Abstract
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults. The standard treatment for GBM consists of surgical resection followed by concurrent chemoradiotherapy and adjuvant temozolomide. O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status is an important prognostic biomarker that predicts the response to temozolomide and guides treatment decisions. At present, the only reliable way to determine MGMT promoter methylation status is through the analysis of tumor tissues. Considering the complications of the tissue-based methods, an imaging-based approach is preferred. This study aimed to compare three different deep learning-based approaches for predicting MGMT promoter methylation status. We obtained 576 T2WI with their corresponding tumor masks, and MGMT promoter methylation status from, The Brain Tumor Segmentation (BraTS) 2021 datasets. We developed three different models: voxel-wise, slice-wise, and whole-brain. For voxel-wise classification, methylated and unmethylated MGMT tumor masks were made into 1 and 2 with 0 background, respectively. We converted each T2WI into 32 × 32 × 32 patches. We trained a 3D-Vnet model for tumor segmentation. After inference, we constructed the whole brain volume based on the patch’s coordinates. The final prediction of MGMT methylation status was made by majority voting between the predicted voxel values of the biggest connected component. For slice-wise classification, we trained an object detection model for tumor detection and MGMT methylation status prediction, then for final prediction, we used majority voting. For the whole-brain approach, we trained a 3D Densenet121 for prediction. Whole-brain, slice-wise, and voxel-wise, accuracy was 65.42% (SD 3.97%), 61.37% (SD 1.48%), and 56.84% (SD 4.38%), respectively.
Keywords: Deep learning, Classification, MGMT, BraTS, Brain tumor
Introduction
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults. In the USA, the median age of diagnosis is 64 years old, and its incidence is rising in the elderly population [1, 2]. The disease is associated with high mortality, morbidity, and disability levels with patients having a median survival of 16 months [3, 4]. The most effective treatment for GBM consists of surgery (aiming for the maximum possible tumor resection) followed by concurrent chemoradiation therapy (CCRT) [5, 6]. In this treatment protocol, temozolomide (TMZ) is administered concurrently with radiotherapy, followed by a number of adjuvant TMZ therapy cycles. This is an effective regimen and has shown an increase in 24-month survival from 10.4 to 26.5% [6].
TMZ is an alkylating agent which causes DNA damage eventually resulting in cell cycle arrest and cell death [7]. However, the O-6-alkylguanine DNA alkyltransferase encoded by the O-6-methylguanine-DNA methyltransferase (MGMT) gene can reverse the DNA damage [8]. Therefore, the MGMT gene expression level can regulate the TMZ therapeutic efficacy on GBM. A hypermethylated MGMT gene promoter region suppresses its expression and results in a lack of MGMT-mediated DNA repair. Hence, cells with methylated MGMT gene promoters are more sensitive to TMZ, leading to the higher effectiveness of this treatment. On the other hand, in malignant cells, the high levels of MGMT activity (low methylation level) blunts the therapeutic effect of alkylating agents, resulting in a resistant phenotype and a significant determinant of treatment failure [9]. For this reason, determining the MGMT promoter methylation status in patients with GBM is a fundamental step to predict patients’ response to therapy.
The current reference standard for assessing the methylation status of MGMT is the epigenetic analysis based on samples obtained from fine-needle aspirations or resected tissue [10]. This conventional invasive tissue-demanding method has major limitations, including the need for large-size tissue samples and the possibility of incomplete biopsy samples due to the spatial heterogeneity of tumors, although this is still debated [11–13]. Several studies have demonstrated that genetic changes can manifest themselves as macroscopic changes that can be detectable by magnetic resonance imaging (MRI) [14, 15]. Different imaging features have been employed to predict MGMT promoter methylation; however, experts have not reached a consensus on the use of features such as tumor texture, size, and location for this classification task [16, 17]. The use of imaging to predict MGMT promoter methylation status can potentially decrease the need for tissue biopsy before resective surgery. It can also result in a less aggressive surgical approach to patients with poor prognosis or tumors near the eloquent cortex. Finally, there are occasional cases where MGMT results cannot be obtained (e.g., inadequate tissue), and having the image-based prediction can help in clinical decision-making.
Numerous research groups evaluated the use of MRI features and machine learning algorithms to predict the status of several gliomas’ molecular markers [14, 18], including the MGMT promoter methylation [15, 19–28]. Most of these studies used data derived either from single centers or publicly available, multi-institutional data provided by The Cancer Imaging Archive (TCIA) and The Cancer Genome Atlas (TCGA) [29, 30]. As proof of the growing interest in trying to predict the MGMT promoter methylation status using MRI, the organizers of the 2021 Brain Tumor Segmentation (BraTS) challenge [31–33] defined it as one of the aims of the competition and released a publicly available dataset of 585 brain MRIs acquired from subjects with GBM including the information regarding the MGMT promoter methylation status [34].
Among the radiogenomics studies focusing on MGMT, only a few studies used the deep learning approach [20, 21, 23, 24].
In this study, we have compared three different deep learning approaches to predict the MGMT promoter methylation status of GBMs from the 2021 BraTS dataset.
Method
Dataset
The BraTS 2021 dataset is a collection of conventional brain MRI sequences of subjects with brain tumors collected at multiple institutions with different acquisition protocols, including (a) native (T1) and (b) post-contrast T1-weighted (T1Gd (gadolinium)), (c) T2-weighted (T2), and (d) T2 fluid attenuated inversion recovery (T2-FLAIR) volumes. The challenge was organized in two tasks: task 1, which focused on brain tumor segmentation, and task 2, focused on the radiogenomic classification of the MGMT promoter methylation. Task 1 dataset contains MRI already skull stripped, co-registered to the same anatomical template, interpolated to the same resolution (1 mm × 1 mm × 1 mm), and with the same size (240, 240, 155). Additionally, the data also include brain tumor segmentation consisting of four masks with different voxel values: 0 for the background, 1 for the necrotic tumor core, 2 for the peritumoral edema/invaded tissue, and 4 for the contrast-enhancing area. Task 2 dataset includes skull stripped, non-co-registered MRIs, and the information on the MGMT promoter methylation status stored as a binary variable. Since we utilized the segmentation masks for voxel-based classification, we used the MRI volumes of task 1 with their corresponding MGMT promoter methylation status from task 2 for the subjects who were present in both cohorts.
We opted to use only T2 images, given that they are routinely acquired, have fairly consistent technique over time and across institutions, and do not rely on the injection of a contrast agent. Another reason is that by using only T2 images, we can compare our results with other studies with acceptable outcomes on other datasets [21, 22].
Dataset Splitting
We split the dataset at the subject level into 5 folds stratified by MGMT status using the Scikit-learn library [35]. We used the same split for all the experiments.
Data Preprocessing and Model Development
We implemented three different approaches, including whole-brain, slice-wise, and voxel-wise approaches, to represent classification, object detection, and segmentation techniques, respectively. For each approach, we discuss the data preprocessing pipeline and model development.
Whole-Brain Approach
In the whole-brain approach, we utilized the whole brain MRI volume as the input to the model.
Data Preprocessing
We standardized T2 intensities to have zero mean and unit standard deviation. To mitigate the mildly imbalanced training dataset, we oversampled the underrepresented class in a 1.1:1 MGMT methylated:MGMT unmethylated ratio [36]. We performed data augmentation using vertical and horizontal flip, rotation, translation, scaling, and Gaussian noise to decrease the risk of overfitting.
Model Development
We initially evaluated several different 3D neural net architectures, including 3D-EfficientNet b0, 3D-EfficientNet b1, 3D-EfficientNet b2, 3D-DenseNet 121, and 3D-DenseNet 169 from the MONAI models package [37]. We found that a 3D-DenseNet121 model performed best during preliminary testing (results not included) and subsequently used only this architecture. We optimized the model hyperparameters using grid search (Table 1). We trained the model for 50 epochs and selected the model weights with the highest accuracy on the validation of each fold. For each fold, we trained three models with different random weight initializations and used majority voting as the model ensemble strategy for final predictions [38].
Table 1.
Optimized object detection model hyperparameters
| Hyperparameter | Value |
|---|---|
| Batch size | 16 |
| Optimizer | Adam |
| Learning rate | 0.0001 |
| First momentum | 0.9 |
| Second momentum | 0.999 |
| Random image rotation (x, y, z, probability) | (15, 15, 12, 0.5) |
| Random image translation (x, y, z, probability) | (15, 15, 5, 0.5) |
| Random image scaling (x, y, z, probability) | (0.05, 0.05, 0.05, 0.5) |
| Random Gaussian noise (mean, standard deviation, probability) | (0, 0.5, 0.2) |
| Random vertical flip (probability) | 0.5 |
| Random horizontal flip (probability) | 0.5 |
For each model in each fold, we reported accuracy, the area under the receiver operating curve (AUCROC), sensitivity, and specificity. Mean value and standard deviation of accuracy, AUCROC, sensitivity, and specificity across the folds are reported.
Slice-Wise Approach
In the slice-wise approach, we used each brain MRI slice as individual input to the model and then obtained a subject-level prediction based on majority voting of the slice predictions for that subject.
Data Preprocessing
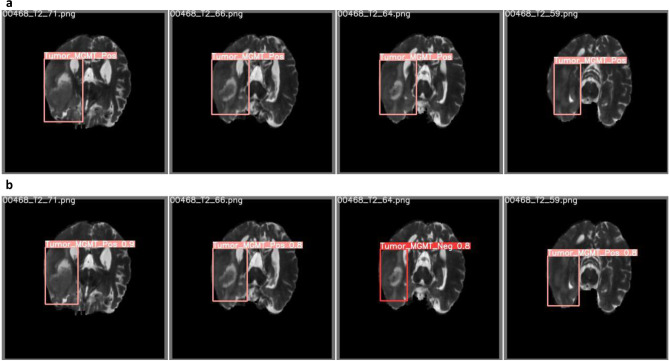
We defined a bounding box around the tumor area for each MRI slice using the segmentation masks as reference: we chose the minimum X and Y coordinates of each segmented area as the X and Y coordinates of the top-left point of the bounding box and the maximum X and Y coordinates as the bottom-right point. We then labeled each bounding box as either methylated or unmethylated based on the subject’s MGMT promoter methylation status. Finally, we converted each slice to 512 × 512 pixels portable graphics format (PNG) (Fig. 1a).
Fig. 1.
T2-weighted brain MRI with bounding boxes and corresponding label. a Ground truth bounding boxes and labels. b Predicted bounding boxes and labels. MGMT, O-6-methylguanine-DNA methyltransferase
Model Development
We initially evaluated different object detection models including YOLOv5s, YOLOv5m, YOLOv5l, and YOLOv5x [39]. We found that a YOLOv5x model performed best during preliminary testing and subsequently used only this architecture. YOLOv5x is pre-trained on the COCO dataset, and the weights were transferred. We trained the model for 50 epochs and selected the model weights with the highest mean average precision (mAP) for each fold on the validation set. Mean average precision is a common performance metric for object detection with 0.5 as the threshold for the intersection over union (mAP0.5). For inference on the test set, we used test time augmentation, an intersection over a union threshold of 0.3, and a confidence level of 0.3, which was determined through optimization during preliminary testing. During inference, the model labels each MRI slice based on the class with the highest confidence. We then obtain subject-level predictions through majority voting of slice labels (Fig. 1b). For each fold, we reported accuracy, AUCROC, sensitivity, and specificity. We also reported the mean value and standard deviation of accuracy, AUCROC, sensitivity, and specificity across the folds [40].
Voxel-Wise Approach
In the voxel-wise approach, we developed our model based on a previously published method [21] in which each tumor voxel is classified as being MGMT methylated or not methylated using as initial inputs patches derived from the whole T2-weighted MRI volume (“T2”).
Data Preprocessing
T2 images were normalized to have zero mean and unit standard deviation. For each subject, we combined the segmentation masks of different tumoral regions (necrotic core, enhancing area, and edema) in a single mask containing the whole lesion. We then assigned each mask a single value based on the MGMT promoter methylation status: one for methylated and two for not methylated tumors [21]. Finally, we divided the T2s and the corresponding masks into patches of size 32 × 32 × 32, with 8 voxels overlapping in each dimension. During training, we only included patches with at least one tumor voxel. However, for inference, all patches, regardless of tumor presence, were fed to the model. We performed data augmentation using flipping, random rotation (± 15°), and zooming (± 0.1) as implemented in the MONAI package [41].
Model Development
A 3D-Vnet was used to segment image patches into corresponding label classes [42]. The encoder had depth of five with 16, 32, 64, 128, and 256 features and two residual units in each level [41]. The network was designed to get a 32 × 32 × 32 patch as input and output a 32 × 32 × 32 volume with three channels: one for background, one for tumor areas showing MGMT promoter methylation, and one for tumor areas showing unmethylated MGMT promoter methylation. We used softmax as the final layer’s activation function. We trained this model with random weight initialization, using Dice-loss for the cost function and a learning rate of 0.00001 for 100 epochs. We used Adam as the network’s optimizer [43]. We used early stopping to terminate training when the validation loss did not improve for more than 10 epochs.
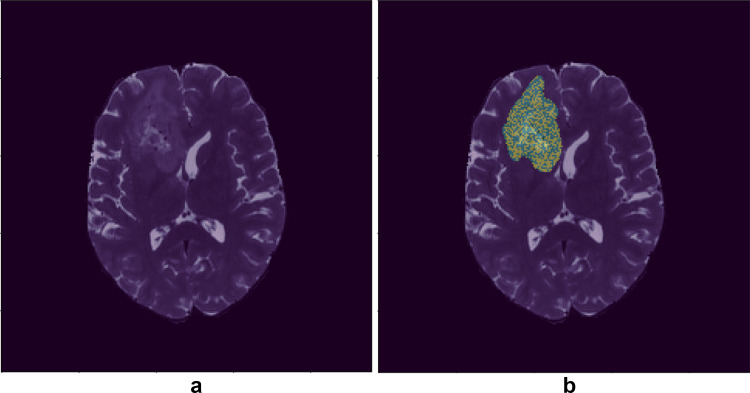
In order to evaluate the final model’s performance, we applied the model to all patches of the validation sets and then combined all the patches of a given subject to get the mask for the whole volume. We applied a threshold of 0.5 on the model’s output to binarize our masks. Finally, the largest connected component was extracted, and the label that made the majority of the component was selected as the label for the whole image (Fig. 2). For each fold, we reported accuracy, AUCROC, sensitivity, and specificity. We also reported the mean value and standard deviation of accuracy, AUCROC, sensitivity, and specificity across the folds [40].
Fig. 2.
T2-weighted brain MRI with and without tumor’s mask. a Brain MRI without tumor mask. b Brain MRI with tumor mask, each mask’s voxel belongs to MGMT methylated or MGMT nonmethylated class. MGMT, O-6-methylguanine-DNA methyltransferase
Results
We identified a total of 576 subjects as the intersection of task 1 and task 2 BraTS datasets: 275 (47.8%) with unmethylated and 301 (52.2%) with methylated MGMT promoter. Since the BraTS dataset consists of deidentified MRIs, the demographic description is not available. After splitting the dataset in 5 folds, folds one to four had 115 subjects, and fold five 116. This resulted in 35,838 unique bounding boxes around the segmentation masks for the slice-wise approach and 1,690,983 3D patches for the voxel-wise approach.
Results of Different Deep Learning Approaches
Whole-Brain Approach
We used majority voting to determine the model performance for the ensemble of results. Using the ensemble method, the mean accuracy among 5 folds was 65.42% (SD 3.97%), the mean AUCROC was 0.6508 (SD 0.03), the mean sensitivity was 71.27% (SD 13%), and the mean specificity was 58.91% (SD 10.06%). Table 2 shows the accuracy, AUCROC, sensitivity, and specificity of each model and each fold.
Table 2.
Results of fivefold cross-validation for the whole-brain approach in discriminating tumors with methylated and unmethylated MGMT promoter. For each fold, we trained three models with different weights initialization and then ensemble. MGMT, O-6-methylguanine-DNA methyltransferase; AUCROC, area under receiver operative characteristic curve
| Model | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | |
|---|---|---|---|---|---|---|
| Accuracy | 1 | 63.79% | 61.20% | 61.40% | 61.40% | 62.93% |
| 2 | 61.20% | 65.51% | 62.28% | 55.26% | 58.62% | |
| 3 | 63.79% | 65.51% | 59.64% | 60.52% | 66.37% | |
| AUCROC | 1 | 0.6360 | 0.5980 | 0.6147 | 0.6052 | 0.6135 |
| 2 | 0.6132 | 0.6569 | 0.6244 | 0.5517 | 0.5877 | |
| 3 | 0.6387 | 0.6461 | 0.6052 | 0.6013 | 0.6597 | |
| Sensitivity | 1 | 67.21% | 86.89% | 59.32% | 35.59% | 91.80% |
| 2 | 59.02% | 62.30% | 57.63% | 57.63% | 55.74% | |
| 3 | 62.30% | 81.97% | 35.59% | 71.19% | 73.77% | |
| Specificity | 1 | 60.00% | 32.73% | 63.64% | 85.45% | 30.91% |
| 2 | 63.64% | 69.09% | 67.27% | 52.73% | 61.82% | |
| 3 | 65.45% | 47.27% | 85.45% | 49.09% | 58.18% |
Slice-Wise Approach
The slice-wise approach achieved 56.84% (SD 4.38%) mean accuracy for all 5 folds, 0.5683 (SD 4.39%) mean AUCROC, 0.5613 (SD 0.07) mean sensitivity, 55.48% (SD 3.95%) mean specificity, and 0.4822 (SD 3.41) mAP. Table 3 summarizes the results for each fold.
Table 3.
Slice-wise approach discrimination among two classes (methylated MGMT promoter vs. unmethylated MGMT promoter) for each fold. MGMT, O-6-methylguanine-DNA methyltransferase; AUCROC, area under receiver operative characteristic curve
| Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | |
|---|---|---|---|---|---|
| Accuracy | 62.06% | 53.91% | 57.89% | 50.00% | 60.34% |
| AUCROC | 0.6236 | 0.5426 | 0.5764 | 0.5023 | 0.5971 |
| Sensitivity | 57.23% | 54.42% | 58.91% | 49.89% | 60.23% |
| Specificity | 52.34% | 51.38% | 59.43% | 53.21% | 61.04% |
| mAP 0.5 | 50.46% | 46.58% | 51.76% | 42.31% | 49.99% |
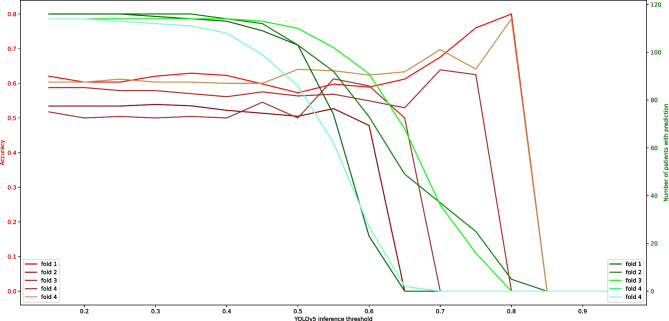
Figure 3 shows the correlation between the YOLO inference threshold and the patient-level accuracy of the model prediction and the total number of patients with the assigned label. On the one hand, by increasing the threshold, fewer labels will be assigned to each patient and, eventually, none for some patients. However, higher confidence will deliver higher accuracy.
Fig. 3.
Correlation between the YOLO inference threshold and the patient-level accuracy of the model prediction and the total number of patients with the assigned label for each fold. YOLO, You Only Look Once
Voxel-Wise Approach
For the voxel-wise approach, the mean accuracy of all 5 folds was 61.37% (SD 1.48%), mean AUCROC was 0.6160 (SD 0.01), mean sensitivity was 65.46% (SD 9.85%), and mean specificity was 51.57% (SD 10.79%). Table 4 summarizes the performance metrics of each fold.
Table 4.
Voxel-wise approach discrimination among two classes (methylated MGMT promoter vs. unmethylated MGMT promoter) for each fold. MGMT, O-6-methylguanine-DNA methyltransferase; AUCROC, area under receiver operative characteristic curve
| Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | |
|---|---|---|---|---|---|
| Accuracy | 62.02% | 63.71% | 61.33% | 59.23% | 60.60% |
| AUCROC | 0.6222 | 0.6314 | 0.6130 | 0.6056 | 0.6078 |
| Sensitivity | 58.23% | 67.30% | 59.22% | 57.96% | 84.62% |
| Specificity | 52.20% | 49.73% | 64.02% | 59.40% | 32.54% |
Discussion
In this study, we aimed to compare three different deep learning approaches to predict the MGMT promoter methylation status in glioblastomas using T2 brain MRIs obtained from the 2021 RSNA-ASNR-MICCAI BraTS challenge dataset [31, 32, 44]. Overall, our results show that the accuracy and AUCROC of the whole-brain approach are higher than those of the voxel-wise approach, and the voxel-wise approach is superior to the slice-wise approach. In light of the fact that the whole-brain and voxel-wise inputs are 3D volumes, the spatial relation between voxels could contain related information and potentially explain the superiority of 3D approaches over the slice-wise method. Considering that the whole-brain approach processes information simultaneously across the brain, it may explain why this approach has higher performance metrics.
Currently, the only way to obtain the MGMT promoter methylation status is to analyze the tumor tissue after a tumor biopsy or resection. Being able to obtain the same information using MRI non-invasively would fundamentally impact clinical practice. For this reason, predicting the MGMT promoter methylation status has been one of the most common tasks in the radiogenomics literature, using both conventional and advanced MRI sequences, with mixed results [15, 19–28, 45, 46].
There are published studies showing promising results in classifying MGMT promoter methylation status on publicly available datasets like the TCIA [20, 21, 24, 47, 48], as well as in private datasets [15, 19, 22, 24–27], and in a combination of public and private datasets [28]. Notably, one study did not find any significant relationship between the MGMT promoter methylation status and histological and radiological features [27]. Nevertheless, the authors limited the analysis of radiological features to tumoral volumes and growth rate as measured using MRI, without performing any radiomics analysis.
Radiomics feature analysis is one of the popular methods for predicting genomic and epigenomic characteristics of GBMs [48–53]. With the use of data characterization algorithms, the authors extracted a large number of features from medical images and performed different methods of feature analysis to predict MGMT methylation status. Jiang et al. reported an 89% accuracy and an AUC of 0.898 (0.786–1.000) on 35 low-grade gliomas from the TCIA validation dataset [50]. Lu et al. combined radiomics features with clinical information and achieved an accuracy of a maximum of 67% on 181 patients from their own center’s private dataset [52]. Hajianfar et al. achieved an AUC of 0.71 on 82 patients from their own centers [51]. Le et al. developed a classifier using XGBoost and by the use of radiomics features of 53 GBM, isocitrate dehydrogenase-1 (IDH) wildtype patients from the TCIA dataset [48]. They achieved an AUC of 0.896. Finally, a study published by Calabrese et al. [53], on 199 GBM patients, found no correlation between MGMT promoter methylation status and any radiomics features. They made their code and radiomics feature data publicly available for further evaluation of other researchers. However, we decided to use deep learning-based models that need no feature engineering techniques.
In this work, besides whole-brain and slice-wise approaches, we implemented the voxel-wise approach with some modifications in architecture and activation functions. We tried to adopt the main backbone of the voxel-wise method and train, validate, and compare to other approaches on the BraTS dataset [21]. While Yogananda et al. used a pre-trained model on a task of IDH mutation status prediction, we did not use the pre-trained model since the IDH mutation status of the patients was not available for the BraTS dataset [21]. In comparison with the Yogananda et al. study done on TCIA T2 images, results on BraTS are not as strong. Also results on the leaderboard of the RSNA-MICCAI brain tumor radiogenomic classification competition in Kaggle, on the BraTS dataset, reveal 62% as the best model’s accuracy. Other studies on TCIA or private datasets achieve higher performance, using T2 MRI [21, 22]. Using only T2 images is a limitation of our study; however, we decided to use T2 images to have a fair comparison to previous studies.
Our findings are in line with other radiogenomics studies performed on the BraTS 2021 datasets. In a recently released preprint article, the authors conducted a large number of experiments with different deep learning architectures and techniques, including 2D and 3D convolutional neural network models, vision transformers, and self-supervision using the BraTS 2021 dataset. They reported that all the experiments yielded an AUCROC that is not significantly better than a random guess for a binary classification problem. They concluded that with the current dataset achieving a medically applicable model is not feasible [45]. Another team proposed a method for MGMT methylation prediction on the BraTS 2021 dataset in a preprint article that combines the use of radiomics with high-level shape features learned by a variational autoencoder. They obtained tumor segmentation by training a segmentation model and then trained a variational autoencoder on masks to learn the tumor’s high-level shape features. Using these features, they trained a random forest classifier. They obtained a validation score (AUCROC) of 0.598 [46].
One hypothesis for poor results on the BraTS dataset with all different models is that the provided MGMT promoter methylation status was determined based on varying methods across the multiple institutions that contributed data, and each institute follows its own methodology [31, 54]. As a result, the labels are noisy due to the heterogeneity of labeling methods. Another hypothesis for poor results on the BraTS dataset is that the outcomes on this multi-institutional data are closer to a generalizable model for MGMT promoter methylation status prediction. In other words, the performance on the BraTS dataset is more reflective of real-world performance than other datasets. While that may be true, it is important to recognize that the BraTS dataset MRIs have already been pre-processed, which limits our options when performing image processing. For example, respacing the images is suboptimal without the original data. This limitation can be solved by sharing the original images.
While the TCIA and BraTS datasets contain only conventional MRI sequences, some authors have explored the possibility to predict the MGMT promoter methylation status including features derived from advanced MRI techniques [15, 25, 26]. Kickingereder et al. [15] tested a series of machine learning models to predict the status of different molecular markers of GBMs, including MGMT, and found that none of the imaging features was associated with the MGMT promoter methylation status. Kihira et al. [25] assessed the advantage of adding diffusion MRI to conventional MRI to predict several molecular markers in gliomas. They reported the highest incremental value for the prediction of MGMT from an AUCROC of 0.64 when using only conventional MRI, to 0.79 when adding diffusion features. Interestingly, the performance they obtained using only conventional MRI is similar to the one we report in our study.
Finally, some groups included only GBMs [15], while others included different tumor grades [21, 23, 25, 26]. While some of these studies reported the distribution of the methylation status across tumor grades [21, 25, 26], others did not [23], making it possible for the model to learn to discriminate grades rather than the MGMT methylation status if, for example, this is unevenly distributed between tumor grades. Given the variable description of the cohorts among different studies, at the moment it is unclear if including different tumor grades might have influenced the models’ performance, and further investigation is needed.
A limitation of our study is that we developed our models using only T2 images. While it is possible that adding more MR sequences could improve the results [55], our main objective in this study was to compare our approaches to others that used only T2 sequences.
In conclusion, we found that the accuracy of MGMT methylation status prediction using the BraTS dataset is inferior to other datasets, and across the whole-brain, slice-wise, and voxel-wise deep learning approaches, the whole-brain approach is the most effective approach for MGMT methylation status prediction on the BraTS dataset.
Acknowledgements
This project was supported in part by funding from the Mayo Clinic Center for Individualized Medicine (CIM).
Abbreviations
- BraTS
Brain Tumor Segmentation
- CCR
Concurrent chemoradiation therapy
- TMZ
Temozolomide
- GBM
Glioblastoma
- MGMT
O-6-methylguanine-DNA methyltransferase
- AUCROC
Area under the receiver operating characteristic curve
- TCIA
The Cancer Imaging Archive
- TCGA
The Cancer Genome Atlas
Code Availability
A link to the code will be shared after peer review.
Data Availability
The raw data required to reproduce the above findings are available to download from http://braintumorsegmentation.org/.
Declarations
Competing Interests
The authors declare no competing interests.
Footnotes
Gian Marco Conte and Bradley J. Erickson are co-senior authors.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tamimi AF, Juweid M. Epidemiology and outcome of glioblastoma. In: De Vleeschouwer S, editor. Glioblastoma. Brisbane (AU): Codon Publications; 2017. [PubMed]
- 2.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20: iv1–iv86. [DOI] [PMC free article] [PubMed]
- 3.Egaña L, Auzmendi-Iriarte J, Andermatten J, Villanua J, Ruiz I, Elua-Pinin A, et al. Methylation of MGMT promoter does not predict response to temozolomide in patients with glioblastoma in Donostia Hospital. Sci Rep. 2020;10:18445. doi: 10.1038/s41598-020-75477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tesileanu CMS, Dirven L, Wijnenga MMJ, Koekkoek JAF, Vincent AJPE, Dubbink HJ, et al. Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: a confirmation of the cIMPACT-NOW criteria. Neuro Oncol. 2020;22:515–523. doi: 10.1093/neuonc/noz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 6.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 7.Hombach-Klonisch S, Mehrpour M, Shojaei S, Harlos C, Pitz M, Hamai A, et al. Glioblastoma and chemoresistance to alkylating agents: involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol Ther. 2018;184:13–41. doi: 10.1016/j.pharmthera.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Vlachostergios PJ, Hatzidaki E, Befani CD, Liakos P, Papandreou CN. Bortezomib overcomes MGMT-related resistance of glioblastoma cell lines to temozolomide in a schedule-dependent manner. Invest New Drugs. 2013;31:1169–1181. doi: 10.1007/s10637-013-9968-1. [DOI] [PubMed] [Google Scholar]
- 9.Hegi ME, Diserens A-C, Gorlia T, Hamou M-F, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 10.Tierling S, Jürgens-Wemheuer WM, Leismann A, Becker-Kettern J, Scherer M, Wrede A, et al. Bisulfite profiling of the MGMT promoter and comparison with routine testing in glioblastoma diagnostics. Clin Epigenetics. 2022;14:26. doi: 10.1186/s13148-022-01244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigliadori G, Goffredo G, Bartolini D, Tosatto L, Gurrieri L, Mercatali L, et al. Influence of intratumor heterogeneity on the predictivity of MGMT gene promoter methylation status in glioblastoma. Front Oncol. 2020;10:533000. doi: 10.3389/fonc.2020.533000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamilton MG, Roldán G, Magliocco A, McIntyre JB, Parney I, Easaw JC. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol. 2011;102:255–260. doi: 10.1007/s11060-010-0307-5. [DOI] [PubMed] [Google Scholar]
- 13.Grasbon-Frodl EM, Kreth FW, Ruiter M, Schnell O, Bise K, Felsberg J, et al. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer. 2007;121:2458–2464. doi: 10.1002/ijc.23020. [DOI] [PubMed] [Google Scholar]
- 14.Singh G, Manjila S, Sakla N, True A, Wardeh AH, Beig N, et al. Radiomics and radiogenomics in gliomas: a contemporary update. Br J Cancer. 2021; 1–17. [DOI] [PMC free article] [PubMed]
- 15.Kickingereder P, Bonekamp D, Nowosielski M, Kratz A, Sill M, Burth S, et al. Radiogenomics of glioblastoma: machine learning-based classification of molecular characteristics by using multiparametric and multiregional MR imaging features. Radiology. 2016;281:907–918. doi: 10.1148/radiol.2016161382. [DOI] [PubMed] [Google Scholar]
- 16.Drabycz S, Roldán G, de Robles P, Adler D, McIntyre JB, Magliocco AM, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage. 2010;49:1398–1405. doi: 10.1016/j.neuroimage.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 17.Ellingson BM, Cloughesy TF, Pope WB, Zaw TM, Phillips H, Lalezari S, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. Neuroimage. 2012;59:908–916. doi: 10.1016/j.neuroimage.2011.09.076. [DOI] [PubMed] [Google Scholar]
- 18.Fathi Kazerooni A, Bakas S, Saligheh Rad H, Davatzikos C. Imaging signatures of glioblastoma molecular characteristics: a radiogenomics review. J Magn Reson Imaging. 2020;52:54–69. doi: 10.1002/jmri.26907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xi Y-B, Guo F, Xu Z-L, Li C, Wei W, Tian P, et al. Radiomics signature: a potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J Magn Reson Imaging. 2018;47:1380–1387. doi: 10.1002/jmri.25860. [DOI] [PubMed] [Google Scholar]
- 20.Han L, Kamdar MR. MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Biocomputing 2018. WORLD SCIENTIFIC; 2017. pp. 331–342. [PMC free article] [PubMed]
- 21.Yogananda CGB, Shah BR, Nalawade SS, Murugesan GK, Yu FF, Pinho MC, et al. MRI-based deep-learning method for determining glioma MGMT promoter methylation status. AJNR Am J Neuroradiol. 2021;42:845–852. doi: 10.3174/ajnr.A7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korfiatis P, Kline TL, Coufalova L, Lachance DH, Parney IF, Carter RE, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med Phys. 2016;43:2835–2844. doi: 10.1118/1.4948668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang P, Grinband J, Weinberg BD, Bardis M, Khy M, Cadena G, et al. Deep-learning convolutional neural networks accurately classify genetic mutations in gliomas. AJNR Am J Neuroradiol. 2018;39:1201–1207. doi: 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korfiatis P, Kline TL, Lachance DH, Parney IF, Buckner JC, Erickson BJ. Residual deep convolutional neural network predicts MGMT methylation status. J Digit Imaging. 2017;30:622–628. doi: 10.1007/s10278-017-0009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kihira S, Tsankova NM, Bauer A, Sakai Y, Mahmoudi K, Zubizarreta N, et al. Multiparametric MRI texture analysis in prediction of glioma biomarker status: added value of MR diffusion. Neurooncol Adv. 2021;3: vdab051. [DOI] [PMC free article] [PubMed]
- 26.Wei J, Yang G, Hao X, Gu D, Tan Y, Wang X, et al. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur Radiol. 2019;29:877–888. doi: 10.1007/s00330-018-5575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikkelsen VE, Dai HY, Stensjøen AL, Berntsen EM, Salvesen Ø, Solheim O, et al. MGMT promoter methylation status is not related to histological or radiological features in IDH wild-type glioblastomas. J Neuropathol Exp Neurol. 2020;79:855–862. doi: 10.1093/jnen/nlaa060. [DOI] [PubMed] [Google Scholar]
- 28.Li Z-C, Bai H, Sun Q, Li Q, Liu L, Zou Y, et al. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: a multicentre study. Eur Radiol. 2018;28:3640–3650. doi: 10.1007/s00330-017-5302-1. [DOI] [PubMed] [Google Scholar]
- 29.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45: 1113–1120. [DOI] [PMC free article] [PubMed]
- 31.Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv [cs.CV]. 2021. Available: http://arxiv.org/abs/2107.02314
- 32.Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) IEEE Trans Med Imaging. 2015;34:1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, et al. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4:170117. doi: 10.1038/sdata.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.RSNA-MICCAI Brain Tumor Radiogenomic Classification. [cited 8 Apr 2022]. Available: https://www.kaggle.com/competitions/rsna-miccai-brain-tumor-radiogenomic-classification/overview
- 35.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 36.Rouzrokh P, Khosravi B, Faghani S, Moassefi M, Vera Garcia DV, Singh Y, et al. Mitigating bias in radiology machine learning: 1. Data handling. Radiology: Artificial Intelligence. 2022;4:e210290. doi: 10.1148/ryai.210290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MONAI Consortium. MONAI: Medical Open Network for AI. Zenodo; 2022.
- 38.Zhang K, Khosravi B, Vahdati S, Faghani S, Nugen F, Rassoulinejad-Mousavi SM, et al. Mitigating bias in radiology machine learning: 2. Model development. Radiology: Artificial Intelligence. 2022;4:e220010. doi: 10.1148/ryai.220010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jocher G, Stoken A, Borovec J, NanoCode, ChristopherSTAN, Changyu L, et al. ultralytics/yolov5: v3.1 - Bug Fixes and Performance Improvements. 2020.
- 40.Faghani S, Khosravi B, Zhang K, Moassefi M, Jagtap JM, Nugen F, et al. Mitigating bias in radiology machine learning: 3. Performance metrics. Radiology: Artificial Intelligence. 2022; e220061. [DOI] [PMC free article] [PubMed]
- 41.The MONAI Consortium. Project MONAI. 2020. 10.5281/zenodo.4323059
- 42.Milletari F, Navab N, Ahmadi S-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. arXiv [cs.CV]. 2016. Available: http://arxiv.org/abs/1606.04797
- 43.Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv [cs.LG]. 2014. Available: http://arxiv.org/abs/1412.6980
- 44.Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv [cs.CV]. 2018. Available: http://arxiv.org/abs/1811.02629
- 45.Saeed N, Hardan S, Abutalip K, Yaqub M. Is it Possible to Predict MGMT Promoter Methylation from Brain Tumor MRI Scans Using Deep Learning Models? arXiv [eess.IV]. 2022. Available: http://arxiv.org/abs/2201.06086
- 46.Pálsson S, Cerri S, Van Leemput K. Prediction of MGMT Methylation Status of Glioblastoma Using Radiomics and Latent Space Shape Features. arXiv [eess.IV]. 2021. Available: http://arxiv.org/abs/2109.12339
- 47.Kanas VG, Zacharaki EI, Thomas GA, Zinn PO, Megalooikonomou V, Colen RR. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput Methods Programs Biomed. 2017;140:249–257. doi: 10.1016/j.cmpb.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 48.Le NQK, Do DT, Chiu F-Y, Yapp EKY, Yeh H-Y, Chen C-Y. XGBoost improves classification of MGMT promoter methylation status in IDH1 wildtype glioblastoma. J Pers Med. 2020;10. 10.3390/jpm10030128 [DOI] [PMC free article] [PubMed]
- 49.Crisi G, Filice S. Predicting MGMT promoter methylation of glioblastoma from dynamic susceptibility contrast perfusion: a radiomic approach. J Neuroimaging. 2020;30:458–462. doi: 10.1111/jon.12724. [DOI] [PubMed] [Google Scholar]
- 50.Jiang C, Kong Z, Liu S, Feng S, Zhang Y, Zhu R, et al. Fusion radiomics features from conventional MRI predict MGMT promoter methylation status in lower grade gliomas. Eur J Radiol. 2019;121:108714. doi: 10.1016/j.ejrad.2019.108714. [DOI] [PubMed] [Google Scholar]
- 51.Hajianfar G, Shiri I, Maleki H, Oveisi N, Haghparast A, Abdollahi H, et al. Noninvasive O6 methylguanine-DNA methyltransferase status prediction in glioblastoma multiforme cancer using magnetic resonance imaging radiomics features: univariate and multivariate radiogenomics analysis. World Neurosurg. 2019;132:e140–e161. doi: 10.1016/j.wneu.2019.08.232. [DOI] [PubMed] [Google Scholar]
- 52.Lu Y, Patel M, Natarajan K, Ughratdar I, Sanghera P, Jena R, et al. Machine learning-based radiomic, clinical and semantic feature analysis for predicting overall survival and MGMT promoter methylation status in patients with glioblastoma. Magn Reson Imaging. 2020;74:161–170. doi: 10.1016/j.mri.2020.09.017. [DOI] [PubMed] [Google Scholar]
- 53.Calabrese E, Villanueva-Meyer JE, Cha S. A fully automated artificial intelligence method for non-invasive, imaging-based identification of genetic alterations in glioblastomas. Sci Rep. 2020;10:11852. doi: 10.1038/s41598-020-68857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malmström A, Łysiak M, Kristensen BW, Hovey E, Henriksson R, Söderkvist P. Do we really know who has an MGMT methylated glioma? Results of an international survey regarding use of MGMT analyses for glioma. Neurooncol Pract. 2020;7:68–76. doi: 10.1093/nop/npz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calabrese E, Rudie JD, Rauschecker AM, Villanueva-Meyer JE, Clarke JL, Solomon DA, et al. Combining radiomics and deep convolutional neural network features from preoperative MRI for predicting clinically relevant genetic biomarkers in glioblastoma. Neurooncol Adv. 2022;4: vdac060. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A link to the code will be shared after peer review.
The raw data required to reproduce the above findings are available to download from http://braintumorsegmentation.org/.