Abstract
Objective
Clinical trials are an essential part of the effort to find safe and effective prevention and treatment for COVID-19. Given the rapid growth of COVID-19 clinical trials, there is an urgent need for a better clinical trial information retrieval tool that supports searching by specifying criteria, including both eligibility criteria and structured trial information.
Materials and Methods
We built a linked graph for registered COVID-19 clinical trials: the COVID-19 Trial Graph, to facilitate retrieval of clinical trials. Natural language processing tools were leveraged to extract and normalize the clinical trial information from both their eligibility criteria free texts and structured information from ClinicalTrials.gov. We linked the extracted data using the COVID-19 Trial Graph and imported it to a graph database, which supports both querying and visualization. We evaluated trial graph using case queries and graph embedding.
Results
The graph currently (as of October 5, 2020) contains 3392 registered COVID-19 clinical trials, with 17 480 nodes and 65 236 relationships. Manual evaluation of case queries found high precision and recall scores on retrieving relevant clinical trials searching from both eligibility criteria and trial-structured information. We observed clustering in clinical trials via graph embedding, which also showed superiority over the baseline (0.870 vs 0.820) in evaluating whether a trial can complete its recruitment successfully.
Conclusions
The COVID-19 Trial Graph is a novel representation of clinical trials that allows diverse search queries and provides a graph-based visualization of COVID-19 clinical trials. High-dimensional vectors mapped by graph embedding for clinical trials would be potentially beneficial for many downstream applications, such as trial end recruitment status prediction and trial similarity comparison. Our methodology also is generalizable to other clinical trials.
Keywords: clinical trial, COVID-19, eligibility criteria, graph representation
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic is an ongoing global pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). As of September 15, 2020, more than 29 million cases had been reported worldwide, resulting in over 900 000 deaths. The United States alone has reported 6.5 million cases, with more than 194 000 deaths.1 Unfortunately, as COVID-19 is very new, there are no drugs or other therapeutics approved by the US Food and Drug Administration so far to treat or prevent COVID-19.2
As a part of essential efforts to identify safe and effective approaches for the prevention of and treatment for COVID-19, clinical trials research, communities are accelerating efforts in response to this pandemic. As of 09-22-2020, more than 3300 clinical trials have been registered at ClinicalTrials.gov for COVID-19. More than half are actively recruiting participants.3 A registered clinical trial typically contains both structured information (eg, recruitment status, study type, intervention/treatment) and unstructured information, including eligibility criteria. Given the high volume and rapid growth of COVID-19 clinical trials, it is critical to have an integrated repository to link clinical trial information to facilitate better trial searching, matching, and accrual.
Existing systems (eg, clinical trials registry,4 ClinicalTrials.gov) to track and manage COVID-19 trials focus more on the retrieval of structural information (eg, title, target condition or disease, study types). Eligibility criteria, which define the subject cohort in each clinical study in the free-text format, however, is often not available for searching in these existing systems. Although ClinicalTrials.gov offers advanced search functions (eg, https://clinicaltrials.gov/ct2/results/refine?cond=COVID-19) for clinical trials, most of its searching abilities focus on structured information. Due to the free-text nature of eligibility criteria, ClinicalTrials.gov provides only very limited search functionality on eligibility criteria, only including age, sex, and whether it accepts healthy volunteers.3 Other important information in the cohort definition, such as existing conditions of potential participants (eg, diabetes, pregnancy), use of prior medications, and lab tests/measurement, are not searchable in ClinicalTrials.gov.
In this study, we built the COVID-19 Trial Graph, a graph-based clinical trial data repository, to link structured and unstructured (ie, eligibility criteria) information for existing registered COVID-19 clinical trials. The COVID-19 Trial Graph supports diverse search queries with a particular focus on eligibility criteria and provides a graph-based visualization of COVID-19 clinical trials. In addition, as one of the first efforts, we trained a clinical trial graph embedding on the COVID-19 Trial Graph. We evaluated the trained trial embedding through visualization and one supervised machine learning task: prediction of trial end recruitment status.
MATERIALS AND METHODS
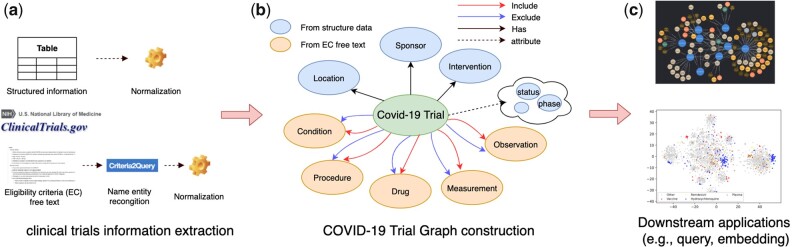
The overview of the study design can be seen in Figure 1. For each COVID-19 clinical trial collected from ClinicalTrials.gov, we first leveraged natural language processing (NLP) tools to extract and normalize structured and unstructured (ie, eligibility criteria) information. Then, we linked the extracted data and entities to construct the COVID-19 Trial Graph. As a part of the evaluation, we further tested some case queries and evaluated the precision and recall of the results. We also trained a clinical trial graph embedding, which can be used to represent clinical trials for downstream applications.
Figure 1.
System workflow for the COVID-19 Trial Graph
COVID-19 clinical trials information extraction
We collected structured data from COVID-19 relevant clinical trials from ClinicalTrials.gov on September 22, 2020. Data included National Clinical Trial ID, title, intervention, study type, location, phase, sponsor, outcome measures, etc. For the intervention target, we manually normalized the most frequent terms (top 300) to their full names, for example, “hcq,” “hydroxychloroquine (hcq),” and “hydroxychloroquine sulfate 200 mg” was mapped to “hydroxychloroquine.” For the location, we leveraged Google Map API to extract the country/region information. For unstructured data in clinical trials eligibility criteria free text, Criteria2query,5 a hybrid information extraction pipeline for parsing eligibility criteria text, was then adopted to extract a variety of named entities from both inclusion and exclusion criteria, including Condition, Drug, Measurement, Procedure, and Observation. All of the extracted entities from eligibility criteria text were then mapped to the OMOP Common Data Model, which is a popular standardized data representation for observational data.6 Specifically, we indexed all the standard concept names and corresponding synonyms of OMOP standard vocabulary using Apache Lucene. Then we performed BM25 algorithm to retrieve candidate concepts from the Lucene index and leverage text processing and rule-based concept reranking functions of CLAMP7 to map to the final standard concept ID.
Linked graph construction and evaluation
Figure 1(b) shows the meta-data level design of the COVID-19 Trial Graph. There are 9 types of concepts (ie, nodes) defined in the graph, including a concept for the clinical trial, 5 concepts for eligibility criteria, and 3 concepts for structured trial information. The 9 types of nodes as well as their statistics can be seen in Table 1. All other relevant information (eg, status, study type, phase) were added as attributes for the clinical trial concept. For each clinical trial and eligibility criteria concept pair, 2 relationships, including both inclusion and exclusion, were defined, depending on where the pair appears. The full list of relationships as well as the statistics can be seen in Table 2. We leveraged Neo4j, a leading graph database, to import all of the graph data and host the COVID-19 Trial Graph.8 Cypher is a declarative graph query language that is supported by Neo4j.9 We designed several Cypher case queries and manually checked the return results and calculated the precision and recall of these queries. The graph data files and Cypher case queries are publically available at https://github.com/UT-Tao-group/clinical_trial_graph.
Table 1.
Node types and the number of unqiue entities in the COVID-19 Trial Graph
| Clinical Trial | Sponsor | Intervention | Location | Condition | Drug | Measurement | Procedure | Observation |
|---|---|---|---|---|---|---|---|---|
| 3392 | 3585 | 3167 | 114 | 3524 | 1754 | 1171 | 538 | 235 |
Table 2.
Relationship types and the number of unqiue relationships in the COVID-19 Trial Graph
| Condition |
Drug |
Measurement |
Observation |
Procedure |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Has location | Has sponsor | Has intervention | Include | Exclude | Include | Exclude | Include | Exclude | Include | Exclude | Include | Exclude |
| 2977 | 6604 | 4148 | 10 304 | 18 912 | 1454 | 6547 | 4014 | 3691 | 797 | 795 | 2328 | 2665 |
Clinical trial embedding
Embedding was first proposed for NLP tasks that map words or phrases in vocabulary to vectors of real numbers based on word–co. co-occurrences, so as to enable the mathematical computation between text pieces.10 Similarly, graph embedding refers to a set of algorithms that transforms graphs or elements in graphs to vectors of real numbers by capturing the topology and node-to-node relationships in the graphs, to facilitate a series of downstream computational operations. In this study, we leveraged node2vec to learn the node representations in the COVID-19 Trial Graph. Node2vec algorithms adopt a random-walk sampling strategy to learn the representation for nodes by optimizing a neighborhood-preserving objective.11,12 We empirically set the embedding dimension as 100. The walk length was set at 10, and the number of walks was set at 2000. The number of iterations was set at 1. We implemented the algorithm from Grover and Leskovec.13
To visualize the trial graph embedding, we first reduced the embedding dimension to 50, using principal component analysis and then further reduced the dimensions to 2, using t-Distributed Stochastic Neighbor Embedding (t-SNE). t-SNE is a machine learning-based algorithm for dimension reduction and has been widely adopted for high-dimensional embedding visualization for biomedical concepts.14,15 The perplexity of t-SNE was set at 50, and the number of iterations was set at 10 000. We implemented t-SNE using the code available at Ulyanov.16
Clinical trial status prediction
To further test whether the learned clinical trial graph embedding can convey critical trial information, we conducted a pilot experiment to evaluate the efficacy of trial graph embedding in the prediction of trial status (ie, whether a trial can complete recruitment successfully). We excluded ongoing clinical trials with incomplete recruiting statuses (eg, status “Not yet recruiting,” “Recruiting,” “Enrolling by invitation,” “Active, not recruiting”) or those where the status is unknown in ClinicalTrials.gov. Based on the recruitment status provided, we split the trials into 2 groups: (1) completed, including the recruitment status “Completed” in ClinicalTrials.gov; and (2) stopped, including the recruitment status “Terminated” (ie, the study stopped early and will not start again), “Withdrawn” (ie, the study stopped early, before enrolling its first participant), and “Suspended” (ie, the study stopped early but may start again). We took Clinical Trial nodes embeddings from the whole trial graph embeddings as the input for each clinical trial and evaluated several machine-learning algorithms to predict the recruitment status from the embedding vectors directly, including logistic regression, extra trees, support vector machines, random forest, and gradient boosting. Ten-fold cross-validations were conducted, and the average accuracy was reported.
RESULTS
A total of 3392 clinical trials from 114 countries or regions that targeted COVID-19 were collected from ClinicalTrial.gov. The COVID-19 Trial Graph currently contains 17 480 nodes in total, with 9 types and 65 236 relationships with 13 types. The statistics of nodes and relationships can be seen in Tables 1 and 2, respectively. Condition is the most prevalent concept extracted from the eligibility criteria of COVID-19-related clinical trials, and Condition concepts appeared more in exclusion criteria texts than in inclusion criteria texts.
Case query evaluation
We designed the following example queries to evaluate whether the COVID-19 Trial Graph can support a diverse search ability across structured information and eligibility criteria. The queries ranged from easy to complicated, depending on the involvement of criteria. As the COVID-19 Trial Graph is hosted on a Neo4j database, we implemented these queries using the Cypher graph query language. We used a hybrid method including keywords search followed by manual review to identify relevant clinical trials to be considered as ground truth for each case query (the details are available in the Supplementary Material S1) and then compared the manually reviewed results with clinical trials retrieved by the queries. The precision and recall scores were calculated based on these gold standards. Table 3 shows precision and recall scores for each query. As we can observe, these case queries have high scores in both precision and recall.
Table 3.
Case query evaluation results
| Query | Total eligible clinical trialsa | Number of retrieved clinical trials | Precision (True positive) | Recall (False negative) |
|---|---|---|---|---|
| Case query 1 | 24 | 24 | 100% (24) | 100% (0) |
| Case query 2 | 11 | 11 | 100% (11) | 100% (0) |
| Case query 3 | 21 | 18 | 88.8% (16) | 76.2% (5) |
| Case query 4 | 10 | 10 | 90% (9) | 90% (1) |
These eligible trials were identified using keywords search followed by manual review.
Case query 1: Retrieve all COVID-19 clinical trials that target “remdesivir” as the intervention.
Case query 2: Retrieve all COVID-19 clinical trials that target “remdesivir” as the intervention but exclude pregnant women [OMOP ID: 4299535] from participating.
Case query 3: Retrieve all COVID-19 clinical trials that target “hydroxychloroquine” as the intervention and allow patients with shortness of breath [OMOP ID: 312437] to participate.
Case query 4: Retrieve all COVID-19 clinical trials in the United States that target “hydroxychloroquine” as the intervention and allow patients with diabetes to participate.
The errors were mainly caused by false negative of the NLP models (ie, not able to identify and normalize specific clinical terms from the eligibility criteria texts). For example, there were 5 false negatives for case query 3. Three of them included phrases like “difficult to breath” or “respiratory distress” in their eligibility criteria texts. However, the existing NLP models were not able to map these phrases to “shortness of breath” (Dyspnea in OMOP).
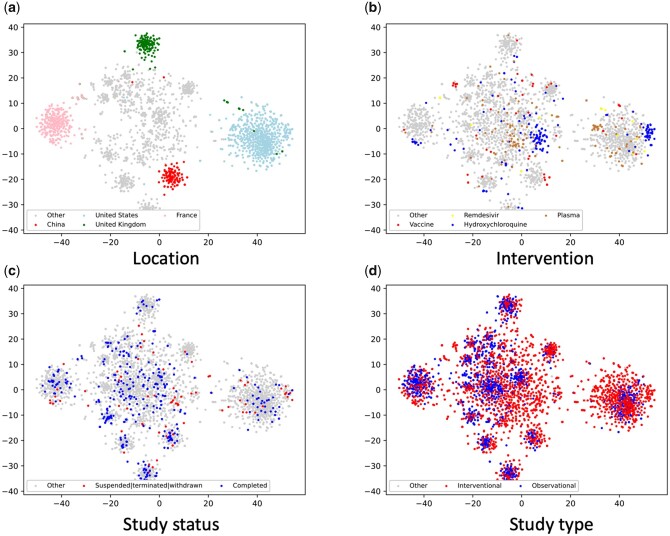
Results for clinical trial embedding
Figure 2 shows the graph embedding of the COVID-19 clinical trials using the first and second components of t-SNE. We directly visualized the embedding by a variety of clinical trial properties, and there is a strong clustering of clinical trials based on locations. The United States has conducted the largest number of COVID-19 clinical trials with an isolated group on the embedding map. We also identified isolated groups for France, the United Kingdom, and China, respectively. There is also clustering for the intervention target. For example, we observed that some clinical trials related to hydroxychloroquine and plasma treatment were clustered. Clustering also can be observed for study status (ie, recruitment status) and study types (ie, interventional versus observational).
Figure 2.
COVID-19 Trial Graph embedding graphs
Recruitment status prediction
In our experiments, we categorized 355 clinical trials to the “completed” group, and 78 trials to the “stopped” group, respectively, based on their status reported at Clinicaltrial.gov. The accuracy of machine-learning algorithms in predicting recruitment status can be seen in Table 4. As this dataset is highly-unbalanced, we defined a baseline accuracy when the classifier assigns a “completed” label to a trial in every prediction, which is 0.820. The embedding generated from the COVID-19 Trial Graph has been found effective in predicting the recruitment status for almost all of the algorithms. Support vector machine with radial basis function kernel achieved the highest accuracy at 0.870 with the COVID-19 Trial Graph embedding, a 5-point increase compared with baseline accuracy. We performed a comparison between graph-embedding vectors and random initialized vectors. The highest accuracy generated by machine-learning algorithms for random vectors was 0.820 (+/−0.018), which is the same as the baseline accuracy.
Table 4.
Comparison of recruitment status prediction using graph embedding and random vector
| Algorithm | Graph embedding | Random vector |
|---|---|---|
| LR | 0.829 (+/−0.103) | 0.781 (+/−0.064) |
| ET | 0.843 (+/−0.059) | 0.820 (+/−0.018) |
| SVM (RBF) | 0.870 (+/−0.087) | 0.799 (+/−0.031) |
| RF | 0.852 (+/−0.078) | 0.820 (+/−0.018) |
| GB | 0.843 (+/−0.093) | 0.802 (+/−0.053) |
Note: Average accuracy from 10-fold validation.
Abbreviations: ET: extra trees; GB: gradient boosting; LR: logistics regression; RBF: radial basis function; RF: random forest; SVM: support vector machine.
DISCUSSION
The rapid growth of COVID-19 clinical trials calls for innovative solutions to organize clinical trials for better trial information retrieval. Eligibility criteria, which define the population for clinical trials, is not very searchable for most of the existing repositories due to their free-text nature. In this study, we leveraged NLP tools to extract information from both ClinicalTrials.gov and eligibility criteria free-form texts and build the COVID-19 Trial Graph. The COVID-19 Trial Graph links structured and unstructured information for 3392 (and growing) registered COVID-19 clinical trials. With a community-support graph database, the COVID-19 Trial Graph allows diverse search queries and provides a graph-based visualization of COVID-19 clinical trials. A graph embedding further identified the clustering of clinical trials and was found to be effective in predicting the recruitment status. Clinical trials were mapped to high-dimensional vectors, which may be useful for many other downstream applications, such as the identification of similar clinical trials.
Information extraction from free-text eligibility criteria is considered a challenging task, as the eligibility criteria descriptions are often arbitrary and ambiguous.5 Although we leveraged state-of-the-art NLP tools to extract the mention of clinical concepts from eligibility criteria, errors could occur in regard to entity recognition and normalization tasks. In addition, the present study also ignores temporal and math operators in eligibility criteria, such as “mechanically ventilated” ≥ “5 days.” Although such information is important to define the study cohort, the accurate recognition and inference of these logic operators are much more complicated and, thus, beyond the scope of this study. Clinical trials also contain other free-text information, such as study description and arms which were not represented in the COVID-19 Trial Graph.
As an early attempt to represent clinical trial information in a graph, there remains some interesting research questions. For example, we used node2vec to generate graph embedding of the clinical trials, but it might be not the optimal graph embedding algorithm, as it ignores the types of relationships. Novel embedding algorithms such as deep learning might be applied.17 In addition, although we demonstrated the potential efficacy of predicting trial recruitment status through graph embedding, it’s unclear which features or data in the clinical trials have a high predictive power through our existing experiments. This would be worth investigating in the future as well.
CONCLUSION
Formal representation is essential for managing the heterogeneity and diversity of data in clinical trials. This study reports our pilot efforts to represent clinical trials, and we used COVID-19 as the use case. The COVID-19 Trial Graph is a linked graph that captured essential information in clinical trials related to COVID-19. Evaluations demonstrated its potential to expand conventional search abilities and to represent clinical trials through graph embedding that could be helpful for downstream tasks, such as predicting recruitment status or finding similar trials. Our methodology is also generalizable to other clinical trials, such as cancer clinical trials.
FUNDING
JD and CT received funding support from the National Institutes of Health under Award Nos. R56AI150272 and R01AI130460. XJ is CPRIT Scholar in Cancer Research (RR180012), and he is supported in part by the Christopher Sarofim Family Professorship, UT Stars award, UTHealth startup, the National Institute of Health (NIH) under award numbers R01AG066749 and R01AG066749-S1.
DISCLAIMER
The National Institutes of Health, and Cancer Prevention and Research Institute of Texas had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
AUTHOR CONTRIBUTIONS
JD and CT have full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept design: JD, XJ, and CT
Data collection: JD and XJ
Design of experiments and results interpretation: JD, XJ, and CT
Neo4j database construction: JD, QW
Concept normalization: JW, PR
Draft of the manuscript: JD, XJ, and CT
Critical revision of the manuscript for intellectual content: JD, XJ, CT, and YX.
DATA AVAILABILITY
The data underlying this article and case queries are available at https://github.com/UT-Tao-group/clinical_trial_graph
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
Jingqi Wang and The University of Texas Health Science Center at Houston have research-related financial interests in Melax Technologies, Inc.
Supplementary Material
REFERENCES
- 1.COVID-19 Map - Johns Hopkins Coronavirus Resource Center. https://coronavirus.jhu.edu/map.htmlAccessed April 13, 2020
- 2.COVID-19 Hub | Covid-19 Treatment Hub. https://covid19.reaganudall.org/Accessed September 15, 2020
- 3.Search of: COVID-19 - Modify Search - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/results/refine?cond=COVID-19Accessed August 19, 2020
- 4.Thorlund K, Dron L, Park J, et al. A real-time dashboard of clinical trials for COVID-19. Lancet Digit Heal 2020; 2 (6): e286–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan C, Ryan PB, Ta C, et al. Criteria2Query: a natural language interface to clinical databases for cohort definition. J Am Med Inform Assoc 2019; 26: 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.OMOP Common Data Model – OHDSI. https://www.ohdsi.org/data-standardization/the-common-data-model/Accessed August 19, 2020
- 7.Soysal E, Wang J, Jiang M, et al. CLAMP: a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc 2018; 25: 331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.neo4j. Neo4j Graph Platform – The Leader in Graph Databases. 2020. https://neo4j.com/Accessed March 30, 2020
- 9.Cypher Query Language – Neo4j Graph Database Platform. https://neo4j.com/developer/cypher/Accessed September 29, 2020
- 10.Mikolov T, Sutskever I, Chen K, et al. Distributed representations of words and phrases and their compositionality. In: Advances in Neural Information Processing Systems 2013: 3111–9. [Google Scholar]
- 11.node2vec. https://snap.stanford.edu/node2vec/Accessed August 19, 2020
- 12.Oniani D, Jiang G, Liu H, et al. Constructing co-occurrence network embeddings to assist association extraction for COVID-19 and other coronavirus infectious diseases. J Am Med Inform Assoc 2020; 27 (8): 1259–67. doi:10.1093/jamia/ocaa117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grover A, Leskovec J.. Node2vec: scalable feature learning for networks. In: proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2016: 855–64. doi:10.1145/2939672.2939754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen LH, Holmes S.. Ten quick tips for effective dimensionality reduction. PLoS Comput Biol 2019; 15 (6): e1006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du J, Jia P, Dai Y, et al. Gene2vec: distributed representation of genes based on co-expression. BMC Genomics 2019; 20 (S1): 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulyanov D. Multicore-TSNE. GitHub Repos; 2016. https://github.com/DmitryUlyanov/Multicore-TSNEAccessed August 19, 2020
- 17.Cai H, Zheng VW, Chang KCC.. A comprehensive survey of graph embedding: problems, techniques, and applications. IEEE Trans Knowl Data Eng 2018; 30 (9): 1616–37. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article and case queries are available at https://github.com/UT-Tao-group/clinical_trial_graph