vivo margin assessment during partial mastectomy breast surgery using Raman spectroscopy. Cancer Res. 2006;66(6):3317–3322. doi: 10.1158/0008-5472.CAN-05-2815. [DOI] [PubMed] [Google Scholar]
- 7.Bellanova L, et al. MRI-based assessment of safe margins in tumor surgery. Sarcoma. 2014;2014:686790. doi: 10.1155/2014/686790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paganelli G, Luini A, Veronesi U. Radioguided occult lesion localization (ROLL) in breast cancer: Maximizing efficacy, minimizing mutilation. Ann Oncol. 2002;13(12):1839–1840. doi: 10.1093/annonc/mdf343. [DOI] [PubMed] [Google Scholar]
- 9.Judy RP, et al. Quantification of tumor fluorescence during intraoperative optical cancer imaging. Sci Rep. 2015;5:16208. doi: 10.1038/srep16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen FT, et al. Intraoperative evaluation of breast tumor margins with optical coherence tomography. Cancer Res. 2009;69(22):8790–8796. doi: 10.1158/0008-5472.CAN-08-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu G, et al. Quantifying Gleason scores with photoacoustic spectral analysis: Feasibility study with human tissues. Biomed Opt Express. 2015;6(12):4781–4789. doi: 10.1364/BOE.6.004781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyle TE, et al. High-frequency ultrasound for intraoperative margin assessments in breast conservation surgery: A feasibility study. BMC Cancer. 2011;11(1):444. doi: 10.1186/1471-2407-11-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakiuchi Y, et al. Role of frozen section analysis of surgical margins during robot-assisted laparoscopic radical prostatectomy: A 2608-case experience. Hum Pathol. 2013;44(8):1556–1562. doi: 10.1016/j.humpath.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Nunez AL, et al. Frozen section evaluation of margins in radical prostatectomy specimens: A contemporary study and literature review. Ann Diagn Pathol. 2016;24:11–18. doi: 10.1016/j.anndiagpath.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Eberlin LS, et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc Natl Acad Sci USA. 2013;110(5):1611–1616. doi: 10.1073/pnas.1215687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberlin LS, et al. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc Natl Acad Sci USA. 2014;111(7):2436–2441. doi: 10.1073/pnas.1400274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberlin LS, et al. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc Natl Acad Sci USA. 2014;111(29):10450–10455. doi: 10.1073/pnas.1409778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry RH, et al. Characterization of MYC-induced tumorigenesis by in situ lipid profiling. Anal Chem. 2013;85(9):4259–4262. doi: 10.1021/ac400479j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberlin LS, et al. Pancreatic cancer surgical resection margins: Molecular assessment by mass spectrometry imaging. PLoS Med. 2016;13(8):e1002108. doi: 10.1371/journal.pmed.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eberlin LS, et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72(3):645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cooks RG. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim Biophys Acta. 2011;1811(11):946–960. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calligaris D, et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci USA. 2014;111(42):15184–15189. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iurlaro R, León-Annicchiarico CL, Muñoz-Pinedo C. Regulation of cancer metabolism by oncogenes and tumor suppressors. In: Lorenzo G, Guido K, editors. Methods in Enzymology. Vol 542. Academic; Oxford: 2014. pp. 59–80. [DOI] [PubMed] [Google Scholar]
- 24.Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 25.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc B. 1996;58(1):267–288. [Google Scholar]
- 26.Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16(4):385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 27.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning: With Applications in R. Springer; New York: 2014. pp. 203–264. [Google Scholar]
- 28.Eberlin LS, et al. Cholesterol sulfate imaging in human prostate cancer tissue by desorption electrospray ionization mass spectrometry. Anal Chem. 2010;82(9):3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dakubo GD. Mitochondrial Genetics and Cancer. Springer; Berlin: 2010. The Warburg phenomenon and other metabolic alterations of cancer cells; pp. 39–66. [Google Scholar]
- 30.Li L, et al. Prostate cancer magnetic resonance imaging (MRI): Multidisciplinary standpoint. Quant Imaging Med Surg. 2013;3(2):100–112. doi: 10.3978/j.issn.2223-4292.2013.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks JD. Epigenetic changes in histologically normal prostate tissues. J Urol. 2013;189(6):2020–2021. doi: 10.1016/j.juro.2013.02.3193. [DOI] [PubMed] [Google Scholar]
- 32.Wiseman JM, Puolitaival SM, Takáts Z, Cooks RG, Caprioli RM. Mass spectrometric profiling of intact biological tissue by using desorption electrospray ionization. Angew Chem Int Ed Engl. 2005;44(43):7094–7097. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- 33.Payton S. Prostate cancer: ‘Galectin signature’ reveals gal-1 as key player in angiogenesis. Nat Rev Urol. 2012;9(12):667. doi: 10.1038/nrurol.2012.216. [DOI] [PubMed] [Google Scholar]
- 34.Koochekpour S, et al. Serum glutamate levels correlate with Gleason score and glutamate blockade decreases proliferation, migration, and invasion and induces apoptosis in prostate cancer cells. Clin Cancer Res. 2012;18(21):5888–5901. doi: 10.1158/1078-0432.CCR-12-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Daniels G, Lee P, Monaco ME. Lipid metabolism in prostate cancer. Am J Clin Exp Urol. 2014;2(2):111–120. [PMC free article] [PubMed] [Google Scholar]
- 36.Costello LC, Franklin RB. The intermediary metabolism of the prostate: A key to understanding the pathogenesis and progression of prostate malignancy. Oncology. 2000;59(4):269–282. doi: 10.1159/000012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costello LC, Franklin RB. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: Connecting the dots. Mol Cancer. 2006;5(1):17. doi: 10.1186/1476-4598-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franklin RB, Costello LC. Zinc as an anti-tumor agent in prostate cancer and in other cancers. Arch Biochem Biophys. 2007;463(2):211–217. doi: 10.1016/j.abb.2007.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costello LC, Franklin RB, Narayan P. Citrate in the diagnosis of prostate cancer. Prostate. 1999;38(3):237–245. doi: 10.1002/(sici)1097-0045(19990215)38:3<237::aid-pros8>3.0.co;2-o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novara G, et al. Systematic review and meta-analysis of studies reporting oncologic outcome after robot-assisted radical prostatectomy. Eur Urol. 2012;62(3):382–404. doi: 10.1016/j.eururo.2012.05.047. [DOI] [PubMed] [Google Scholar]
- 41.Silberstein JL, Eastham JA. Significance and management of positive surgical margins at the time of radical prostatectomy. Indian J Urol. 2014;30(4):423–428. doi: 10.4103/0970-1591.134240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eberlin LS. DESI-MS imaging of lipids and metabolites from biological samples. In: Raftery D, editor. Mass Spectrometry in Metabolomics: Methods and Protocols. Springer; New York: 2014. pp. 299–311. [DOI] [PubMed] [Google Scholar]
- 43.Eberlin LS, et al. Nondestructive, histologically compatible tissue imaging by desorption electrospray ionization mass spectrometry. ChemBioChem. 2011;12(14):2129–2132. doi: 10.1002/cbic.201100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banerjee S, Mazumdar S. Electrospray ionization mass spectrometry: A technique to access the information beyond the molecular weight of the analyte. Int J Anal Chem. 2012;2012:282574. doi: 10.1155/2012/282574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman J, Hastie T, Simon N, Tibshirani R. glmnet: Lasso and Elastic-Net Regularized Generalized Linear Models. 2013 Available at https://cran.r-project.org/web/packages/glmnet/index.html. Accessed September 24, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.