Abstract
Objective:
This study aimed to identify sleep disturbance subtypes (“phenotypes”) among Latinx adults based on objective sleep data using a flexible unsupervised machine learning technique.
Methods:
This study was a secondary analysis of data from three cross-sectional studies of the Precision in Symptom Self-Management Center at Columbia University. All studies focused on sleep health in Latinx adults at increased risk for sleep disturbance. Data on total sleep time (TST), time in bed (TIB), wake after sleep onset (WASO), sleep efficiency (SE), number of awakenings (NOA) and the mean length of nightly awakenings were collected using wrist-mounted accelerometers. Cluster analysis of the sleep data was conducted using an unsupervised machine learning approach that relies on mixtures of multivariate generalized linear mixed models.
Results:
The analytic sample included 494 days of data from 118 adults (Ages 19–77). A 3-cluster model provided the best fit based on deviance indices (i.e., DΔ~ −75 and −17 from 1- and 2- to 3-cluster models, respectively) and likelihood ratio (Pdiff ~ 0.93). Phenotype 1 (n=64) was associated with greater likelihood of overall adequate SE and less variability in SE and WASO. Phenotype 2 (n=11) was characterized by higher NOAs, and greater WASO and TIB than the other phenotypes. Phenotype 3 (n=43) was characterized by greater variability in SE, bed times and awakening times.
Conclusion:
Robust digital data-driven modeling approaches can be useful for detecting sleep phenotypes from heterogenous patient populations, and have implications for designing precision sleep health strategies for management and early detection of sleep problems.
Keywords: digital phenotyping, minority health, actigraphy, machine learning, mixture models
1. Background
Sleep health is recognized as a public health concern in the United States [1]. Inadequate sleep duration and poor sleep quality are associated with incident obesity, impaired physical function, hypertension, and cancer[2, 3]. Risk of poor sleep health is higher among certain populations, such as individuals with chronic conditions associated with circadian rhythm disturbance (e.g., HIV)[4, 5]. In addition, stigmatized populations, such as racial, ethnic, or sexual and gender minority (SGM) adults, may experience poor sleep health due to chronic exposure to social stressors (e.g., perceived or anticipated rejection, discrimination)[6–9]. Such physiological and psychological factors reportedly contribute to a disproportionately higher prevalence of sleep problems in these individuals[6, 9]. However, there is scarce research using objectively-estimated (e.g., accelerometer-based) data to investigate sleep health patterns and potential between-individual heterogeneity that could be indicative of sleep disturbance subtypes (i.e., “phenotypes”). Identification of possible sleep phenotypes constitutes a starting point for developing individualized prevention and treatment strategies, providing precedence for undertaking this investigation.
An increased prevalence of sleep disturbance across various groups (e.g., older adults, transgender individuals), and during life transitions (e.g., menopause), as well as those with chronic co-morbidities (e.g., HIV, disorders) is well-documented.[10–15] However, existing studies evaluating sleep disturbances in minority populations have been mostly limited to self-report [15, 16], which has low concordance with objectively-estimated sleep measures[17], and detection of sleep pathologies (e.g., sleep apnea) in samples with a formal diagnosis of sleep disorders [18]. However, investigation of sleep phenotypes based on sleep disturbance among heterogenous, at-risk individuals who have not yet received a formal diagnosis can aid in detection during earlier stages of their sleep disturbance. Such phenotyping investigations can further help elucidate the degree to which treatments or interventions for sleep problems should be tailored. For example, should we identify that sleep health patterns vary within an at-risk population but clusters of patients with similar patterns can be identified, tailoring screening and intervening approaches at the sub-population level might be suitable.
The lack of research in this area might in part be due to the complex structure of objectively-estimated sleep data and the limited applicability of traditional clustering methods for such data.[19] Most traditional clustering techniques (e.g., k-means) cannot accommodate longitudinal data that have complex covariance structures with correlated measurements across time points, and vary in type (e.g., continuous, binary, multinomial) and time intervals of measurement.[20] They require certain assumptions to be met in the structure of the data, therefore they are sensitive to outliers, and can fail to identify the optimum structure of the clusters.[21] As the data size increases, exhausting all partitioning possibilities becomes computationally prohibitive.[22] Importantly, certain clustering methods assume that each data point (e.g., participant) belongs exclusively to a single latent group,[19, 23] which is an unrealistic assumption in most scenarios involving patient-oriented research.[23, 24] In such instances, a probabilistic approach that quantifies the likelihood of a participant belonging to any of the clusters present in the dataset by considering the entire probability distribution of membership assignments is more suitable. This allows assessment of the relative probabilities, and therefore degree of confidence with which the assignment is made.[25, 26] Flexible and dynamic methods that simultaneously consider uncertainty whilst delineating variability in sleep patterns are thus needed to improve applicability in real-life settings.
Accordingly, this study investigates the question, “Are there homogeneous sub-groups of individuals with distinct sleep patterns at risk for worsened sleep outcomes but heterogeneous with respect to age, co-morbidities, lifestyle, and self-reported health?” To address the challenges related to digital sleep data described earlier, we implement an unsupervised machine learning technique that can accommodate mixed membership structures within the data.[25, 27] This approach relies on the assumption that data come from several subpopulations and that the overall population is a mixture of these subpopulations, and further allows multivariate response data. This provides an ideal phenotype analysis framework to meet the objectives of the present study.
2. Methods
This study is an analysis of objectively-estimated sleep data collected to identify determinants of sleep disturbances in Latinx adults as a part of the Precision in Symptom Self-Management (PriSSM) Center (P30NR016587) at Columbia University. The PriSSM Center aims to advance the science of symptom self-management among the Latinx population through a social ecological lens that accounts for variability in individual, interpersonal, organizational, and environmental factors across the life course. All procedures were approved by the institutional review boards of the Columbia University Irving Medical Center and the New York State Psychiatric Institute. Informed consent was obtained from all participants.
2.1. Study Sample.
The study sample included Latinx adults enrolled into one of three pilot studies of PriSSM Center. One of the studies investigated sleep health and disturbance among women ages 40 or older (N=53). The second study investigated sleep health outcomes associated with exposure to discrimination-related stress among SGM adults 18 or older (N=39). The third study focused on improving sleep disturbance and fatigue among adults 50 or over (N=26) living with HIV through a fatigue self-management program. We used the baseline (i.e., pre-intervention) data from this sample. Complete inclusion and exclusion criteria are included in Supplemental file 1. For all studies, Latinx self-identification and ability to provide informed consent were inclusion criteria. The three study samples were combined for the analysis to allow heterogeneity in the overall sample motivated by the a priori research question.
2.2. Data Collection.
Demographic data were collected using self-report. Objective sleep data were collected using Actilife Actigraph (wGT3X-BT) accelerometers, worn on participants’ non-dominant wrist. Participants were asked to keep a log of the times they went to bed and got out of bed each day. Raw sleep data were collected in 60-sec epochs, validated and processed using the Actilife software and the Cole-Kripke algorithm. We primarily relied on the bed times and awakening times detected by the Actilife software and confirmed against the participant-reported times and Actigraph’s light sensor as recommended [28] for discrepancies. For determination of daytime sleep episodes (i.e., non-nocturnal sleep periods outside of the participants’ main sleep bout), we used methods followed by other similar studies[29, 30] and supplemented Actilife sleep scoring by sleep logs and accelerometer data on illumination (Lux) and activity.
2.2.1. Objective Sleep Variables.
Objectively-estimated sleep variables considered for cluster-relevant predictors included total sleep time (TST), time spent in bed (TIB), bed time, awakening time, wake after sleep onset (WASO), number of nightly awakenings (NOA) and the mean length of nightly awakenings after sleep onset, sleep efficiency (SE; 100×sleep duration/the time between sleep onset and awakening time), and daytime sleep.
2.2.2. Subjective Sleep Disturbance.
Self-reported sleep disturbance was measured using the 6-item Patient-Reported Outcomes Measurement Information System (PROMIS)[31] scale that assesses presence of sleep disturbance or sleep-related impairment in the past 7 days, and has good psychometric properties[31, 32]. Sample items include: “I had difficulty sleeping” and “My sleep was restless.” The raw scores were converted to standardized T-scores where 50 represents the population norm average with a standard deviation (SD) of 10. Higher scores indicate higher sleep disturbance[32].
2.3. Data Analytic Approach.
While there is no gold standard to determine the optimal set of predictors and number of clusters in unsupervised learning, we followed previously recommended methods[33–35] to identify the most parsimonious set of independent predictors that inform clustering behavior with the greatest capacity for discriminating between phenotypes. There are three steps involved in this methodological framework: 1) Dimension reduction and selection of predictors most associated with the principal dimensions, which indicate variables that explain the largest amount of variability in the data, 2) Estimation of the phenotypes through iterative model fitting with k number of mixtures to identify the model with the best fit, and 3) Comprehensive evaluation of the profiles of the identified phenotypes through significance tests of differences for all sleep variables in consideration. This step also includes evaluating variables used in the estimation models for confirming substantial differences amongst the mixtures. The following sections describe each of these steps in further detail.
2.3.1. Dimension Reduction and Selection of Predictors.
We first conducted a principal component analysis (PCA) on the standardized data using the singular value decomposition (SVD) method.[36, 37] Given the high correlation between several sleep variables (e.g., TST and TIB, SE and WASO), PCA is particularly useful as an exploratory technique to determine cluster-specific variable relevance and to identify dimensions along which the variation in the data is maximal.[36, 38] To determine how many PCs to consider for subsequent selection of the most contributing variables in the chosen component space, we relied on the combinatory metrics of >70% of the cumulative variance explained and the minimum fraction of the variance explained per PC (i.e., >(100/k)% where k is the number of sleep variables under consideration, thus 11.1% for the 9 sleep variables).[38] We then inspected variable contribution (%), correlation (i.e., “coordinates”) and cosine 2 values (quality of the representation) within each of these three PCs. Further details of the PCA steps are provided in Supplemental File 2.
The variables explaining the most variability from the PCA were then used in the subsequent mixture estimation models to estimate sleep phenotypes. In instances where two predictors were similarly associated with the PC but highly collinear (i.e., WASO and SE), we inspected each predictor’s contribution to between-cluster distance (which provides a measure of distinction) during the mixture estimation step, as well as practical feasibility (i.e., whether the model is able to partition the data into specified number of clusters) to determine predictor retention.
2.3.2. Clustering Analysis and Estimation of Sleep Phenotypes.
Cluster analysis was conducted using an unsupervised machine learning approach that relies on the mixture of multivariate generalized linear mixed models (MMGLMMs)[25, 27]. MMGLMMs assume that each individual is characterized by values of a set of latent random effects (i.e., predictors) and their probability of class membership (i.e., “individual component probability”; ICP) is estimated using Markov Chain Monte Carlo (MCMC)-based Bayesian inference. This consists of specifying a prior distribution for the model parameters and then basing the inference on the posterior sample distribution, estimated from the MCMC simulation. Consequently, ICPs and thus the maximum likelihood of being classified in a cluster is based on these resulting posterior median probabilities that are obtained by combining the prior distribution and the likelihood estimated from the MCMC simulation.
MMGLMMs accommodate different types of data that are irregularly sampled at multiple time points, and correlations between repeated observations of a marker measured on the same individual through inclusion of random effects. The model does not require classical normality and instead assumes different normal mixtures in the distribution of these random effects for different clusters, which improves model fit and robustness against misspecification of the random effects distribution. All of these factors collectively make MMGLMM a particularly useful technique in the context of our phenotyping problem. We used the “MixAK” library in R statistical software for the analyses [25]. Further details of the MMGLMM are described elsewhere [25, 27].
2.3.3. Assessment of Model Fit.
We assessed overall model fit based on the penalty of expected deviance (PED) index and the posterior distribution of model deviances (D) as recommended for MMGLMMs [25, 39]. PED is the sum of the expected model deviance and the model complexity penalty term optimism, p(opt)[40]. Similar to Akaike Information Criterion (AIC), smaller PED values indicate improvement in model fit by comparison of multiple MCMC-estimated models, thus informing model choice based on minimum loss. PED has been demonstrated to be robust across a range of sample sizes and parameter specifications and used for similar phenotyping problems in healthcare contexts (e.g., spatial mapping for diseases)[40]. Similarly, we inspected model deviance distributions to select between competing models following recommendations [41]. We quantified the certainty of posterior probabilities of models with 1–4 clusters, and used likelihood ratios of the posterior probabilities (Pdiff) for model comparison[39]. While there are no established Pdiff cut-off values, higher Pdiff values indicate better model fit and a ratio of > 0.9 (i.e., 90% model likelihood) has been suggested to indicate very strong confidence with which to choose a model.[39]
2.3.3.1. Model Convergence Diagnostics.
Convergence of the MCMC simulations of the model was assessed using the split- and the effective sample size (ESS), following published guidelines [42, 43]. The split- statistic quantifies whether the Markov chains mix well and a score of ~1.1 or less indicates desirable convergence to same stationary distribution [42]. The ESS is a measure of adequacy of the sample size of the iterations in the MCMC chains for obtaining a stable estimate [43] ESS values of >100 indicate adequate estimator accuracy.
2.3.4. Evaluation of Estimated Phenotype Profiles.
We conducted t-tests adjusted for number of comparisons to: 1) confirm inter-cluster differentiation in the predictor variables, and 2) assess phenotype-level differences in the sleep variables not used in the mixture estimation models, within-individual variability in the sleep variables, as well as age and sex assigned at birth (i.e., female or male sex). This allows for a comprehensive evaluation of the identified sleep phenotype profile with respect to all the sleep variables under consideration, not just those used in the phenotyping algorithm.
3. Results
3.1. Sample Characteristics.
Sample characteristics are provided in Table 1. The final sample included 494 person-level days of data from 118 participants, with a normally-distributed age range (skew=−0.53). Participants on average had 4.2 days of data (SD=1.47; Range=2–8). Two instances (from different participants) where TST values were uncharacteristically longer than the participants’ averages (i.e., ~12 hours vs averages of ~5.5 hours) were removed before analyses. Sample multivariate correlations between the sleep parameters are provided in Table 2. Of note, TST was highly correlated with TIB (r=0.93), and WASO was highly correlated with SE (r=−0.82).
Table 1.
Study sample characteristics (N=118).
| Outcome | Median (MAD) / Frequency (%) | Range |
|---|---|---|
| Age (Years) | 55.50 (15.57) | 19–77 |
| Female Sex at Birth | 70% (N=83) | NA |
| Ethnicity | Latinx/Hispanic | NA |
| Weight (lbs.) | 161 (31.13) | 102–335 |
| Sleep Parameter | Median (MAD) | Range |
| TST | 382.50 (91.92) | 175.0 – 587.0 |
| TIB | 437.00 (108.23) | 223.0 – 665.0 |
| WASO | 37.25 (20.02) | 3.0 – 201.0 |
| PROMIS-T | 45.30 (9.34) | 26.90 – 71.30 |
| Length of nightly awakenings | 2.72 (0.87) | 1–15.12 |
| SE | 90.78 (4.70) | 63.67 – 98.56 |
| NOA | 14.50 (6.67) | 3.0 – 32.0 |
| BED TIME | 03:00 (4.45) | 00.00 – 23.00 |
| AWAKENING TIME | 08:00 (1.48) | 02:00 – 19:50 |
MAD= Median absolute deviation. TST= Total sleep time, TIB= Time in bed, WASO= wake after sleep onset, PROMIS-T = PROMIS sleep disturbance T-score, SE= sleep efficiency, NOA= number of awakenings.
Table 2.
Pearson’s rho correlation coefficients for the multivariate correlation analysis among sleep parameters (N=494 person-level days).
| TST | TIB | WASO | Length of nightly awakenings | SE | NOA | Bed time | Awakening Time | |
|---|---|---|---|---|---|---|---|---|
| TST | 1 | |||||||
| TIB | 0.938*** | 1 | ||||||
| WASO | 0.222*** | 0.518*** | 1 | |||||
| Length of nightly awakenings | −0.080 | 0.214*** | 0.675*** | 1 | ||||
| SE | 0.156* | −0.181*** | −0.821*** | −0.634*** | 1 | |||
| NOA | 0.395*** | 0.565*** | 0.658*** | 0.026 | −0.507*** | 1 | ||
| Bed time | 0.249*** | 0.318*** | 0.270*** | 0.20*** | −0.191*** | 0.159*** | 1 | |
| Awakening Time | 0.292*** | 0.285*** | 0.052 | −0.06 | 0.103* | 0.168*** | −0.219*** | 1 |
p<0.05,
p<0.001,
<0.0001.
3.2. PCA, dimension and variable reduction.
Results of the PCA indicated the first three dimensions (i.e., principal components; PCs) individually explained >11.1% of the total variance and collectively explained >70% of the total variance. Results of the total contribution of each sleep variable under consideration, quality of their contributions, and the individual coordinates (i.e., correlations) within each PC are provided in Supplemental Table 2. Sleep parameters that explained the largest variance within these PCs included TIB, SE, WASO, TST, and TIB. In the dataset, TST and TIB were almost perfectly collinear (rho=0.94), and WASO was highly correlated to SE (rho=−0.82). Upon model comparisons, inclusion of SE versus WASO led to the same pattern of data partitioning; however, SE was associated with lower model deviance (i.e., PEDΔ =~30) indicating model improvement. Similarly, TIB was more discriminant compared to TST, where correct partitions were not achieved with TST (thus further penalizing the model). Consequently, TIB, SE, and bed time were included in the MMGLMMs for phenotype estimation. (See Supplemental File 2 for full PCA results).
3.3. MMGLMM Fit Characteristics.
A three-cluster resolution provided the best fit for the data based on the combinatory model fit indices of PED, posterior distributions (D), and the Pdiff values. There was a significant improvement from a single- to three-cluster model (i.e., PEDΔ ~35 and (D)Δ~74.55) and small improvement from the two- to three-cluster model (i.e., PEDΔ ~1 and (D) Δ~17.15). Similarly, the Pdiff scores were well above the cut-off value of 0.9 (i.e., 0.99 and 0.93 for 1- to 3- and 2- to 3-cluster models), collectively indicating evidence for up to 3 mixtures in the data. Overall cluster membership probability was high (Median ICP=0.98, MAD=0.03), indicating more than half of the participants were assigned into their respective cluster with at least 98% certainty. Model convergence diagnostics indicated good convergence to stable equilibrium based on the split- of 1.01 and the ESS of 3632.47 (See Supplemental File 3 for further model results).
3.4. Sleep Phenotypes.
Descriptive statistics for the sleep parameters across the three phenotypes are provided in Table 3 and Figure 1. Results of t-tests comparing the phenotypes are provided in Table 4. Differences between phenotypes for all model predictors were statistically significant, providing further support for model-estimated mixtures.
Table 3.
Sleep parameter summary scores and demographic variables for the estimated phenotypes (N1=64, N2=11, N3=43).
| Sleep Parameter | Median (MAD) | Range |
|---|---|---|
| Total Sleep Time (hours) | ||
| Phenotype 1 | 5.71 (1.76) | 3.14–8.8 |
| Phenotype 2 | 6.38 (0.83) | 4.36–7.38 |
| Phenotype 3 | 7.03 (1.07) | 2.91–9.78 |
| Total Time in Bed (hours) | ||
| Phenotype 1 | 6.35 (1.76) | 3.71–9.63 |
| Phenotype 2 | 9.0 (0.74) | 6.13–10.00 |
| Phenotype 3 | 7.90 (1.23) | 3.78–11.08 |
| Wake After Sleep Onset (minutes) | ||
| Phenotype 1 | 32.50 (12.62) | 10.0–81.50 |
| Phenotype 2 | 113 (38.55) | 56–201 |
| Phenotype 3 | 48 (23.72) | 3–134 |
| Number of Awakenings (minutes) | ||
| Phenotype 1 | 12.0 (5.56) | 3–32 |
| Phenotype 2 | 22.0 (2.97) | 14–27 |
| Phenotype 3 | 16.0 (6.67) | 3–30 |
| Length of Nightly Awakening (minutes) | ||
| Phenotype 1 | 2.60 (0.69) | 1.29–4.75 |
| Phenotype 2 | 2.79 (1.11) | 1–15.12 |
| Phenotype 3 | 5.14 (1.13) | 3.21–9.13 |
| Sleep Efficiency | ||
| Phenotype 1 | 92.18 (3.64) | 81.89–98.04 |
| Phenotype 2 | 75.19 (6.01) | 63.67–86.77 |
| Phenotype 3 | 89.51 (5.39) | 69.69–98.56 |
| Bed time (24 hour) | ||
| Phenotype 1 | 01:30 (1.48) | 00:00–11:30 |
| Phenotype 2 | 22:00 (1.48) | 20:25–23:00 |
| Phenotype 3 | 20:00 (4.45) | 00:00–23:35 |
| Awakening time (24 hour) | ||
| Phenotype 1 | 8:00 (2.22) | 04:00–13:00 |
| Phenotype 2 | 7:00 (0.34) | 02:00–09:00 |
| Phenotype 3 | 7:00 (1.28) | 02:20–19:36 |
| Daytime Sleep Episodes (minutes) | ||
| Phenotype 1 | 0 (0) | 0–78 |
| Phenotype 2 | 0 (0) | 0–0 |
| Phenotype 3 | 0 (0) | 0–61.14 |
| PROMIS Sleep Disturbance T-score | ||
| Phenotype 1 | 45.80 (8.45) | 26.90–71.30 |
| Phenotype 2 | 46.4 (10.53) | 35.70–65.60 |
| Phenotype 3 | 45.20 (10.45) | 26.90–71.30 |
| Demographics | ||
| Age (years) | ||
| Phenotype 1 | 53.5 (15.56) | 19–77 |
| Phenotype 2 | 63.0 (10.38) | 46–72 |
| Phenotype 3 | 57.0 (14.08) | 24–77 |
| Weight (lbs.) | ||
| Phenotype 1 | 165.50 (31.13) | 102–335 |
| Phenotype 2 | 163 (28.17) | 125–200 |
| Phenotype 3 | 159 (31.13) | 110–213 |
Panel Figure 1 Legend.
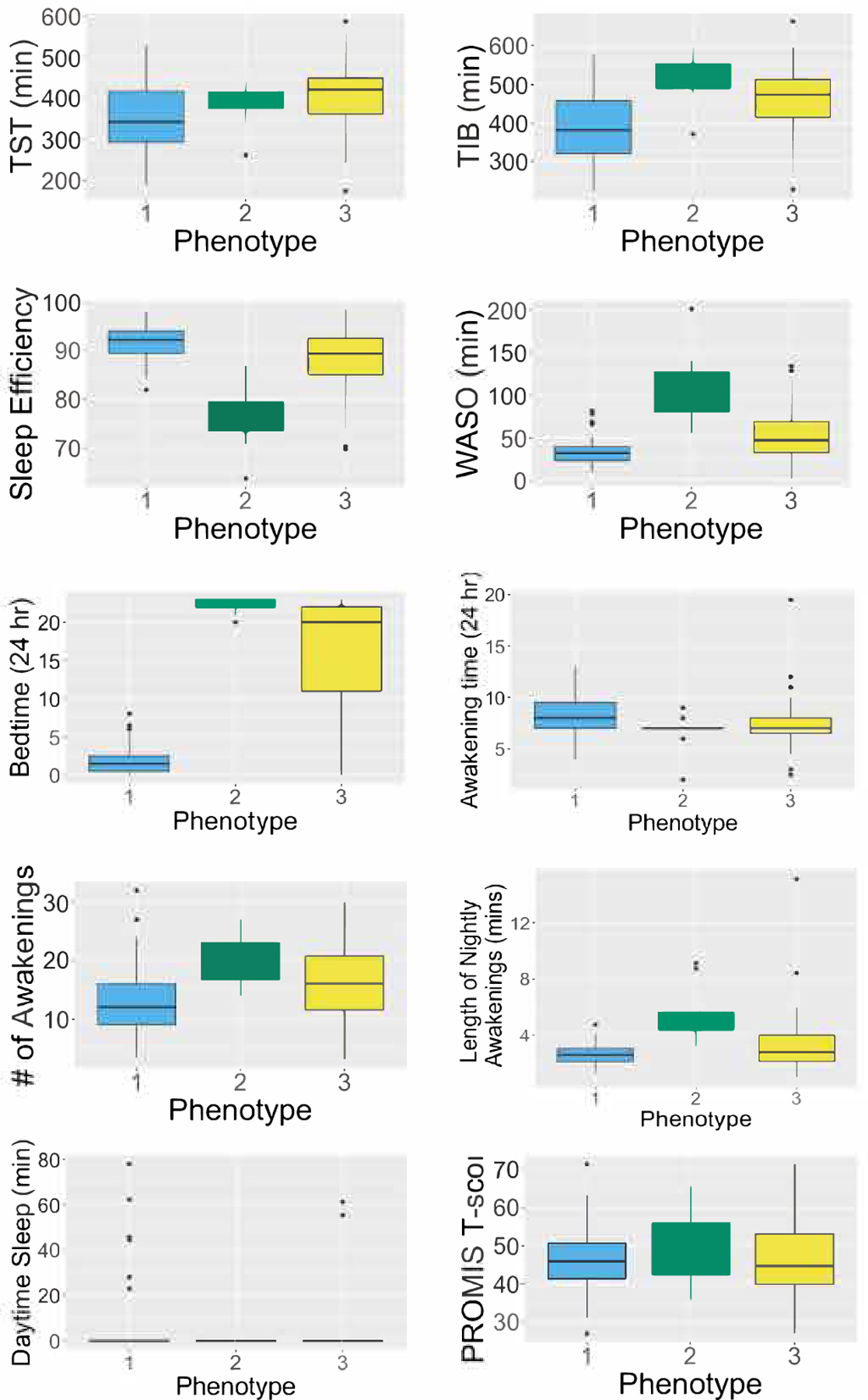
Distribution of sleep parameters in the sample based on person-level medians (N=118).
Table 4.
Results of the t-tests adjusted for number of comparisons in person-level medians and variability of sleep parameter between the three identified phenotypes (N1=64, N2=11, N3=43). Bonferroni-adjusted p-values are used.
| Sleep Parameter | t-statistic (df) | p-value | Sleep Parameter | t-statistic (df) | p-value |
|---|---|---|---|---|---|
| TST | TST SD | ||||
| Phenotypes 1–2 | −1.63 (21.5) | 0.238 | Phenotypes 1–2 | 6.60 (42.4) | <0.0001 |
| Phenotypes 1–3 | −2.98 (92.7) | 0.011 | Phenotypes 1–3 | 0.65 (99.9) | 0.566 |
| Phenotypes 2–3 | −0.95 (25.3) | 0.351 | Phenotypes 2–3 | −5.81 (39.1) | <0.0001 |
| TIB | TIB SD | ||||
| Phenotypes 1–2 | −5.98 (18.5) | <0.0001 | Phenotypes 1–2 | 6.11 (26.3) | <0.0001 |
| Phenotypes 1–3 | −3.97 (91.2) | 0.0002 | Phenotypes 1–3 | 0.65 (96.1) | 0.514 |
| Phenotypes 2–3 | 2.58 (22.2) | 0.017 | Phenotypes 2–3 | −5.26 (28.8) | <0.0001 |
| WASO | WASO SD | ||||
| Phenotypes 1–2 | −6.35 (10.5) | 0.0002 | Phenotypes 1–2 | −2.56 (12.8) | 0.048 |
| Phenotypes 1–3 | −3.86 (55.1) | 0.0006 | Phenotypes 1–3 | −2.56 (63.3) | 0.038 |
| Phenotypes 2–3 | 4.41 (13.5) | 0.0006 | Phenotypes 2–3 | 0.518 (21.9) | 0.610 |
| SE | SE SD | ||||
| Phenotypes 1–2 | 7.27 (12.1) | <0.0001 | Phenotypes 1–2 | −2.29 (10.8) | 0.086 |
| Phenotypes 1–3 | 3.04 (54.0) | 0.004 | Phenotypes 1–3 | −2.87 (62.7) | 0.017 |
| Phenotypes 2–3 | −4.85 (18.8) | 0.0002 | Phenotypes 2–3 | 0.96 (13.5) | 0.352 |
| Length of nightly awakening | Bed time SD | ||||
| Phenotypes 1–2 | −4.95 (10.4) | 0.002 | Phenotypes 1–2 | 0.84 (12.8) | 0.414 |
| Phenotypes 1–3 | −2.17 (46.6) | 0.035 | Phenotypes 1–3 | −6.88 (82.0) | <0.0001 |
| Phenotypes 2–3 | 3.06 (18.6) | 0.013 | Phenotypes 2–3 | −4.66 (15.7) | <0.001 |
| NOA | Awakening Time SD | ||||
| Phenotypes 1–2 | −5.05 (17.8) | 0.0002 | Phenotypes 1–2 | 4.57 (34.8) | 0.0002 |
| Phenotypes 1–3 | −2.47 (79.8) | 0.031 | Phenotypes 1–3 | 0.81 (96.9) | 0.419 |
| Phenotypes 2–3 | 2.52 (26.4) | 0.031 | Phenotypes 2–3 | −3.56 (36.0) | 0.002 |
| Bed time | PROMIS T-score | ||||
| Phenotypes 1–2 | −56.7 (23.8) | <0.0001 | Phenotypes 1–2 | −0.85 (11.6) | 1 |
| Phenotypes 1–3 | −10.2 (44.5) | <0.0001 | Phenotypes 1–3 | 0.13 (69.5) | 1 |
| Phenotypes 2–3 | 5.66 (45.9) | <0.0001 | Phenotypes 2–3 | 0.86 (15.8) | 1 |
| Awakening time | |||||
| Phenotypes 1–2 | 2.76 (13.8) | 0.046 | |||
| Phenotypes 1–3 | 1.75 (67.4) | 0.168 | |||
| Phenotypes 2–3 | −1.17 (23.1) | 0.199 | |||
Phenotype 1 (n=64, 61% female sex) constituted the largest cluster and was characterized by shorter TIB and WASO compared to the other two phenotypes (p<0.001 for both variables). These individuals were also more likely to be short sleepers (TST<6 hours/night), with half meeting criteria for insufficient TST[2]. Phenotype 1 was also associated with the highest overall SE values (i.e., ~92%) and more than 90% of the participants with this phenotype had average SE values greater than 85%, which is considered adequate SE. Other characteristics included decreased likelihood of NOA and later bed times than the other phenotypes (Table 4), as well as greater likelihood of within-person variability in TST and TIB. Daytime sleep episodes were more likely in phenotype 1, especially compared to phenotype 2, though the results were not statistically significant.
Phenotype 2 (n=11, 100% female sex) constituted the smallest cluster and was characterized by increased overall sleep impairment, with a median SE score of ~76% and 113 minutes of WASO. Similarly, more frequent nighttime awakenings were associated with this cluster, based on the significantly higher NOAs in comparison to phenotypes 1 and 3 (i.e., 22 vs 12 and 16, See Tables 3 and 4). Consequently, all participants clustered into this mixture had WASO values over 55 minutes, which is higher than various thresholds (e.g., >30 mins, >60mins) in the literature reportedly indicative of insomnia [44, 45]. While the median age was higher than that of phenotype 1, this difference was not statistically significant after accounting for sex. Earlier bed times and awakening times were also associated with phenotype 2, though the differences were small and only significant in comparison to phenotype 1 (See Table 3).
Phenotype 3 constituted ~36% of the sample (n=43, 76% female sex) and was characterized by overall adequate SE(~89%) but still greater NOA and mean length of awakenings compared to phenotype 1. This cluster was associated with the highest TST values overall, which was statistically significant when compared to phenotype 1 (Table 4). With a median TST of ~7 hours, members of this phenotype were more likely meet the recommended amount for adults. Within-person variability in TST, TIB and bed time was also higher in these individuals, compared to those in other phenotypes. When awakening times were compared using hourly intervals instead of decimal times, phenotype 3 was associated with greater variability in awakening times compared to phenotype 2 but less variability in comparison to phenotype 1.
Self-reported sleep disturbance (PROMIS) across phenotypes was comparable with no significant differences (Table 4). Similarly, weight did not differ among the three phenotypes (t=−0.15–1.61, p>0.05 for all three comparisons). Though the mean age between phenotypes 1 and 2 differed significantly based on the t-tests (t=−3.36, p=0.003), this effect was explained away when adjusted for sex (i.e., log odds of 6.64 and 0.55 for likelihood of phenotype 2 vs 1 and 3 vs 1, respectively; z-scores of 0.53 and 1.18, and p values of 0.59 and 0.23). Finally, the number of participants from the three studies within each phenotype are provided in Supplemental Table 5.
4. Discussion
This study investigated possible subgroups (“phenotypes”) based on objectively-estimated sleep parameters among individuals at increased risk for sleep disturbance. Using an unsupervised learning approach that accommodates inherent data complexities and sample heterogeneity, we identified three distinct sleep phenotypes that were best estimated through TIB, SE, and bed time. To our knowledge, this study is the first to investigate digital sleep phenotyping leveraging accelerometry and mixture models an unsupervised learning approach.
Our analyses indicated three distinct clusters (i.e., mixtures) to be the most suitable based on model fit, which were not independently associated with age or weight. Phenotype 1 constituted the largest cluster (N=64, 56% of the sample) and was associated with significantly greater likelihood of overall adequate SE and less variability in SE and WASO. Later bed times and shorter TST were also associated with this cluster, which were statistically significant in adjusted t-tests (See Table 4). Phenotype 2 constituted the smallest cluster (N=11) and had the highest likelihood of substantial and consistent sleep disturbance characterized by lower SE, more frequency awakenings NOAs, and greater WASO and TIB than the other phenotypes. The median age associated with this phenotype was also older than those of others though it was only significant when compared to phenotype 1 (i.e., 65.0 vs 56.0 years). Phenotype 3 (N=43) was characterized by a more variable sleep profile, with lower SE compared to phenotype 1, but also lower sleep disturbance compared to phenotype 2 as determined by fewer NOAs (t=−2.52, p=0.018) and shorter WASO (t=−4.41, p<0.001). Self-reported sleep disturbance scores across the three phenotypes were comparable and within ½ SD of the population-normalized mean. This suggests low concordance between scores from self-reported vs accelerometer-based sleep measures. This is consistent with previous studies,[17, 46] and supports the need for phenotyping based on multiple variables rather than using a single score when identifying individuals at risk for sleep problems.
Previous studies have reported shorter sleep duration with increasing age.[47] However, in this study phenotype 1 was associated with shorter TST but also significantly younger age compared to phenotype 2. While this finding might appear discrepant, our results indicated a trend where phenotype 1 was also more likely to engage in daytime sleep episodes (p=0.046). Combined with the relatively greater variability in TST linked to phenotype 1, it is possible that these characteristics collectively point to a sleep profile that is flexible in nature. These “short-but-efficient” sleepers might be better at adapting their sleep behaviors as needed according to changes in their daily schedules. While the lack of contextual information on these daytime sleep periods hinders a more comprehensive interpretation of daytime sleep episodes,[48] we note their significantly longer duration (i.e., ~2.5 hours on average) than what has been recommended and considered as beneficial.[49] Prior evidence on daytime sleep architecture is mixed,[50] with some studies indicating greater daytime sleep episodes are associated with increasing age,[51] while others report age-related changes in the timing but not the duration of daytime sleep episodes.[29, 47] There were 13 daytime sleep episodes from 9 participants in total, suggesting that these individuals might not necessarily be habitual daytime nappers based on the existing definitions of habitual napping (e.g., every day or at least twice/week).[49] It is possible that the individuals in phenotype 2 have difficulty initiating sleep onset in general or have daily schedules that are not amenable to daytime naps or longer sleep periods in general, which could explain their overall shorter sleep durations. Ultimately, daytime sleep architecture differs from that of the main nocturnal sleep period and their implications are still under debate[48, 52–54]. Our findings warrant further investigation into how these daytime sleep episodes might be related to different sleep phenotypes.
Our results indicated a trend (p-adjusted=0.07) where phenotype 3 was characterized by greater variability in both bedtimes and waketimes when using hourly intervals (i.e., p-adjusted<0.0001 for both pairwise comparisons). Regularity in bed times and waketimes has been suggested to promote adequate sleep by synchronizing sleep drive and circadian rhythm.[55] Similarly, greater variability in bed and wake time has been associated with increased likelihood of insomnia.[56] This is somewhat interesting given that this phenotype was not associated with the lowest sleep quality or older age. On the other hand, phenotype 2 was characterized by shorter but less variable TST, lower SE, and increased WASO, as well as earlier bed times and older age. Variability in bed times and waketimes has been found to decrease with increasing age, [57] therefore it is possible that these older participants had well-established sleep schedules. However, future studies with large sample sizes and longer data collection periods from each individual are needed to delineate whether the within-person variability associated with these phenotypes is stable over time.
SE has been recommended as a valid and robust indicator of sleep quality,[48] and its patterns have been studied by others across the life span.[30] Increased prevalence of sleep disturbance among women, SGM adults, and older adults have been well-documented in the literature, though mostly limited to data from population-based self-reported data.[8, 11, 58, 59] For instance, Zheng et al.[12] reported naturally occurring cyclical fluctuations in SE in menstruating midlife women (mean age=51.5). The identified profile characteristics of phenotypes 2 and 3 are consistent with these previous findings with respect to increased likelihood of variable SE, TIB, bed time and awakening time, as well as female sex. However, we also identified variable TST associated with phenotype 1 and similar age ranges compared to phenotype 2 (i.e., 19–77 for phenotype 1 vs 24–77 phenotype 2). To our knowledge, this study is the first to quantify these phenotypic characteristics using objective sleep data within a sample comprising ages 19–77, 32% transgender individuals, and 22% older adults with HIV. That we were able to identify these sleep phenotypes described in the literature in a mixed sample of individuals relying on ~4 days of data underscores the promise of unsupervised machine learning techniques to better understand sleep health phenotypes with relatively minimal participant burden.
Although validating the potential prognostic value of these phenotypes is beyond the scope of this study, these preliminary findings have possible implications from a precision sleep health perspective. For example, phenotype 3 may represent an “at risk” category where someone with these characteristics could be in the early stages of chronic sleep disturbance but perhaps not as severe as someone of phenotype 2. Individuals with different sleep phenotypes would likely benefit from different sleep health interventions (e.g., increasing TST for phenotype 2 individuals vs. establishing consistent a sleep schedule for phenotype 3). There are currently no established guidelines on how such sleep health prescriptions can or should be individually tailored in clinical settings, or which sleep parameters constitute optimal intervention targets. Moreover, the lack of established sleep parameter cut-off values for determination of sleep disturbance [44, 45, 60] could partly be because using a single cut-off across all patient populations is unsuitable. Instead, multiple cut-off values depending on patient phenotypes might be a more suitable approach. Similarly, integration of variability metrics and composite scores derived from frequently-sampled and/or longitudinal data are unique advantages of not only research-oriented devices such as that used in these studies, but increasingly of consumer devices such as fitness trackers and mobile phone apps (mHealth)that can augment clinical data.[61, 62] To this end, there is great promise in interdisciplinary efforts that employ mHealth devices for data collection and machine learning techniques for optimizing and designing scalable sleep solutions.[63]
Inclusion of participants at greater risk for sleep problems across the adult life span [6, 7, 64] further expands the existing literature on sleep health parameters within marginalized populations.[65, 66] For example, despite evidence that Latinx SGM adults report a higher prevalence of short sleep duration (≤6 hours per night), evidence on sleep health in SGM adults is limited to largely White samples. Similarly, cohort studies with racial/ethnic minorities and HIV-positive patients indicate that sleep disturbances and apnea are common[67], yet largely undiagnosed and untreated [5]. A previous study reported that despite having poorer self-reported sleep quality, Black women were less likely than other women to report their sleep disturbances to a physician [68]. Evidence indicates SGM patients face significant discrimination within healthcare settings,[69, 70] which could raise hesitancy to seeking counseling for sleep issues. Passively-collected and sleep data might therefore be particularly useful as it circumvents this challenge, as well as various others including inadequate clinician documentation of patient-reported sleep problems,[71] and providers’ implicit and explicit attitudes in clinical settings [72] which can impact clinical decision-making.[73] These collectively underscore the advantages of this low-effort, low-cost, and time-efficient method for delineating sleep problems among patient populations who might be harder to engage in clinical care.
There are several methodological strengths of our study. First, MCMC-based MMGLMMs consider each participant’s probability of membership to all existing clusters in the dataset to decide on the optimal classification, based on the maximal value of the posterior component probabilities. Next, they can accommodate sample heterogeneity and do not require classical normality in the random effects distribution. Instead, they assume a normal mixture and captures any possible dependence across values of different biomarkers through a joint distribution. Thus MMGLMMs improve clustering quality when using data with repeated measures, and have been demonstrated to be superior to averaging over the repeated measurements for producing relatively more stable and accurate clusters, and increase robustness against noise [74].
5. Limitations
We acknowledge limitations of this study. First, we had a relatively modest sample size of Latinx adults with more female than male participants. Therefore, generalizability of some of these findings might be limited and we cannot ascertain if the number and size of clusters, and cluster-relevant predictors might differ if the sample included other races and ethnicities. Similarly, temporal stability of the identified clusters across time is another interesting question this study did not investigate. Next, accelerometry cannot detect sleep apnea, which can be common in HIV, and contributes to poor sleep quality. The relationship of non-nocturnal sleep to overall sleep health remains an open research question [48], and whether they provide clinically relevant information on phenotype membership warrants further investigation. Our study sample included a wide age range cross the lifespan, which allows evaluation of variability in bed time and wake time in relation to key transition periods (e.g., young adulthood).[57] However, five participants only had 2 days of data, which limits our ability to interpret within-person variability outcomes, as well as to compare differences between week days vs weekend days. Though the median number of available days was 4 for all 3 clusters, it is possible that we might not have had sufficient days of data to capture the true variability for some of the participants. We similarly did not measure chronotype (i.e., morningness-eveningness preference), which warrants further investigation. An interesting future direction would be to conduct a phenotyping analysis that assesses the stability of the chronotypes associated with each phenotype.
6. Conclusion
In conclusion, digital data-driven approaches integrating robust data analytic techniques can aid in early detection and management of sleep problems. Our limited understanding of how sleep patterns unfold among different patient populations hinders the development of tailored interventions (e.g., behavioral intervention for promoting patient engagement in their care to attenuate the negative effects of discrimination-related stress on sleep health). These results have implications for informing individually-tailored strategies and precision sleep health applications across diverse patient populations.
Supplementary Material
Highlights.
Flexible unsupervised learning techniques can delineate sleep disturbance patterns
We identify 3 distinct mixtures “phenotypes” in a heterogenous Latinx sample
One phenotype was characterized by more efficient but shorter sleep patterns
Another phenotype was associated with overall variable sleep quality and bedtime
Sleep disturbance patterns might be identified through few mHealth-based variables
Role of the funding source
Funding for the work is provided by a fellowship from the Data Science Institute at Columbia University (Ensari) and an award from the National Institutes of Health (P30NR016587, MPI: Bakken).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
All authors report no actual or potential conflict of interest.
References
- 1.Office of Disease Prevention and Health Promotion 2020 [October 16, 2020]. Available from: https://health.gov/healthypeople/objectives-and-data/browse-objectives/sleep#cit2.
- 2.St-Onge MP, Grandner MA, Brown D, Conroy MB, Jean-Louis G, Coons M, et al. Sleep Duration and Quality: Impact on Lifestyle Behaviors and Cardiometabolic Health: A Scientific Statement From the American Heart Association. Circulation. 2016;134(18):e367–e86. Epub 2016/11/02. doi: 10.1161/cir.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin J, Jin X, Shan Z, Li S, Huang H, Li P, et al. Relationship of Sleep Duration With All-Cause Mortality and Cardiovascular Events: A Systematic Review and Dose-Response Meta-Analysis of Prospective Cohort Studies. J Am Heart Assoc. 2017;6(9). Epub 2017/09/11. doi: 10.1161/jaha.117.005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee KA, Gay C, Portillo CJ, Coggins T, Davis H, Pullinger CR, et al. Types of sleep problems in adults living with HIV/AIDS. Journal of Clinical Sleep Medicine. 2012;8(1):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutierrez J, Tedaldi EM, Armon C, Patel V, Hart R, Buchacz K. Sleep disturbances in HIV-infected patients associated with depression and high risk of obstructive sleep apnea. SAGE open medicine. 2019;7:2050312119842268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler ES, McGlinchey E, Juster R-P. Sexual and gender minority sleep: A narrative review and suggestions for future research. Journal of Sleep Research. 2020;29(1):e12928. doi: 10.1111/jsr.12928. [DOI] [PubMed] [Google Scholar]
- 7.Caceres BA, Hickey KT, Heitkemper EM, Hughes TL. An intersectional approach to examine sleep duration in sexual minority adults in the United States: findings from the Behavioral Risk Factor Surveillance System. Sleep Health. 2019;5(6):621–9. Epub 2019/08/05. doi: 10.1016/j.sleh.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caceres BA, Hickey KT. Examining Sleep Duration and Sleep Health Among Sexual Minority and Heterosexual Adults: Findings From NHANES (2005–2014). Behav Sleep Med. 2020;18(3):345–57. Epub 2019/03/28. doi: 10.1080/15402002.2019.1591410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slopen N, Lewis TT, Williams DR. Discrimination and sleep: a systematic review. Sleep Medicine. 2016;18:88–95. doi: 10.1016/j.sleep.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rock P, Goodwin G, Harmer C, Wulff K. Daily rest-activity patterns in the bipolar phenotype: A controlled actigraphy study. Chronobiology International. 2014;31(2):290–6. doi: 10.3109/07420528.2013.843542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai H, Hao J. Sleep Deprivation and Chronic Health Conditions Among Sexual Minority Adults. Behavioral Sleep Medicine. 2019;17(3):254–68. doi: 10.1080/15402002.2017.1342166. [DOI] [PubMed] [Google Scholar]
- 12.Zheng H, Harlow SD, Kravitz HM, Bromberger J, Buysse DJ, Matthews KA, et al. Actigraphy-defined measures of sleep and movement across the menstrual cycle in midlife menstruating women: Study of Women’s Health Across the Nation Sleep Study. Menopause. 2015;22(1):66–74. doi: 10.1097/GME.0000000000000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kravitz HM, Zhao X, Bromberger JT, Gold EB, Hall MH, Matthews KA, et al. Sleep disturbance during the menopausal transition in a multi-ethnic community sample of women. Sleep. 2008;31(7):979–90. [PMC free article] [PubMed] [Google Scholar]
- 14.Kolp H, Wilder S, Andersen C, Johnson E, Horvath S, Gidycz CA, et al. Gender minority stress, sleep disturbance, and sexual victimization in transgender and gender nonconforming adults. Journal of Clinical Psychology. 2020;76(4):688–98. doi: 10.1002/jclp.22880. [DOI] [PubMed] [Google Scholar]
- 15.Bowen AE, Staggs S, Kaar J, Nokoff N, Simon SL. Short sleep, insomnia symptoms, and evening chronotype are correlated with poorer mood and quality of life in adolescent transgender males. Sleep Health. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauderdale DS, Knutson KL, Rathouz PJ, Yan LL, Hulley SB, Liu K. Cross-sectional and longitudinal associations between objectively measured sleep duration and body mass index: the CARDIA Sleep Study. American journal of epidemiology. 2009;170(7):805–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Den Berg JF, Van Rooij FJ, Vos H, Tulen JH, Hofman A, Miedema HM, et al. Disagreement between subjective and actigraphic measures of sleep duration in a population-based study of elderly persons. J Sleep Res. 2008;17(3):295–302. Epub 2008/03/07. doi: 10.1111/j.1365-2869.2008.00638.x. [DOI] [PubMed] [Google Scholar]
- 18.Tachikawa R, Minami T, Matsumoto T, Murase K, Tanizawa K, Inouchi M, et al. Changes in Habitual Sleep Duration after Continuous Positive Airway Pressure for Obstructive Sleep Apnea. Ann Am Thorac Soc. 2017;14(6):986–93. Epub 2017/03/23. doi: 10.1513/AnnalsATS.201610-816OC. [DOI] [PubMed] [Google Scholar]
- 19.Xu D, Tian Y. A Comprehensive Survey of Clustering Algorithms. Annals of Data Science. 2015;2(2):165–93. doi: 10.1007/s40745-015-0040-1. [DOI] [Google Scholar]
- 20.Everitt BS. A finite mixture model for the clustering of mixed-mode data. Statistics & Probability Letters. 1988;6(5):305–9. doi: 10.1016/0167-7152(88)90004-1. [DOI] [Google Scholar]
- 21.Raykov YP, Boukouvalas A, Baig F, Little MA. What to Do When K-Means Clustering Fails: A Simple yet Principled Alternative Algorithm. PLOS ONE. 2016;11(9):e0162259. doi: 10.1371/journal.pone.0162259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong S, Deb S, Yang X-S, Zhuang Y. Towards Enhancement of Performance of K-Means Clustering Using Nature-Inspired Optimization Algorithms. The Scientific World Journal. 2014;2014:564829. doi: 10.1155/2014/564829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erosheva EA, Fienberg SE. Bayesian mixed membership models for soft clustering and classification. Classification—The Ubiquitous Challenge: Springer; 2005. p. 11–26. [Google Scholar]
- 24.Airoldi EM, Blei DM, Erosheva EA, Fienberg SE. Introduction to Mixed Membership Models and Methods. Handbook of mixed membership models and their applications. 2014;100:3–14. [Google Scholar]
- 25.Komárek A A new R package for Bayesian estimation of multivariate normal mixtures allowing for selection of the number of components and interval-censored data. Computational Statistics & Data Analysis. 2009;53(12):3932–47. doi: 10.1016/j.csda.2009.05.006. [DOI] [Google Scholar]
- 26.Hughes DM, Komárek A, Czanner G, Garcia-Fiñana M. Dynamic longitudinal discriminant analysis using multiple longitudinal markers of different types. Stat Methods Med Res. 2018;27(7):2060–80. Epub 2016/10/30. doi: 10.1177/0962280216674496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komárek A, Komárková L. Clustering for multivariate continuous and discrete longitudinal data. The Annals of Applied Statistics. 2013;7(1):177–200. [Google Scholar]
- 28.Martin JL, Hakim AD. Wrist actigraphy. Chest. 2011;139(6):1514–27. doi: 10.1378/chest.10-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon IY, Kripke DF, Youngstedt SD, Elliott JA. Actigraphy suggests age-related differences in napping and nocturnal sleep. J Sleep Res. 2003;12(2):87–93. Epub 2003/05/20. doi: 10.1046/j.1365-2869.2003.00345.x. [DOI] [PubMed] [Google Scholar]
- 30.Dashti HS, Zuurbier LA, de Jonge E, Voortman T, Jacques PF, Lamon-Fava S, et al. Actigraphic sleep fragmentation, efficiency and duration associate with dietary intake in the Rotterdam Study. Journal of sleep research. 2016;25(4):404–11. Epub 2016/02/09. doi: 10.1111/jsr.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu L, Buysse DJ, Germain A, Moul DE, Stover A, Dodds NE, et al. Development of short forms from the PROMIS™ sleep disturbance and Sleep-Related Impairment item banks. Behavioral sleep medicine. 2011;10(1):6–24. doi: 10.1080/15402002.2012.636266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buysse DJ, Yu L, Moul DE, Germain A, Stover A, Dodds NE, et al. Development and validation of patient-reported outcome measures for sleep disturbance and sleep-related impairments. Sleep. 2010;33(6):781–92. Epub 2010/06/17. doi: 10.1093/sleep/33.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yau C, Holmes C. Hierarchical Bayesian nonparametric mixture models for clustering with variable relevance determination. Bayesian analysis. 2011;6(2):329–52. doi: 10.1214/11-BA612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maugis C, Celeux G, Martin-Magniette ML. Variable selection for clustering with Gaussian mixture models. Biometrics. 2009;65(3):701–9. Epub 2009/02/13. doi: 10.1111/j.1541-0420.2008.01160.x. [DOI] [PubMed] [Google Scholar]
- 35.Guan WJ, Jiang M, Gao YH, Li HM, Xu G, Zheng JP, et al. Unsupervised learning technique identifies bronchiectasis phenotypes with distinct clinical characteristics. Int J Tuberc Lung Dis. 2016;20(3):402–10. Epub 2016/04/06. doi: 10.5588/ijtld.15.0500. [DOI] [PubMed] [Google Scholar]
- 36.Ringnér M What is principal component analysis? Nature biotechnology. 2008;26(3):303–4. [DOI] [PubMed] [Google Scholar]
- 37.Nakatsukasa Y, Higham NJ. Stable and Efficient Spectral Divide and Conquer Algorithms for the Symmetric Eigenvalue Decomposition and the SVD. SIAM Journal on Scientific Computing. 2013;35(3):A1325–A49. doi: 10.1137/120876605. [DOI] [Google Scholar]
- 38.Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374(2065):20150202-. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aitkin M, Liu CC, Chadwick T. Bayesian model comparison and model averaging for small-area estimation. The Annals of applied statistics. 2009;3(1):199–221. [Google Scholar]
- 40.Plummer M Penalized loss functions for Bayesian model comparison. Biostatistics. 2008;9(3):523–39. doi: 10.1093/biostatistics/kxm049. [DOI] [PubMed] [Google Scholar]
- 41.Raftery AE, Dean N. Variable selection for model-based clustering. Journal of the American Statistical Association. 2006;101(473):168–78. [Google Scholar]
- 42.Vehtari A, Gelman A, Simpson D, Carpenter B, Bürkner P-C. Rank-normalization, folding, and localization: An improved $\widehat {R} $ for assessing convergence of MCMC. Bayesian Analysis. 2020. [Google Scholar]
- 43.Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB. Bayesian data analysis: CRC press; 2013. [Google Scholar]
- 44.Lichstein KL, Durrence HH, Taylor DJ, Bush AJ, Riedel BW. Quantitative criteria for insomnia. Behaviour Research and Therapy. 2003;41(4):427–45. doi: 10.1016/S0005-7967(02)00023-2. [DOI] [PubMed] [Google Scholar]
- 45.Lineberger MD, Carney CE, Edinger JD, Means MK. Defining insomnia: quantitative criteria for insomnia severity and frequency. Sleep. 2006;29(4):479–85. [DOI] [PubMed] [Google Scholar]
- 46.Kreutz C, Müller J, Schmidt ME, Steindorf K. Comparison of subjectively and objectively assessed sleep problems in breast cancer patients starting neoadjuvant chemotherapy. Supportive Care in Cancer. 2021;29(2):1015–23. doi: 10.1007/s00520-020-05580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Campbell SS, Murphy PJ. The nature of spontaneous sleep across adulthood. J Sleep Res. 2007;16(1):24–32. Epub 2007/02/21. doi: 10.1111/j.1365-2869.2007.00567.x. [DOI] [PubMed] [Google Scholar]
- 48.Ohayon M, Wickwire EM, Hirshkowitz M, Albert SM, Avidan A, Daly FJ, et al. National Sleep Foundation’s sleep quality recommendations: first report. Sleep health. 2017;3(1):6–19. [DOI] [PubMed] [Google Scholar]
- 49.Milner CE, Cote KA. Benefits of napping in healthy adults: impact of nap length, time of day, age, and experience with napping. Journal of sleep research. 2009;18(2):272–81. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Vitiello MV, Gooneratne NS. Sleep in Normal Aging. Sleep Med Clin. 2018;13(1):1–11. Epub 2017/11/21. doi: 10.1016/j.jsmc.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furihata R, Kaneita Y, Jike M, Ohida T, Uchiyama M. Napping and associated factors: a Japanese nationwide general population survey. Sleep Med. 2016;20:72–9. Epub 2016/06/19. doi: 10.1016/j.sleep.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Goldman SE, Hall M, Boudreau R, Matthews KA, Cauley JA, Ancoli-Israel S, et al. Association between nighttime sleep and napping in older adults. Sleep. 2008;31(5):733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDevitt EA, Alaynick WA, Mednick SC. The effect of nap frequency on daytime sleep architecture. Physiol Behav. 2012;107(1):40–4. Epub 2012/05/31. doi: 10.1016/j.physbeh.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee T-Y, Chang P-C, Tseng I-J, Chung M-H. Nocturnal sleep mediates the relationship between morningness–eveningness preference and the sleep architecture of afternoon naps in university students. PLOS ONE. 2017;12(10):e0185616. doi: 10.1371/journal.pone.0185616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irish LA, Kline CE, Gunn HE, Buysse DJ, Hall MH. The role of sleep hygiene in promoting public health: A review of empirical evidence. Sleep medicine reviews. 2015;22:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stepanski EJ, Wyatt JK. Use of sleep hygiene in the treatment of insomnia. Sleep medicine reviews. 2003;7(3):215–25. [DOI] [PubMed] [Google Scholar]
- 57.Duncan MJ, Kline CE, Rebar AL, Vandelanotte C, Short CE. Greater bed- and wake-time variability is associated with less healthy lifestyle behaviors: a cross-sectional study. Z Gesundh Wiss. 2016;24(1):31–40. Epub 2015/10/20. doi: 10.1007/s10389-015-0693-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madrid-Valero JJ, Martínez-Selva JM, Ribeiro do Couto B, Sánchez-Romera JF, Ordoñana JR. Age and gender effects on the prevalence of poor sleep quality in the adult population. Gaceta Sanitaria. 2017;31(1):18–22. doi: 10.1016/j.gaceta.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 59.Tang J, Liao Y, Kelly BC, Xie L, Xiang Y-T, Qi C, et al. Gender and Regional Differences in Sleep Quality and Insomnia: A General Population-based Study in Hunan Province of China. Scientific Reports. 2017;7(1):43690. doi: 10.1038/srep43690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vgontzas AN, Liao D, Bixler EO, Chrousos GP, Vela-Bueno A. Insomnia with Objective Short Sleep Duration is Associated with a High Risk for Hypertension. Sleep. 2009;32(4):491–7. doi: 10.1093/sleep/32.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ensari I, Pichon A, Lipsky-Gorman S, Bakken S, Elhadad N. Augmenting the Learning Health System for Enigmatic Diseases: a Cross-Sectional Study of Clinical Documentation and Self-Tracking Data in Endometriosis. . Applied Clinical Informatics, under review. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ensari I, Elhadad N. mHealth For Research: Participatory Research Applications to Gain Disease Insights: Elsevier; 2021. [Google Scholar]
- 63.Goldstein CA, Berry RB, Kent DT, Kristo DA, Seixas AA, Redline S, et al. Artificial intelligence in sleep medicine: background and implications for clinicians. Journal of Clinical Sleep Medicine. 2020;16(4):609–18. doi: 10.5664/jcsm.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whinnery J, Jackson N, Rattanaumpawan P, Grandner MA. Short and Long Sleep Duration Associated with Race/Ethnicity, Sociodemographics, and Socioeconomic Position. Sleep. 2014;37(3):601–11. doi: 10.5665/sleep.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan JY, Baig AA, Chin MH. High Stakes for the Health of Sexual and Gender Minority Patients of Color. Journal of General Internal Medicine. 2017;32(12):1390–5. doi: 10.1007/s11606-017-4138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Powell Sears K Improving cultural competence education: the utility of an intersectional framework. Medical Education. 2012;46(6):545–51. doi: 10.1111/j.1365-2923.2011.04199.x. [DOI] [PubMed] [Google Scholar]
- 67.Chen X, Wang R, Zee P, Lutsey PL, Javaheri S, Alcántara C, et al. Racial/Ethnic Differences in Sleep Disturbances: The Multi-Ethnic Study of Atherosclerosis (MESA). Sleep. 2015;38(6):877201388. Epub 2014/11/20. doi: 10.5665/sleep.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amyx M, Xiong X, Xie Y, Buekens P. Racial/Ethnic Differences in Sleep Disorders and Reporting of Trouble Sleeping Among Women of Childbearing Age in the United States. Matern Child Health J. 2017;21(2):306–14. Epub 2016/07/22. doi: 10.1007/s10995-016-2115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ayhan CHB, Bilgin H, Uluman OT, Sukut O, Yilmaz S, Buzlu S. A systematic review of the discrimination against sexual and gender minority in health care settings. International Journal of Health Services. 2020;50(1):44–61. [DOI] [PubMed] [Google Scholar]
- 70.Howard SD, Lee KL, Nathan AG, Wenger HC, Chin MH, Cook SC. Healthcare Experiences of Transgender People of Color. Journal of General Internal Medicine. 2019;34(10):2068–74. doi: 10.1007/s11606-019-05179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alegría M, Nakash O, Lapatin S, Oddo V, Gao S, Lin J, et al. How missing information in diagnosis can lead to disparities in the clinical encounter. J Public Health Manag Pract. 2008;14Suppl(Suppl):S26–S35. doi: 10.1097/01.PHH.0000338384.82436.0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anastas TM, Miller MM, Hollingshead NA, Stewart JC, Rand KL, Hirsh AT. The Unique and Interactive Effects of Patient Race, Patient Socioeconomic Status, and Provider Attitudes on Chronic Pain Care Decisions. Ann Behav Med. 2020;54(10):771–82. doi: 10.1093/abm/kaaa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young T, Hutton R, Finn L, Badr S, Palta M. The Gender Bias in Sleep Apnea Diagnosis: Are Women Missed Because They Have Different Symptoms? Archives of Internal Medicine. 1996;156(21):2445–51. doi: 10.1001/archinte.1996.00440200055007. [DOI] [PubMed] [Google Scholar]
- 74.Yeung KY, Medvedovic M, Bumgarner RE. Clustering gene-expression data with repeated measurements. Genome Biol. 2003;4(5):R34–R. Epub 2003/04/25. doi: 10.1186/gb-2003-4-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


