Abstract
Coronavirus disease 2019 (COVID‐19) epidemic has devastating effects on personal health around the world. It is significant to achieve accurate segmentation of pulmonary infection regions, which is an early indicator of disease. To solve this problem, a deep learning model, namely, the content‐aware pre‐activated residual UNet (CAPA‐ResUNet), was proposed for segmenting COVID‐19 lesions from CT slices. In this network, the pre‐activated residual block was used for down‐sampling to solve the problems of complex foreground and large fluctuations of distribution in datasets during training and to avoid gradient disappearance. The area loss function based on the false segmentation regions was proposed to solve the problem of fuzzy boundary of the lesion area. This model was evaluated by the public dataset (COVID‐19 Lung CT Lesion Segmentation Challenge—2020) and compared its performance with those of classical models. Our method gains an advantage over other models in multiple metrics. Such as the Dice coefficient, specificity (Spe), and intersection over union (IoU), our CAPA‐ResUNet obtained 0.775 points, 0.972 points, and 0.646 points, respectively. The Dice coefficient of our model was 2.51% higher than Content‐aware residual UNet (CARes‐UNet). The code is available at https://github.com/malu108/LungInfectionSeg.
Keywords: area loss function, computed tomography (CT) image segmentation, COVID‐19, deep learning, pre‐activated residual block
1. INTRODUCTION
Since the outbreak of COVID‐19, it has been endangering the world, and seriously affecting economic development and social life. 1 The disease was caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), which mainly affecting the human respiratory system. As of January 2, 2022, nearly 289 million cases were reported worldwide, of which 5.4 million people died. 2
Due to the rapid spread and strong infectivity of COVID‐19, rapid and accurate disease diagnosis become an urgent need. According to clinical recommendations, virus real‐time reverse transcription‐polymerase chain reaction (RT‐PCR) detection can effectively diagnose SARS‐CoV‐2 infection. 3 However, due to difficulties in sample collection and transportation, low sensitivity and long time consuming, 4 RT‐PCR detections delay diagnosis and treatment of many patients with COVID‐19 and increase the harm to society.
COVID‐19 lung CT lesion segmentation is the most useful as a preliminary step in identifying the degree of disease. 5 However, it is extremely time‐consuming and laborious for radiologists to manually segment virus infection regions in chest CT images, and the subjective factors of radiologists also have a certain impact on the segmentation results. 6 Therefore, automatic segmentation of lesions is necessary for clinical practice. Over the years, with the rapid development of information technology, deep learning methods have been generally applied in medical image processing tasks 7 , 8 especially algorithms based on the convolutional neural network have achieved good performance. 9 , 10 , 11 In the clinical diagnosis and detection of COVID‐19, it is necessary to classify and segment images when evaluating the risk of progression or deterioration of the disease, in which deep learning methods play an important role with their powerful feature extraction ability. 12 , 13
Many researchers have designed varieties of deep learning algorithms to process COVID‐19 images so far, and have achieved good results. One is COVID‐19 classification detection, and the other is lesion segmentation. Some of them simultaneously completed the two tasks of COVID‐19 detection and lesion segmentation. 14 , 15 , 16 Gao et al. 14 proposed a dual‐branch combined network for the diagnosis of COVID‐19, which could realize classification and lesion segmentation simultaneously. The algorithm first used UNet to extract the accurate lung region, and then used the lesion attention (LA) module to complete the slice classification and segmentation of CT images at the same time. A final slice probability mapping method was proposed to synthesize the slice results and obtained the final classification results. Pezzano et al. 15 designed a U‐shaped network structure with a multi‐convolution layer structure, realized automatic COVID‐19 disease detection and lesion segmentation, and proposed a new loss function for balancing accuracy and sensitivity. Wu et al. 16 proposed a sequence region generation network (SRGNet) for joint detection and segmentation of COVID‐19 lesions. The context enhancement module and a new edge loss function were designed to augment the context information of the images for generating segmented images of different scales.
The CT image segmentation of lesions has attracted much attention. Xie et al. 17 proposed a new coronary pneumonia lesion segmentation network named DUDA‐Net based on UNet. First, the coarse segmentation network was used for extracting lung regions from chest CT images; Second, the dilated convolution attention (DCA) module was added to the fine segmentation network to increase the network's capacity to segment fine areas of the lesion. Bizopoulos et al. 18 compared several deep learning models for COVID‐19 lesions segmentation, and mainly tested four network structures (UNet, LinkNet, Feature Pyramid Network, and Pyramid Scene Parsing Network) along with 25 encoders for segmenting lung lesions. Pei et al. 19 designed a multipoint monitoring network (MPS‐Net) for lesion segmentation in CT images to settle the matter of various infected areas. Zhang et al. 20 designed a network named COVSeg‐NET for lesion segmentation according to a fully convolutional neural network structure. Yu et al. 21 proposed a COVID‐19 CT images segmentation model that used a pyramid attention module to extract multiscale context information and a residual convolution module to improve the capacity of network identification. An edge loss function was also used to extract the edge features of images effectively. Saeedizadeh et al. 22 designed a network structure similar to UNet called TV‐UNet and introduced a regularization term based on two‐dimensional anisotropic total variation in the loss function to complete the lesion area segmentation on a voxel level. Zhao et al. 23 proposed a dilated dual attention UNet, namely D2A U‐Net, to segment COVID‐19 lesions in CT images. Wang et al. 24 proposed a noise‐robust learning algorithm for the segmentation of COVID‐19 lung lesions. The method combined maximum pooling and average pooling for downsampling and used the bridge connection instead of the jump connection in the UNet structure. In addition, they proposed a noise‐robust Dice loss function for training network. Compared with UNet, the pneumonia lesion area could be effectively segmented from the noise label. Rajamani et al. 25 proposed a new dynamic deformable attention network (DDANet), which could calculate the feature information more accurately. Based on the classical UNet network, this method added a deformable criss‐cross attention block (CCA) to the connection between encoder and decoder that can effectively improve the performance of the model. Stefano et al. 26 proposed a customized ENET for real‐time and automatic recognition and segmentation of computer tomography images infected with COVID‐19 disease and achieved good results. Xu et al. 27 proposed a content‐aware residual network (CARes‐UNet) to segment COVID‐19 lesions from chest CT images. In the network, the content‐aware upsampling module was introduced to enhance model performance.
Some researchers have used transfer learning methods to study COVID‐19 image processing. Lu et al. 28 used transfer learning to implement image‐level representation (ILR) learning, and proposed neighboring aware representation to extract the neighboring information among the ILRs. Finally, they proposed neighboring aware graph neural network (NAGNN) for COVID‐19 classification. Liu et al. 29 proposed a novel model named nCoVSegNet for segmenting COVID‐19 lesions in CT slices. In detail, it used attention perception feature fusion to segment and utilized a two‐stage cross‐domain transfer learning method to pre‐train the network. The performance results of the abovementioned methods are presented in Table 1.
TABLE 1.
The performance results of the abovementioned COVID‐19 segmentation methods
| Study | Method | Dataset | Dice |
|---|---|---|---|
| Gao et al. 14 | DCN | Private dataset | 0.8351 |
| Wu et al. 16 | SRGNet | Private dataset | 0.83 |
| Xie et al. 17 | DUDA‐Net | Public database from Radiopaedia | 0.8706 |
| Pei et al. 19 | MPS‐Net | COVID‐19 CT segmentation dataset | 0.8325 |
| Zhang et al. 20 | COVSeg‐NET | Kaggle's COVID‐19 CT image segmentation competition | 0.561 |
| Yu et al. 21 | Multi‐class COVID‐19 segmentation network | COVID‐SemiSeg dataset | 0.779 |
| Saeedizadeh et al. 22 | COVID TV‐Unet | COVID‐19 CT segmentation dataset | 0.861 |
| Zhao et al. 23 | D2A U‐Net | COVID‐19 CT segmentation dataset | 0.7298 |
| Wang et al. 24 | COPLE‐Net | Private dataset | 0.8029 |
| Rajamani et al. 25 | DDANet | COVID‐19 CT segmentation dataset | 0.743 |
| Stefano et al. 26 | C‐ENET | COVID‐19 CT lesion segmentation Challenge‐2020 dataset | 0.7483 |
| Xu et al. 27 | CARes‐UNet | COVID‐SemiSeg dataset | 0.731 |
| Liu et al. 29 | nCoVSegNet | MosMedData | 0.6843 |
Although these studies have obtained fine segmentation results, the segmentation of pulmonary lesions is still a challenging task. The main reasons are as follows. First, the size and location of the infection area are various between different infection periods and patients. Second, the lesion features are complex and diverse, such as ground glass shadow, consolidation shadow, and infiltration shadow; in addition, the low contrast and blurred boundary between the lesion areas and the healthy areas increase difficulties of automatic segmentation of lesions. 6 To address these problems, in this study, we design a content‐aware pre‐activated residual network (CAPA‐ResUNet) to segment COVID‐19 lesions.
The contribution of this article is to improve the network structure and apply the mathematical optimization method to the loss function to obtain better training and segmentation results. In the network, a pre‐activated residual block was proposed for down sampling and utilized content‐aware up‐sampling algorithm for up sampling. The area loss function based on the error segmentation region was proposed, and carried out the network optimization together with Combo loss. 30 We also performed comparative studies and ablation experiments to prove the feasibility and effectiveness of our method.
The rest of this article is as follows. Section 2 explains the methods in detail. Section 3 offers the experiment environment, settings, and the experiment study. Finally, Section 4 provides the conclusion and future work.
2. METHODS
2.1. Network structure
Figure 1 shows the structure of the proposed CAPA‐ResUNet. Inspired by the variant of UNet 9 and the encoder‐decoder structure 31 , 32 we use a similar frame, mainly including a down‐sampling path and an up‐sampling path. While the down‐sampling path expands the receptive field, it reduces the computational cost, and the up‐sampling path restores the missing feature information in the down‐sampling path. All encoder modules are composed of 3 × 3 convolution operations with stride 1, batch normalization (BN) layer, 33 rectified linear unit (ReLU) activation function 34 and residual block. 35 The down‐sampling block is consisted of a 3 × 3 convolution operation with stride 2, a BN layer, a ReLU activation function and a pre‐activated residual block, 36 and makes channel numbers remain unchanged. The structure of the residual block and the pre‐activated residual block is explained in Figure 2. In E1 (Figure 1), the first convolution operation changes channel numbers from 1 to 64, and the latter two convolution operations only extract feature information without changing channel numbers. Similarly, in other layers from E2 to E5, the number of channels turns 128, 256, 512, and 1024. When the number of channels is 1024, the feature information is up‐sampling through the content‐aware sampling operation, 37 , 38 , 39 and then the feature in the down‐sampling path is connected with those in the up‐sampling path by jumping connection for supplementing feature information. From D4 to D1, the number of channels decreases to 512, 256, 128, and 64. When the number of channels is 64, 1 × 1 convolution operation and Sigmoid activation function are utilized to convert feature maps of final segmentation results into probability maps. Next, we will introduce the details of the network and loss function.
FIGURE 1.
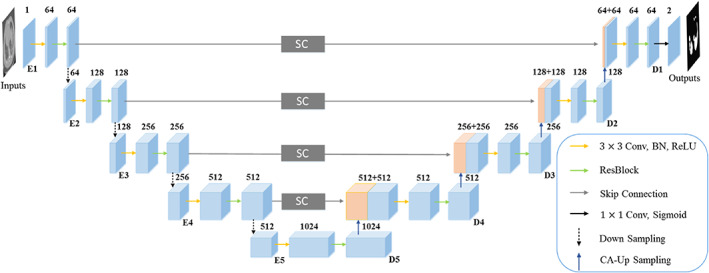
The network architecture of the proposed CAPA‐ResUNet.
FIGURE 2.
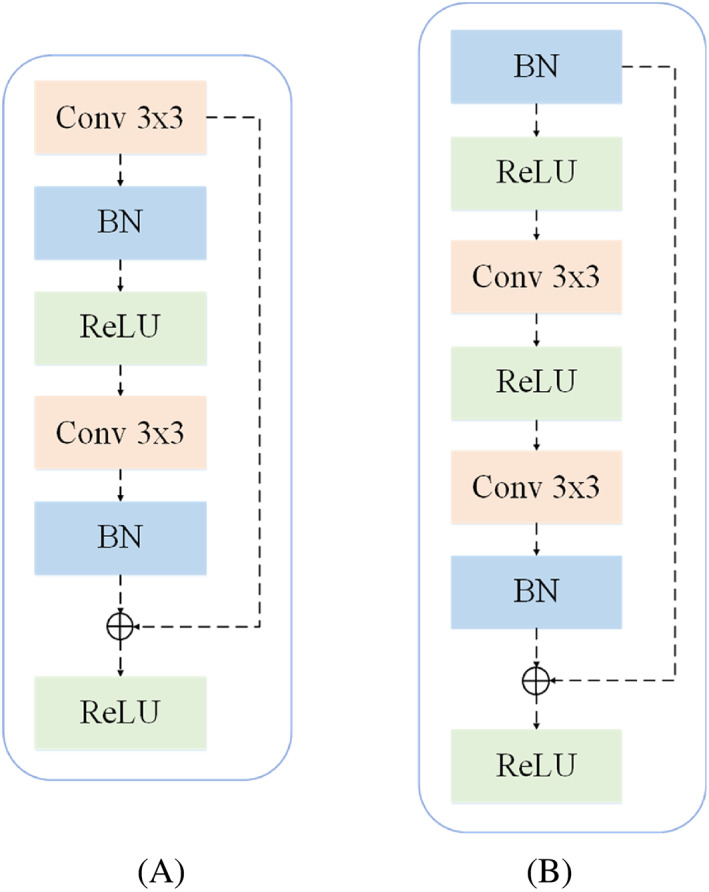
Modules illustration. (A) The structure of the residual block. (B) The structure of the proposed pre‐activated residual block. The dotted line on the right is residual connection.
2.2. Residual block
In our model, the residual block is applied to the encoder module to extract feature information, as shown in the residual block in Figures 1 and 2A, and explicated in Figure 2A. In the residual block, the first is two 3 × 3 convolution layers with equal output channel numbers, and each convolution operation is followed by a BN layer and a ReLU activation function. To alleviate the gradient disappearance problem, we introduce the residual connection 40 and append inputs to the final ReLU activation function.
Assuming that the input is , the ideal mapping that the model needs to fit is , and this mapping is an identity mapping. However, due to the effect of multiple nonlinear layers, it is difficult to directly fit the identity mapping. After adding the residual connection, the model output is . To make an identity mapping, we only need to set the weights and bias of multiple nonlinear layers to 0, that is, , and the mapping obtained at this time is an identity mapping. In the model training, the residual connection is easier to be optimized, 40 and the gradient disappearance problem is also alleviated.
2.3. Pre‐activated residual block
Since the foreground distribution of COVID‐19 lesion segmentation data is complex, for instance, shapes, sizes, and locations of lesions are different, which has a certain impact on the training results of the network. In addition, during the training, the changes of network parameters at each layer will result in changes in data distributions at the latter layer.
For that matter, inspired by Lu et al., 36 we designed the pre‐activated residual block by using BN layer, and applied the pre‐activated residual block to the down‐sampling block, as shown in Figure 2B. In a word, adding pre‐activated ResBlock before inputting at each layer, and then enter for learning. Inputs of each layer are preceded by a pre‐activated residual block, which is then input for learning.
2.4. Content‐aware up‐sampling module
Feature sampling is an important operation in image processing. At present, there are three main up‐sampling methods. The first one is a bilinear interpolation, but it only considers the adjacent sub‐voxel space, it cannot obtain sufficient semantic information. The second is deconvolution, which extends dimensions by utilizing convolution layers. However, deconvolution uses the same convolution for the whole image, limiting the capture of local information changes. It not only increases the parameters but also reduces the computational efficiency. The third is the feature up‐sampling operator content‐aware reassembly of features (CARAFE), 37 which is mainly divided into two parts, namely, kernel prediction module and content‐aware reassembly module. First, the kernel prediction module is used to predict the sampling convolution kernel, and then take advantage of the content‐aware reassembly module completing the sampling and obtaining the feature maps. In this article, we adopt the third method above for up sampling (Algorithm 1). The specific steps are as follows:
ALGORITHM 1. The content‐aware up‐sampling algorithm.
Given the feature map with the shape of and the up‐sampling rate ;
- Generate the reassembly kernels (Kernel Prediction Module).
- The channels numbers of the feature map are compressed from to by convolution;
- The compressed feature map is used to convert the number of channels from to by convolution of , and then expanded in the spatial dimension to obtain the up‐sampling kernel of .
- Normalize the up‐sampling kernel by the Softmax function to make their weights add up to 1.
-
Reassemble the features within a local region (Content‐aware Reassembly Module).
For each position of the output feature maps, map it to the input feature maps, and then select a neighborhood and the upper sampling kernel as the inner product to get new feature maps.
Output the final feature map.
2.5. Loss function
In the early stage of COVID‐19 infection, the proportion of lesion area in the whole image is small, which may lead to the prediction results of the network being more biased towards the background. Inspired by Taghanaki et al., 30 we have effectively overcome this problem by using similar combo loss forms to balance the proportion of foreground and background.
| (1) |
where is the total number of voxels in the images. is the probability set of prediction voxels, is the probability set of corresponding label voxels.
Another important problem is that the boundaries of COVID‐19 lesion regions are fuzzy and locations are scattered and irregular, which makes the prediction results of the network more prone to misclassification. To solve this problem, we construct a loss function based on the area of misclassified regions. As shown in Figure 3, and are misclassified regions, and is the correct segmented region. Taking as an example, the purpose is to continuously reduce the area of enclosed by and . We use the idea of converting curve integral into double integral to construct the loss function of this part, as shown in Equation (2).
| (2) |
FIGURE 3.
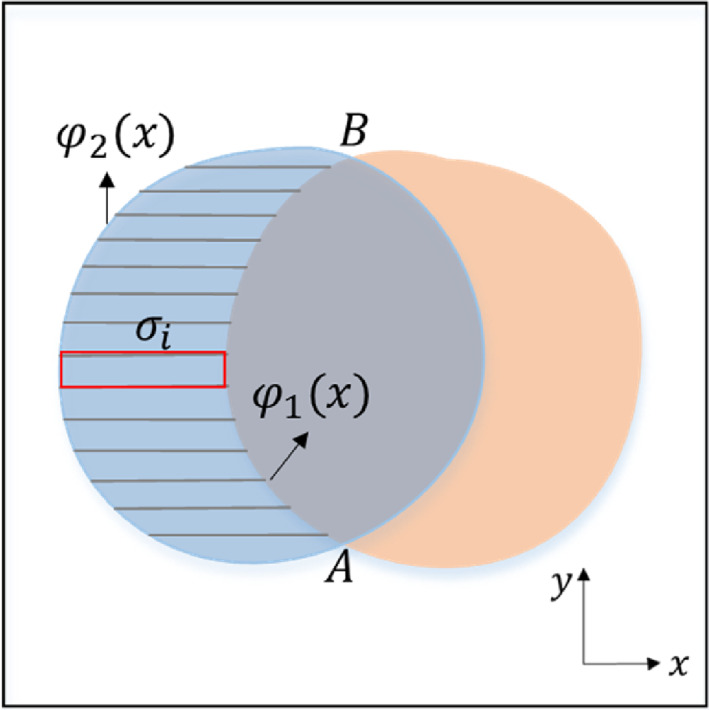
The proposed loss function schematic diagram. and are masks, and are predictions. The area of and are calculated by using the concept of mathematical calculus. Our purpose is to continuously reduce the area of and .
Since the integral form is difficult to realize and optimize in network training, we intend to discrete Equation (2). First, the region is divided into rectangles, and then the area of rectangles is summed to obtain the approximate value of the area of the region . To simplify the calculation, we further zoom it into the product of the maximum distance between and and the distance from point A to point B, as shown in Equation (3).
| (3) |
where and are vertical coordinates of point A and point B, respectively, and is the area of the red rectangle in Figure 3. Finally, the loss function based on the misclassified area is shown in Equation (4).
| (4) |
Finally, the total loss function in this article is the linear combination of and , as shown in Equation (5). Experimental results demonstrate that the linear combination of and is effective to train model.
| (5) |
where denotes the weight of and .
3. EXPERIMENTS AND RESULTS
3.1. Datasets
The public dataset 41 was used in our experiment, which is the COVID‐19 Lung CT Lesion Segmentation Challenge—2020 42 dataset, in which the CT data were provided by The Multi‐national NIH Consortium through the NCI TCIA public website. 43 All of the CT data were positive for SARS‐CoV‐2 RT‐PCR, and the COVID‐19 lesions were labeled by the gold standard.
Since the challenge has ended, we only got part of the dataset, namely, we got a total of 13 595 CT slices. In this article, the dataset was grouped into the train set and the test set according to the ratio of 3:1. Specifically, the train set includes 10 876 CT slices and the test set contains 2719 CT slices. Before the training, we preprocessed the dataset, such as automatically cutting the image according to the boundary of the lung area and reshaping the image size into 256 × 256. In addition, we performed data augmentation by using the Transforms method in Pytorch, mainly including horizontal or vertical flip, random rotation 90°.
3.2. Training details
The experiments in this article are implemented using Pytorch 44 and a server with two NVIDIA PH402 GPUs 32 GB card. Ranger optimizer 29 was used to train the model. Ranger optimizer was the combination of RAdam 45 and Lookahead. 46 On the one hand, we used RAdam to reduce the variance at the beginning of training and alleviate the problem of easily converging to the local optimal solution. On the other hand, we enhanced training stability and decreased the training variance by using Lookahead.
Hyper‐parameters were tuned by performing five‐fold validation on the train set. All the control experiments used the same parameters. In our experiments, the number of training epochs was 60. Our proposed loss function was combined with the Ranger optimizer and weight decay 0.0001. The learning rate was initialized to and decayed 0.1 times at the 40th epoch, once every 5 epochs. Also, we used a batch size of 4. In our loss function, we set to 0.5.
3.3. Evaluation metrics
To assess the performance of all models, five different metrics were used, namely, dice coefficient (Dice), sensitivity (Sen), specificity (Spe), precision (PC), and intersection over union (IoU).
Dice: Dice measures the similarity between the model segmentation results and the ground truth. Dice is defined as,
| (6) |
-
2
Sen: Sen represents the proportion of the infected areas that are accurately predicted. Its definition is as follows,
| (7) |
-
3
Spe: Spe represents the proportion of non‐infected areas which are accurately predicted and is defined as,
| (8) |
-
4
PC: PC represents the probability of pulmonary infections which are accurately predicted, which is formulated as,
| (9) |
-
5
IoU: IoU is the overlapping areas divided by the joint areas between segmentation results and the ground truth. Its definition is as follows,
| (10) |
where denote the true positive, false positive, true negative, and false negative between segmentation results and ground truth, respectively.
3.4. Comparative experiments
To evaluate the effect of our model, some comparative experiments were carried out. Concretely, CAPA‐ResUNet was compared with various advanced deep learning models, including UNet, 9 SegNet, 11 CopleNet, 24 DDANet, 25 and CARes‐UNet. 27 All models were trained from the open‐source codes in the article. In our experiments, all networks shared the same dataset to ensure fairness.
Compared with the other models mentioned above, the influence of label data complexity on experiments was reduced by adding pre‐activated residual blocks in our network. And considering the expansion of the wrong regions due to blurring lesion boundary, the area loss function was proposed to reduce the area of the false positive regions. Experiment results of all models on the COVID‐19 Lung CT Lesion Segmentation Challenge—2020 dataset are shown in Table 2.
TABLE 2.
Segmentation results of different models in test set
| Model | Dice | Sen | Spe | PC | IoU |
|---|---|---|---|---|---|
| UNet | 0.743 | 0.471 | 0.971 | 0.526 | 0.609 |
| SegNet | 0.730 | 0.461 | 0.972 | 0.527 | 0.583 |
| CopleNet | 0.722 | 0.460 | 0.972 | 0.526 | 0.577 |
| DDANet | 0.723 | 0.482 | 0.97 | 0.529 | 0.573 |
| CARes‐UNet | 0.756 | 0.475 | 0.971 | 0.527 | 0.628 |
| CAPA‐ResUNet | 0.775 | 0.530 | 0.972 | 0.533 | 0.646 |
Note: All metrics are larger, the better the segmentation effect is. The best results are marked by boldface. CAPA‐ResUNet is the model proposed in this article.
Our model obtains the best results in all models, especially in Dice, Sen, Spe, PC, and IoU. Specifically, in terms of Dice, it is 2.51% higher than the second‐best model CARes‐UNet and 4.31% higher than the classic UNet model, while Dice was the significant evaluation index in image segmentation. Furthermore, while maintaining a high Spe, Sen also significantly increased by 11.6%. Compared with CARes‐UNet, the PC value of the model also increased from 0.527 to 0.533, which increased by 1.14%, and the IoU value of the model increased from 0.628 to 0.646, an increase of 2.87%. The effectiveness of the proposed model is fully proved.
Analysis of variance (ANOVA) based on Dice was calculated to test statistical differences between methods. When the p‐value is less than 0.05, it indicates a significant difference. The experimental results are shown in Table 3.
TABLE 3.
ANOVA on the dice showed statistical differences between segmentation methods
| ANOVA | F value | p‐value |
|---|---|---|
| Unet and CAPA‐ResUNet | 0.7172 | 4.4621E−08 |
| SegNet and CAPA‐ResUNet | 0.6771 | 2.1639E−09 |
| CopleNet and CAPA‐ResUNet | 0.3294 | 2.1499E−08 |
| DDANet and CAPA‐ResUNet | 0.1108 | 1.9966E−10 |
| CARes‐UNet and CAPA‐ResUNet | 0.1575 | 0.000199319 |
We selected CT images with different degrees of infection as presentations. The results of CT images segmentation are shown in Figure 4. It can be observed from Figure 4 that CAPA‐ResUNet was more accurate than any other model in the segmentation of COVID‐19 lesions.
FIGURE 4.
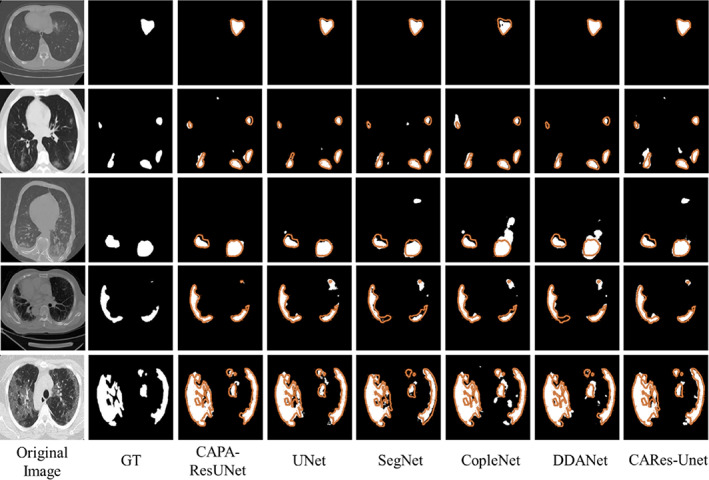
CT image segmentation results of all models. The first column is the original CT images, and the second column is their corresponding masks. Columns from the third to the last are predicted results of the different models. Orange outlines indicate ground truth contours.
First, for CT images with lighter COVID‐19 infection, the lesion area is relatively small. As shown in the first row of Figure 4, the segmentation results of CAPA‐ResUNet model are closer to ground truth, while the lesion areas segmented by UNet, SegNet, and CopleNet are smaller.
Second, for CT images with deep infection, the range of lesion area is expanded, the distribution is dispersed, and the shapes are complex and different, as shown in the CT images from the second to fourth rows in Figure 4. For the segmentation of such CT images, the results of CAPA‐ResUNet model show that the false‐positive and false‐negative regions are small, which are more accurate than those of other comparative models. In particular, the comparison of the image results of the third and fourth lines is the most obvious. The lesion regions segmented by SegNet, CopleNet, DDANet, and CARes‐Unet are much larger than those segmented by ground truth, and the lesion regions segmented by UNet are smaller than those segmented by ground truth.
Finally, for CT images with the deepest infection, as shown in the fifth row of CT images in Figure 4, most areas in the lungs have been infected. It can be seen from the segmentation results that the CAPA‐ResUNet model has better segmentation effect. In summary, it can be concluded from the segmented CT images that CAPA‐ResUNet model can better deal with CT images with different infection levels.
3.5. Ablation study
In this section, we performed a few experiments to verify the effectiveness of different components in our model, including pre‐activated residual block and area loss function based on the error segmentation regions. We showed the results in Figure 5. For the basic network, our network adopted the CARes‐UNet framework, using combo loss for training. As can be seen from Figure 5, all the proposed modules had great contributions to the performance of CARes‐UNet.
FIGURE 5.
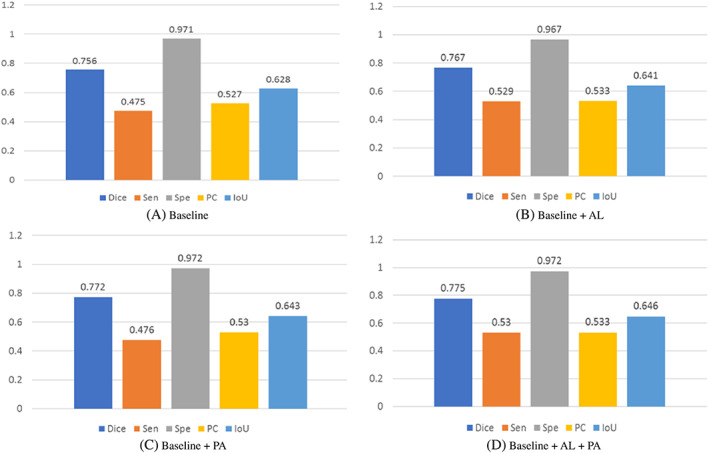
Experimental results of ablation studies. “Baseline,” namely CARes‐UNet, is trained with only the combo loss, and “AL” represents the area loss based on error segmentation regions. “PA” represents the pre‐activated residual block module. “Baseline + AL + PA” is our CAPA‐ResUNet.
3.5.1. Effectiveness of pre‐activated residual block
To study the effectiveness of pre‐activated residual blocks, we carried out two control experiments. One was the basic network CARes‐UNet whose down‐sampling is composed of the residual block, and the other was that the down‐sampling composed of pre‐activated residual blocks. Figure 5 shows the results of the two groups of comparative tests. The results show that the network performance using pre‐activated residual block for down‐sampling was better, and its Dice value increased by 2.11%.
3.5.2. Effectiveness of area loss function
To study the effectiveness of area loss function based on the error segmentation regions, two control experiments were also designed. One was the basic network CARes‐UNet, which was an adopted combo loss function. The other was also the basic network CARes‐UNet, and the loss function adopted the linear combination of the combo loss and area loss function based on error segmentation regions. The results are shown in Figure 5. The results indicate that the training effect of the latter was better than that of the former, and the Dice value was increased by 1.46%.
3.5.3. Effectiveness of module combination
At last, we studied the effectiveness of the combination of modules. As shown in Figure 5, the performance of the module combination was better than that of the corresponding version using a single module. It's shown that the module combination was valid, especially the Dice value was increased by 2.51% and the IoU value was increased by 2.87%.
3.6. Discussion on hyper‐parameters
Furthermore, we also studied the effect of hyper‐parameter on the predicted results, as shown in Table 4. The hyper‐parameter was set to 0.4, 0.45, 0.5, 0.55, 0.6, and 0.7 in turn, and six groups of experiments were carried out. As can be seen from Table 4, when λ was set to 0.5, the best result was achieved with Dice up to 0.767. This demonstrated that the combo loss and area loss based on error segmentation regions were equally important in the model training. That is to say, for the segmentation of the pulmonary infection area, we should not only consider the imbalance of foreground and background categories but also consider the fuzzy boundaries of lesion regions to obtain better segmentation results.
TABLE 4.
Experimental results of parameter adjustment
|
|
Dice | Sen | Spe | PC | IoU | |
|---|---|---|---|---|---|---|
|
|
0.751 | 0.476 | 0.97 | 0.53 | 0.618 | |
|
|
0.765 | 0.477 | 0.971 | 0.53 | 0.633 | |
|
|
0.767 | 0.529 | 0.967 | 0.533 | 0.641 | |
|
|
0.759 | 0.499 | 0.969 | 0.531 | 0.632 | |
|
|
0.757 | 0.512 | 0.968 | 0.532 | 0.630 | |
|
|
0.753 | 0.478 | 0.968 | 0.53 | 0.621 |
Note: The best results are in boldface.
4. CONCLUSION
In this article, we proposed a content‐aware pre‐activated residual network (CAPA‐ResUNet) to segment the pulmonary infection area of COVID‐19 from CT images. We reconstructed the CARes‐UNet network structure and added a pre‐activated residual block to solve problems of uneven sample distribution in datasets. We also proposed a loss function based on the area of misclassified regions and combined it with the combo loss function for training to deal with the problems of the large difference in the proportion of prospects in different infection periods and blurred boundary of lesion regions. To prove the effectiveness and accuracy of the proposed model, we conducted comparative studies and ablation experiments. Results showed that our method was superior to several other advanced baseline models.
Although the network structure and loss function of the model are improved considering the problems of complex foreground distribution and blurred lesion boundary in the data, the processing of some small lesion areas is not perfect enough. In the future, the multi‐dimensional extraction of image feature information can be attempted in the network structure, which may have a certain effect.
AUTHOR CONTRIBUTIONS
Lu Ma: Conceptualization, Methodology, Software, Formal analysis, Writing—Original Draft and Editing. Shuni Song: Formal analysis, Writing—Review and Editing, Funding acquisition. Liting Guo: Writing—Review and Editing. Wenjun Tan: Formal analysis, Writing—Review and Editing. Lisheng Xu: Formal analysis, Validation, Writing—Review and Editing, Funding acquisition.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 61971118, No. 61773110, and No. 11801065), the Natural Science Foundation of Liaoning Province (No. 20170540312 and No. 2021‐YGJC‐14), the Basic Scientific Research Project (Key Project) of Liaoning Provincial Department of Education (LJKZ00042021), and Fundamental Research Funds for the Central Universities (No. N2119008). This work was also supported by the Shenyang Science and Technology Plan Fund (No. 21‐104‐1‐24, No. 20‐201‐4‐10, and No. 201375), and the Member Program of Neusoft Research of Intelligent Healthcare Technology, Co. Ltd. (No. MCMP062002).
Ma L, Song S, Guo L, Tan W, Xu L. COVID‐19 lung infection segmentation from chest CT images based on CAPA‐ResUNet . Int J Imaging Syst Technol. 2023;33(1):6‐17. doi: 10.1002/ima.22819
Funding information Basic Scientific Research Project (Key Project) of Liaoning Provincial Department of Education, Grant/Award Number: LJKZ00042021; Fundamental Research Funds for the Central Universities, Grant/Award Number: N2119008; National Natural Science Foundation of China, Grant/Award Numbers: 61971118, 61773110, 11801065; Natural Science Foundation of Liaoning Province, Grant/Award Numbers: 20170540312, 2021‐YGJC‐14; Shenyang Science and Technology Plan Fund, Grant/Award Numbers: 21‐104‐1‐24, 20‐201‐4‐10, 201375; the Member Program of Neusoft Research of Intelligent Healthcare Technology, Co. Ltd., Grant/Award Number: MCMP062002
Contributor Information
Shuni Song, Email: [email protected].
Lisheng Xu, Email: [email protected].
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are openly available at https://covid-segmentation.grand-challenge.org/Download/.
REFERENCES
- 1. Lancet. 2020;395(10223):470‐473. [DOI] [PMC free article] [PubMed]
- 2. Organization WH . Weekly epidemiological update on COVID‐19. 2022. Accessed January 6, 2022. https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019/situation‐reports.
- 3. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing for coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32‐E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karam M, Althuwaikh S, Alazemi M, et al. Chest CT versus RT‐PCR for the detection of COVID‐19: systematic review and meta‐analysis of comparative studies. JRSM Open. 2021;12(5):20542704211011837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stefano A, Gioè M, Russo G, et al. Performance of radiomics features in the quantification of idiopathic pulmonary fibrosis from HRCT. Diagnostics. 2020;10(5):306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Subramaniam S, Jayanthi KB, Rajasekaran C, Kuchelar R. Deep learning architectures for medical image segmentation. 2020 IEEE 33rd International Symposium on Computer‐Based Medical Systems (CBMS); 2020:579‐584.
- 8. Wang EK, Chen C‐M, Hassan MM, Almogren AS. A deep learning based medical image segmentation technique in internet‐of‐medical‐things domain. Future Gener Comput Syst. 2020;108:135‐144. [Google Scholar]
- 9. Ronneberger OFP, Brox T. U‐Net: convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer‐Assisted Intervention; 2015:234‐241; Springer International Publishing.
- 10. Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: a nested U‐Net architecture for medical image segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support. 2018;11045:3‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder‐decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481‐2495. [DOI] [PubMed] [Google Scholar]
- 12. Zhang K, Liu X, Shen J, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID‐19 pneumonia using computed tomography. Cell. 2020;182(5):1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi F, Wang J, Shi J, et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID‐19. IEEE Rev Biomed Eng. 2021;14:4‐15. [DOI] [PubMed] [Google Scholar]
- 14. Gao K, Su J, Jiang Z, et al. Dual‐branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID‐19 using CT images. Med Image Anal. 2021;67:101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pezzano G, Diaz O, Ripoll VR, Radeva P. CoLe‐CNN+: context learning—convolutional neural network for COVID‐19‐ground‐glass‐opacities detection and segmentation. Comput Biol Med. 2021;136:104689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu J, Zhang S, Li X, et al. Joint segmentation and detection of COVID‐19 via a sequential region generation network. Pattern Recognit. 2021;118:108006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie F, Huang Z, Shi Z, et al. DUDA‐Net: a double U‐shaped dilated attention network for automatic infection area segmentation in COVID‐19 lung CT images. Int J Comput Assist Radiol Surg. 2021;16(9):1425‐1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bizopoulos PA, Vretos N, Daras P. Comprehensive comparison of deep learning models for lung and COVID‐19 lesion segmentation in CT scans. ArXiv. 2020;abs/2009.06412.
- 19. Pei HY, Yang D, Liu GR, Lu T. MPS‐Net: multi‐point supervised network for CT image segmentation of COVID‐19. IEEE Access. 2021;9:47144‐47153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X, Wang G, Zhao SG. COVSeg‐NET: a deep convolution neural network for COVID‐19 lung CT image segmentation. Int J Imaging Syst Technol. 2021;31(3):1071‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu F, Zhu Y, Qin X, Xin Y, Yang D, Xu T. A multi‐class COVID‐19 segmentation network with pyramid attention and edge loss in CT images. IET Image Process. 2021;15:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saeedizadeh N, Minaee S, Kafieh R, Yazdani S, Sonka M. COVID TV‐Unet: segmenting COVID‐19 chest CT images using connectivity imposed Unet. Comput Methods Programs Biomed Update. 2021;1:100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao X, Zhang P, Song F, et al. D2A U‐Net: automatic segmentation of COVID‐19 CT slices based on dual attention and hybrid dilated convolution. Comput Biol Med. 2021;135:104526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang G, Liu X, Li C, et al. A noise‐robust framework for automatic segmentation of COVID‐19 pneumonia lesions from CT images. IEEE Trans Med Imaging. 2020;39(8):2653‐2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rajamani KT, Siebert H, Heinrich MP. Dynamic deformable attention network (DDANet) for COVID‐19 lesions semantic segmentation. J Biomed Inform. 2021;119:103816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stefano A, Comelli A. Customized efficient neural network for COVID‐19 infected region identification in CT images. J Imaging. 2021;7(8):131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu X, Wen Y, Zhao L, et al. CARes‐UNet: content‐aware residual UNet for lesion segmentation of COVID‐19 from chest CT images. Med Phys. 2021;48(11):7127‐7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu S, Zhu Z, Gorriz JM, Wang SH, Zhang YD. NAGNN: classification of COVID‐19 based on neighboring aware representation from deep graph neural network. Int J Intell Syst. 2022;37(2):1572‐1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J, Dong B, Wang S, et al. COVID‐19 lung infection segmentation with a novel two‐stage cross‐domain transfer learning framework. Med Image Anal. 2021;74:102205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taghanaki SA, Zheng Y, Kevin Zhou S, et al. Combo loss: handling input and output imbalance in multi‐organ segmentation. Comput Med Imaging Graph. 2019;75:24‐33. [DOI] [PubMed] [Google Scholar]
- 31. Oktay O, Schlemper J, Folgoc LL, et al. Attention U‐Net: learning where to look for the pancreas. ArXiv. 2018;abs/1804.03999.
- 32. Chen LCE, Zhu YK, Papandreou G, Schroff F, Adam H. Encoder‐decoder with atrous separable convolution for semantic image segmentation. Lecture Notes in Computer Science; 2018;11211: 833‐851.
- 33. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Proceedings of Machine Learning Research; 2015: 37, 448‐456.
- 34. Eckle K, Schmidt‐Hieber J. A comparison of deep networks with ReLU activation function and linear spline‐type methods. Neural Netw. 2019;110:232‐242. [DOI] [PubMed] [Google Scholar]
- 35. Yi Y, Zhang Z, Zhang W, Zhang C, Li W, Zhao T. Semantic segmentation of urban buildings from VHR remote sensing imagery using a deep convolutional neural network. Remote Sens. 2019;11:1774. [Google Scholar]
- 36. Lu S, Wang SH, Zhang YD. Detection of abnormal brain in MRI via improved AlexNet and ELM optimized by chaotic bat algorithm. Neural Comput Appl. 2021;33(17):10799‐10811. [Google Scholar]
- 37. Wang J, Chen K, Xu R, Liu Z, Loy CC, Lin D. CARAFE: Content‐Aware ReAssembly of FEatures. IEEE/CVF International Conference on Computer Vision (ICCV) 2019: 3007‐3016.
- 38. Wang J, Chen K, Xu R, Liu Z, Loy CC, Lin D. CARAFE++: Unified Content‐Aware. ReAssembly of FEatures. IEEE Trans Pattern Anal Mach Intell. 2021;44:4674‐4687. [DOI] [PubMed] [Google Scholar]
- 39. Wang Q, Wu B, Zhu P, Li P, Zuo W, Hu Q. ECA‐Net: efficient channel attention for deep convolutional neural networks. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020:11531‐11539.
- 40. Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception‐v4, inception‐ResNet and the impact of residual connections on learning. Proceedings of the Thirty‐First AAAI Conference on Artificial Intelligence; San Francisco, CA; 4278‐4284.
- 41. An PXS, Harmon SA, Turkbey EB, et al. CT images in Covid‐19 [dataset]. Cancer Imaging Archive; 2020.
- 42. Roth H, Xu Z, Diez CT, et al. Rapid artificial intelligence solutions in a pandemic—the COVID‐19‐20 lung CT lesion segmentation challenge. Res Sq. 2021;82:102605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clark K, Vendt B, Smith K, et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paszke A, Gross S, Massa F, et al. Pytorch: an imperative style, high‐performance deep learning library. Advances in Neural Information Processing Systems 2019;32:8026‐8037. [Google Scholar]
- 45. Liu L, Jiang H, He P, et al. On the variance of the adaptive learning rate and beyond. ArXiv. 2020;abs/1908.03265.
- 46. Zhang MR, Lucas J, Hinton GE, Ba J. Lookahead optimizer: k steps forward, 1 step back 2019. arXiv preprint arXiv:1907.08610.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are openly available at https://covid-segmentation.grand-challenge.org/Download/.


