Abstract
To suppress the spread of COVID-19, accurate diagnosis at an early stage is crucial, chest screening with radiography imaging plays an important role in addition to the real-time reverse transcriptase polymerase chain reaction (RT-PCR) swab test. Due to the limited data, existing models suffer from incapable feature extraction and poor network convergence and optimization. Accordingly, a multi-stage residual network, MSRCovXNet, is proposed for effective detection of COVID-19 from chest x-ray (CXR) images. As a shallow yet effective classifier with the ResNet-18 as the feature extractor, MSRCovXNet is optimized by fusing two proposed feature enhancement modules (FEM), i.e., low-level and high-level feature maps (LLFMs and HLFMs), which contain respectively more local information and rich semantic information, respectively. For effective fusion of these two features, a single-stage FEM (MSFEM) and a multi-stage FEM (MSFEM) are proposed to enhance the semantic feature representation of the LLFMs and the local feature representation of the HLFMs, respectively. Without ensembling other deep learning models, our MSRCovXNet has a precision of 98.9% and a recall of 94% in detection of COVID-19, which outperforms several state-of-the-art models. When evaluated on the COVIDGR dataset, an average accuracy of 82.2% is achieved, leading other methods by at least 1.2%.
Keywords: COVID-19, chest x-ray imaging, MSRCovXNet, feature enhancement module, ResNet-18
I. Introduction
On January 30, 2020, the World Health Organization (WHO) formally announced the novel coronavirus pneumonia (COVID-19) as a global health emergency [1], and from March 31, 2020, this was declared as a pandemic [2]. With millions of infected cases and deaths reported in the world [3], COVID-19 has rapidly spread to hundreds of countries and regions. As reported in [4], [5], it has caused more deaths, than the previous coronavirus strains, for instance, the Middle East Respiratory Syndrome (MERS) and the Severe Acute Respiratory Syndrome (SARS). By the end of 2020, the CVOID-19 pandemic has taken massive losses, with respect to the population health [6] and economic recession [7], from many countries. Apart from the prediction model of epidemiological trends [8], it becomes crucial to develop useful tools for fast and effective diagnoses and triaging of patients with suspected COVID-19 symptoms.
Currently, there are two ways in diagnosing of COVID-19, i.e., polymerase chain reaction (RT-PCR) swab test [9] and chest radiography imaging (CRI). RT-PCR swab test, which detects the viral RNA from sputum or nasopharyngeal swab, is now most popularly used for diagnosing COVID-19. However, it may introduce false detections or missing detections, regardless a lengthy waiting time for the results to be released. Studies in [10] have found that false negative rate of the RT-PCR swab test is high, which requires repeated tests for a reliable diagnosis.
As a useful supplementary to the RT-PCR swab test, CRI based diagnostic diagnoses the patients with suspected COVID-19 symptoms through visual analysis of the thoracic lesions on the computed tomography (CT) or chest X-ray (CXR) screening [11]. Compared with CXR imaging, CT is found to be more suitable for COVID-19 detection [12]. As CT imaging is generally more expensive and time-consuming, CXR imaging is thus more popularly used in detecting COVID-19, though with a loss of image resolution and contrast [13]. For both CRI methods, however, key radiological features found in COVID-19 cases, including ground-glass opacities, bilateral involvement, peripheral distributions and crazy-paving patterns, are also partially presented in MERS and SARS [14]. Furthermore, clinical symptoms of COVID-19, such as fever and cough, are similar to viral pneumonia [14]. With a limited period to gain relevant experience, it is a challenging task for the radiologists to discriminate COVID-19 from other pneumonias. With the increased cases of infections, the pressure on health services keeps rising. It is therefore essential to develop a robust and effective computer-aided diagnosis systems to reduce the diagnostic period and alleviate the burden on the clinical staff.
In recent years, deep convolutional neural networks (DCNN) are validated as an effective tool for multiple medical image processing tasks, such as classification, lesion segmentation, and reconstruction. Before DCNN, traditional machine learning (ML) models detect diseases based on extraction of hand-crafted features, which is time-consuming and lack of generalizability. Surpassing over traditional ML approaches, DCNN enables automatic feature extraction during the training, hence it is more efficient on feature searching and more robust on testing on the new data. For the sake of robustness and efficiency, DCNN has been successfully applied in many tasks of medical image analysis, such as detecting retinal diseases [15], breast cancer lesions [16], and brain tumours [17]. For the applications in terms of thoracic imaging, one study [18] has empirically validated that the DCNN can outperform experienced radiologists on classification of 14 thoracic diseases. In the context of COVID-19 diagnosis, existing DCNN based methods can also well address this challenge by extending the depth of network [5] or adopting the model assembling [19].
Although increasing the number of layers (i.e., using a deeper network) can improve the capability of feature extraction, this requires the dataset to be sufficiently large (e.g., millions of images, the similar scale as the ImageNet [20]). Due to the limited availability of the COVID-19 data, the efficacy of the existing deep learning models is severely affected, resulting in less capable feature extraction and difficulty of network convergence and optimization. To tackle these issues, in this paper, a multi-stage residual network, MSRCovXNet, is proposed for effective detection of COVID-19 from the CXR images. We aim to derive highly discriminative features from a shallow network, where the number of samples are limited. To achieve this, a ResNet-18 [21] is used as the feature extractor, which is optimized by the fusion of features from multiple stages for improved classification and decision-making.
The major contributions of this paper can be summarized as follows:
-
i.
Taking the ResNet-18 as the feature extractor, a shallow yet effective COVID-19 classifier, MSRCovXNet, is proposed for effective detection of COVID-19 under limited training samples;
-
ii.
A single stage feature enhancement module (SSFEM) is proposed to enhance the feature representation of low-level features, whilst a multi-stage feature enhancement module (MSFEM) is proposed to obtain highly discriminative features fused from multiple stages;
-
iii.
Without ensembling other deep learning models, the proposed MSRCovXNet has a precision of 98.9% and a recall of 94% in the detection of COVID-19 cases, achieving state-of-the-art performance on the COVIDx dataset. When evaluated on the COVIDGR dataset, an average accuracy of 82.2% is achieved, leading other methods by at least 1.2%. When compared with other CNN models trained on different datasets, the proposed method still shows superior performance.
The remaining parts of this paper are organized as follows. Section II briefly introduces the related work. The architecture of the proposed method and the experimental results are detailed in Section III and Section IV, respectively. Finally, some concluding remarks are given in Section V.
II. Related Work
Since the outbreak of the COVID-19, a number of DCNN-based models have been developed for the detection of COVID-19 from CT and CXR images. At first, many people focused on a two-category classification, in which the COVID-19 cases were distinguished from either healthy cases [30], [31], or other lung infections diseases, such as viral pneumonia [32], [33], [34], [35] and others [36], [37], [38], [39]. Most of these methods report seemingly impressive results, where performances in the range of 90-100% are not uncommon. However, since doctors not only need to determine whether their patient has COVID-19 or not, but to also identify whether a patient with suspected COVID-19 symptoms does indeed have COVID-19 or a similarly presented infection, the two-class approaches over-simplify the detection problem. Without taking into account the possibility of patients having a healthy image, or being unable to distinguish between various pneumonias, the proposed models most likely encourage a greater degree of overfitting to the training data. As a result, there has been an increased trend in the literature towards adopting a three-class approach, where models are trained to detect healthy patients, as well as discriminating between images of COVID-19 from other pneumonias.1 This will improve the model’s diagnostic sensitivity, and additionally help doctors have a better understanding of what separates COVID-19 images from other pneumonias presenting similar features.
For this reason, our work strives to contribute to the body of work addressing the three-class problem for detecting COVID-19 from CXR images, where many methods have been proposed. In Wang and Wong [40], a deep CNN model, namely COVID-Net, is proposed, which is actually one of the first three-class deep learning models on CXR diagnosis. The three categories as defined in COVIDx dataset [40], [41] include COVID-19, pneumonia, and healthy cases, respectively. For performance assessment, the COVIDx dataset is collected, which includes 8066, 5551 and 386 normal, bacterial pneumonia (containing both viral pneumonia and bacterial pneumonia), and COVID-19 patients, respectively. A F1 score of 95.9% has been reported on the testing set.
In comparison to a single model used in Wang and Wong [40], Karim et al. [19] have proposed an ensemble of DenseNet-161 and VGG-19 and form the DeepCOVIDExplainer model. Experiments on a dataset with 11,896 images in total have achieved a precision of 89.61% and a recall of 83% on 77 COVID-19 test samples, where the categories are the same as in Chattopadhay et al. In order to provide an interpretable evidence to the clinical staff, the class-discriminating pixels on the test images are visualized using the Grad-CAM++ method [42].
To cope with the high degree of imbalance within the categories of the collected COVID-19 samples, Bassi and Attux [22] have applied the transfer learning to pretrain the proposed CheXNet [18] on the dataset of 112,120 CXR images with 14 thoracic diseases, including the pneumonia samples. For easy adopting of the pretrained model on the target dataset, the output of the last fully-connected layer is reduced to 3 to coping with the three categories of cases. The dataset used for training in [22] contains 127, 1285 and 1281 COVID-19, pneumonia, and normal CXR images, respectively. With the help of data augmentation, an average classification accuracy of 97.8% on a testing set of 180 images is achieved.
In [23], Zhang et al., domain shift between datasets, namely COVID-DA, is attempted to improve the classification accuracy under a semi-supervised framework, where both labeled and unlabeled data are utilized for learning the model. The training set is composed of 8154, 2306 and 258 normal, pneumonia, and COVID-19 CXR images, respectively. However, the testing set has only two categories, i.e., 885 normal and 60 COVID-19 images, respectively, where an F1-score of 92.98% and AUC of 0.985 are reported.
In [24], similar to Misra et al., a three ResNets were ensemble with each subnet being trained for classifying a single category, dividing the three-category classification task into three binary classification tasks. The three binary classifiers are first trained using normal (1579 cases) vs diseased (4429 cases), pneumonia (4245 cases) vs non-pneumonia (1763 cases), and COVID-19 (184 cases) vs non-COVID-19 (5824 cases), respectively. Afterwards, the three ensembled ResNets are fine-tuned on another dataset with 1579 normal, 4245 pneumonia, and 184 COVID-19 cases, respectively. Eventually, a precision of 94% and a recall of 100% are achieved.
In El Asnaoui and Chawki [26], the pneumonia is split into bacterial pneumonia and viral pneumonia before conducting a comprehensive study on several popularly used models, such as VGG, ResNet, DenseNet, Inception-ResNet, Inception-V3, and MobileNet-V2. The dataset used contains 1583 normal, 2780 bacterial, 1493 viral, and 231 COVID-19 cases, where 80% of the samples are used for training and the remaining 20% for testing. Finally, they have found that the Inception-ResNet-V2 can outperform all others with an overall accuracy and F1-score of 92.18% and 92.07%, respectively. In [43], an ensemble of DNNs is proposed for COVID-19 prediction.
In a recent study [44], a quantification method is proposed for quantifying the level of COVID-19 infection severity, which is adopted from a previously defined Radiographic Assessment of Lung Edema (RALE) score [45]. By measuring the extent of the lesion features on each lung, the severity is quantified in a range of 0 to 8, based on which the severity is further defined in 4 levels, i.e., Normal 0, Mild 1-2, Moderate 3-5 and Severe 6-8 [46]. A two-class COVIDGR dataset is collected, containing 426 Normal images and 426 COVID-19 images, in which the positive cases include 76 Normal-PCR+, 100 Mild, 171 Moderate, and 79 Severe cases, respectively. Specifically, Normal-PCR+ indicates that the radiologist does not find any visual lesion regions, although the PCR test is positive. By conducting a 5-fold cross-validation testing with 5 runs on the COVIDGR, an average accuracy of 81.00% can be achieved by the proposed COVID-SDNet [46].
For a more concise comparison of the three-class CXR literature, we have listed the discussed literature along with other relevant papers, together with their respective methods and results, in Table I. There have also been similarly conducted studies implementing deep learning-based classifiers on CT images [47], [48], [49] and lung ultrasonography [50], however we forego discussing them in any detail since our focus is with CXR-based models. A more detailed review of recent CT based deep learning models can be found here [13], [51].
TABLE I. Summary of Methods and Findings for Three-Class (Normal, Pneumonia, and COVID-19) Approaches on Chest X-Ray Images.
| Reference | Dataset | Method | Results |
|---|---|---|---|
| Wang [19] | COVID-19 386 Pneum. 5551 Normal 8066 |
CNN | 95.9% (F1 score) |
| Karim [20] | COVID-19 259 Pneum. 8614 Normal 8066 |
DenseNet, ResNet, VGG19 (ensemble) | 89.1% (Prec.) 83.0% (Rec.) |
| Bassi [23] | COVID-19 219 Vir. Pneum. 1345 Normal 1341 |
Pre-trained CheXNet (DenseNet-121) | 98.3% (Acc.) |
| Zhang [24] | Training Testing COVID-19 258 60 Pneum. 2306 n/a Normal 8154 885 |
Semi-supervised domain adaption. | 93.0% F1 score |
| Kim [25] | COVID-19 184 Pneum. 4245 Normal 1579 |
Ensemble three ResNets | 94% (Prec.) 100% (Rec.) |
| Yamac [26] | COVID-19 462 Bact. Pneum. 2760 Vir. Pneum. 1485 Normal 1579 |
Pre-trained CheXNet (DenseNet121) | 95.6% (Acc) 93.9% (Rec.) 95.8% (Spec.) |
| Chawki [27] | COVID-19 231 Bact. Pneum. 2780 Vir. Pneum. 1493 Normal 1583 |
Inception-ResNetV2 | 92.2% (Acc) |
| Rahimzadeh [28] | COVID-19 180 Pneum. 6054 Normal 8851 |
Xception, ResNetV2 (ensemble) | 99.5% (Acc. COVID-19) 91.4% (Acc. all classes) |
| Togacar [29] | COVID-19 295 Pneum. 98 Normal 65 |
MobileNetv2, SqueezeNet SVM as classifier | 100.0% (Acc. COVID-19) 99.3% (Acc. Normal and Pneum.) |
| Lv [30] | Dataset 1 Bact. Pneum. 2772 Vir. Pneum. 1493 Normal 1591 |
Cascade network of SEME-ResNet50 and SEME-DenseNet169. | 97.1% (Acc. COVID-19) 85.6% (Acc. Pneum.) |
| Dataset 2 COVID-19 125 other viruses 316 |
In this paper, we aim to improve the feature representation for a shallow network, which is suitable for training on a small number of samples, without using the ensemble strategy.
III. Proposed Method
In this section, we will discuss the proposed method in details, including, the architecture of the proposed MSRCovXnet, especially the implementation of the multi-stage feature enhancement module, which is proposed for improving the feature representation. After that, the training hyperparameters will also be introduced.
A. Network Architecture
The overall architecture of the proposed MSRCovXNet is shown in Fig. 1. At the current stage, as shown in Table I, there is no large dataset (as the ImageNet) for COVID-19 classification. Under limited samples, a shallow network may perform better than a deep network [21], which is also verified by the experiments in this paper. Thus, the ResNet-18 [21], which is pretrained on the ImageNet [20], is used as the feature extractor. We denote the output of the blocks “conv2_x”, “conv3_x”, “conv4_x”, and “conv5_x” as “C2”, “C3”, “C4” and “C5”, respectively, where the total stride with respect to these four blocks are 4, 8, 16, and 32. The prediction
 of the original ResNet can be expressed as below
of the original ResNet can be expressed as below
 |
where
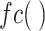 and
and
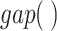 are the fully connected layer (FC) and the global average pooling layer (GAP), respectively.
are the fully connected layer (FC) and the global average pooling layer (GAP), respectively.
Fig. 1.
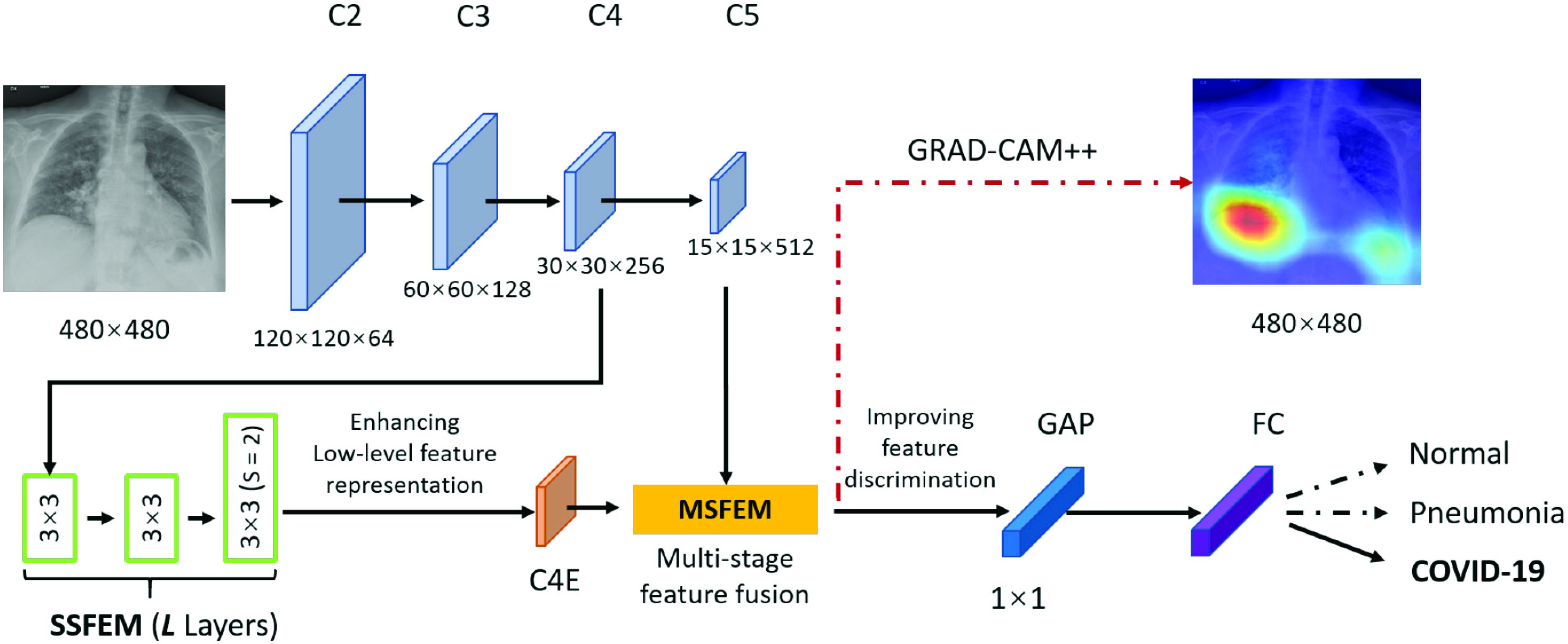
Architecture of the proposed MSRCovXNet.
To enhance the capability of feature extraction, feature maps derived from multiple stages, rather than a single stage as the original ResNet-18, are combined in our model. In this paper, we adopt both the C4 and C5 blocks for the lateral classification procedure. Feature maps before C4 are not utilized, simply because the object (“lung” for this task) in the x-ray image is still too large in the previous blocks. As low-level features are sensitive to small objects [52], this affects their impact on the classification task, which is also verified by the experimental results in this paper. For the feature map C4, a single-stage feature enhancement module (SSFEM), which is a subnet with
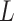 convolution layers of size
convolution layers of size
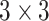 , is assigned to enhance the lower-level feature extraction (denoted by green rectangles in Fig. 1). The size of C4 is reduced, with the number of channels doubled, in the last layer of the subnet to keep the size of the output (denoted as “C4E”) the same as C5.
, is assigned to enhance the lower-level feature extraction (denoted by green rectangles in Fig. 1). The size of C4 is reduced, with the number of channels doubled, in the last layer of the subnet to keep the size of the output (denoted as “C4E”) the same as C5.
The capability of feature extraction can be further enhanced by using a feature fusion module [53], [54], which is applied to fuse features from different stages. Thus, we present a multi-stage feature enhancement module (MSFEM) to further enhance the extracted feature, which is shown in Fig. 2. The architecture of MSFEM will be detailed in the next subsection. In the end of the network, the enhanced multi-stage feature will be fed to a global average pooling layer, and the final prediction is conducted by a fully connected layer, which is the same as the previous methods [21], [55], [56]. In summary, the prediction
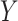 of the proposed method can be expressed as below:
of the proposed method can be expressed as below:
 |
where
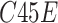 is the feature map extracted using the proposed SSFEM (
is the feature map extracted using the proposed SSFEM (
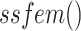 ) and MSFEM(
) and MSFEM(
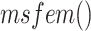 ):
):
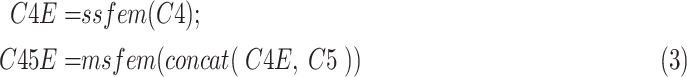 |
where concat is the short of concatenation.
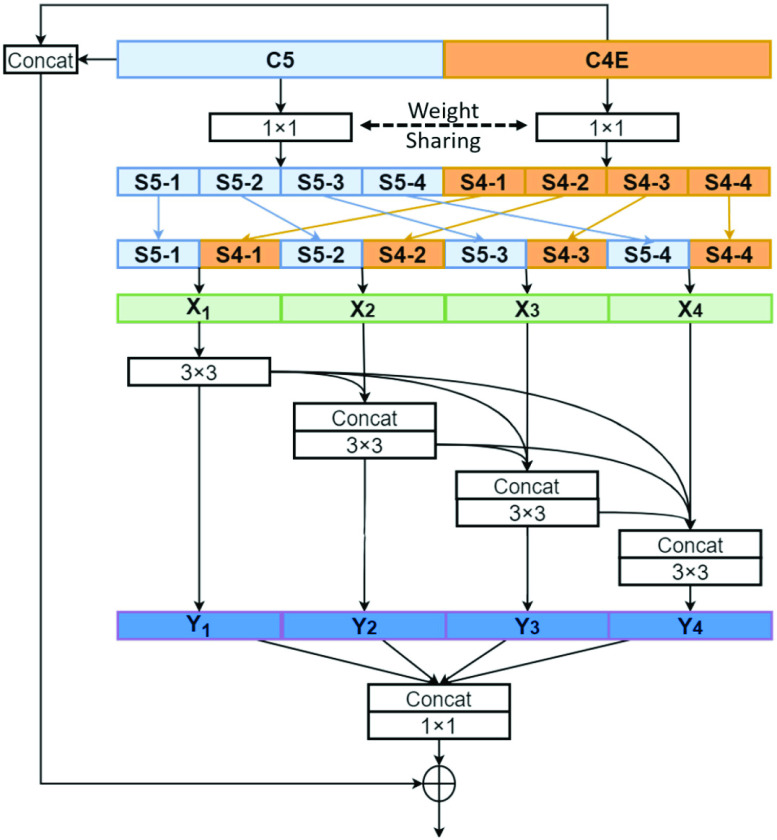
Fig. 2.
Architecture of the proposed multi-stage feature enhancement module.
B. Multi-Stage Feature Enhancement Module
As shown in Fig. 2, The proposed MSFEM adopts the residual learning [21] for feature fusion and feature refinement, which can be mathematically expressed by:
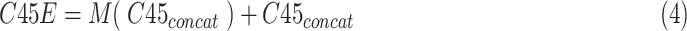 |
where
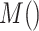 and
and
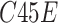 are respectively present the convolution layers in the proposed MSFEM and the feature concatenation of
are respectively present the convolution layers in the proposed MSFEM and the feature concatenation of
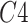 and
and
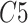 .
.
First of all, two
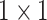 convolution layers are applied to C5 and C4E for adjusting the number of channels, which can be expressed as below:
convolution layers are applied to C5 and C4E for adjusting the number of channels, which can be expressed as below:
 |
where
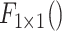 indicates the two
indicates the two
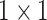 convolution layer. As to the number of layers in terms of C4E and C5 are the same, the weights of two
convolution layer. As to the number of layers in terms of C4E and C5 are the same, the weights of two
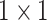 convolution layers are shared for reducing the number of training parameters. The effectiveness of such implementation is also validated during our experiments: compared with non-weight-sharing, the F1 score of the network using weight-sharing could be slightly increased by about 0.1%. As suggested in [56], [57], [58], multi-scale representations, which are acquired in a large range of receptive fields, bring benefits for more accurate prediction. Thus, we utilize multiple subnets, consisting of a variety range of convolutional layers, to achieve multi-scale receptive fields in the proposed MSFEM. However, it will remarkably increase the number of training parameters, if directly taking the fused feature maps as the input of those subnets. As presented above, this will then increase the difficulty of the network optimization, causing the reduction on the detection accuracy. It is also validated in Section IV, where the proposed outperforms methods with deeper networks. Thus, in the proposed MSFEM, only a part of input channels is fed to each subnet, instead of all channels. To achieve this, after normalizing the number of channels, feature maps from C4E and C5 are divided into
convolution layers are shared for reducing the number of training parameters. The effectiveness of such implementation is also validated during our experiments: compared with non-weight-sharing, the F1 score of the network using weight-sharing could be slightly increased by about 0.1%. As suggested in [56], [57], [58], multi-scale representations, which are acquired in a large range of receptive fields, bring benefits for more accurate prediction. Thus, we utilize multiple subnets, consisting of a variety range of convolutional layers, to achieve multi-scale receptive fields in the proposed MSFEM. However, it will remarkably increase the number of training parameters, if directly taking the fused feature maps as the input of those subnets. As presented above, this will then increase the difficulty of the network optimization, causing the reduction on the detection accuracy. It is also validated in Section IV, where the proposed outperforms methods with deeper networks. Thus, in the proposed MSFEM, only a part of input channels is fed to each subnet, instead of all channels. To achieve this, after normalizing the number of channels, feature maps from C4E and C5 are divided into
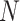 splits. Let
splits. Let
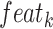 be the
be the
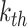 channel in terms of the input feature map feat (
channel in terms of the input feature map feat (
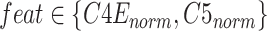 ). The
). The
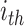 split
split
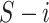 can be the acquired by:
can be the acquired by:
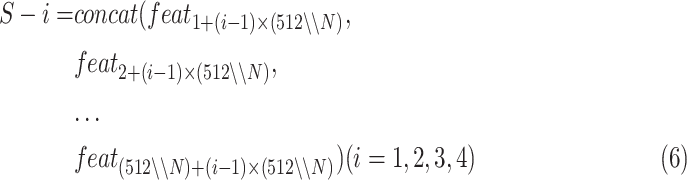 |
as suggested in Res2Net [56], we assign
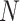 = 4 as the number of splits and utilize the number of channels with respect to each split as 208 in this paper. Splits from C4E and C5 are concatenated by orders, which can be mathematically expressed as follows:
= 4 as the number of splits and utilize the number of channels with respect to each split as 208 in this paper. Splits from C4E and C5 are concatenated by orders, which can be mathematically expressed as follows:
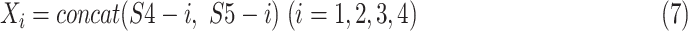 |
where
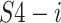 ,
,
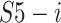 are the splits from
are the splits from
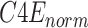 and
and
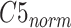 , respectively; concat is the short of concatenation;
, respectively; concat is the short of concatenation;
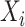 is the input split of the following Densely connected block [55].
is the input split of the following Densely connected block [55].
For each split
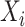 , the input is the concatenation of all the outputs from the previous layer, as well as the
, the input is the concatenation of all the outputs from the previous layer, as well as the
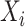 , which can be represented as:
, which can be represented as:
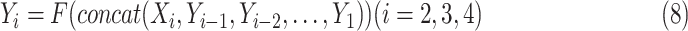 |
where
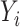 is the output with respect to
is the output with respect to
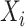 ,
,
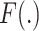 denotes the convolution layer. Specially, for
denotes the convolution layer. Specially, for
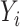 , the input is
, the input is
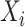 , because there is no output “
, because there is no output “
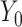 ” before it. In the end, the splits are re-concatenated, and the number of channels is adjusted via a
” before it. In the end, the splits are re-concatenated, and the number of channels is adjusted via a
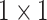 convolution layer. For the ResNet-18 based network, the number of channels are 1024 (C5:512, C4E:512) [21].
convolution layer. For the ResNet-18 based network, the number of channels are 1024 (C5:512, C4E:512) [21].
IV. Experimental Results
A. Experimental Settings
1). Dataset:
Experiments in this paper are conducted on the COVIDx dataset [40], which is currently the largest publicly available dataset with respect to the number of COVID-19 cases. COVIDx dataset consists of 13703 images for training and 300 images for testing, where the COVID-19 samples are collected from more than 266 COVID-19 patients. Images in the COVIDx dataset are labeled in three classes: normal, non-COVID19 infection (pneumonia), and COVID-19. The training and testing sets are randomly divided according to the patient ID, which means for a particular patient, the associated data will be used either for training or testing, hence there is no overlapped data in this context. Details of the sample distribution in terms of each class are shown in Table II. We train our models on the training dataset and evaluate the performance on the testing dataset.
TABLE II. Data Distribution of the COVIDx Dataset.
| Normal | Pneumonia | COVID-19 | Total | |
|---|---|---|---|---|
| Train | 7966 | 5451 | 286 | 13703 |
| Test | 100 | 100 | 100 | 300 |
Tabik et al. [46] argued that the majority COVID-19 cases in COVIDx dataset is at severe level. It is therefore essential to validate the proposed MSRCovXNet on another dataset for validating the performance in early-stage diagnosis. To this end, we also evaluated the proposed MSRCovXNet on the COVIDGR 1.0 dataset [46]. As a two-class dataset, COVIDGR includes 426 samples in each class. The severity distribution of the positive cases is: 76 Normal-PCR+, 100 Mild, 171 Moderate, and 79 Severe, respectively.
2). Implementation Details and Evaluation Metrics:
The proposed method is implemented on PyTorch [61]. The input image is resized to
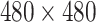 for efficiency. Following the settings in [24], [25], [29], [40], we adopt the Adam optimizer [62] for its promising performance on weight optimization. The initial learning rate is set to 1e-4, which is decreased via the cosine annealing schedule [63]. The batch size is set to 60 on 3 GPUs. The network is trained by 22 epochs. Moreover, we adopt the data augmentation methods as suggested in COVID-Net [40] for a fair comparison, which include: intensity shift, translation, rotation, horizontal flip, and random resizing. For performance evaluation, we report the results using the F1 score (%), as it considers both the recall and precision for an overall assessment of false alarms and missed detections.
for efficiency. Following the settings in [24], [25], [29], [40], we adopt the Adam optimizer [62] for its promising performance on weight optimization. The initial learning rate is set to 1e-4, which is decreased via the cosine annealing schedule [63]. The batch size is set to 60 on 3 GPUs. The network is trained by 22 epochs. Moreover, we adopt the data augmentation methods as suggested in COVID-Net [40] for a fair comparison, which include: intensity shift, translation, rotation, horizontal flip, and random resizing. For performance evaluation, we report the results using the F1 score (%), as it considers both the recall and precision for an overall assessment of false alarms and missed detections.
When training on the COVIDGR dataset, we decrease the training epoch to 15, while all other hyperparameters remain the same as on the COVIDx dataset. Images are cropped to normalize the position of lung as suggested in [46]. Following the same evaluation method in COVIDGR, we conducted 5 different 5-fold cross validations with multiple metrics, including the sensitivity, specificity, precision, F1 and Accuracy. Results of each metric are reported using the average values and the standard deviation over the five runs. As the Normal-PCR+ may impede the overall performance [46], such cases are excluded in our experiments.
B. Ablation Study
In this section, we conduct an ablation study to examine how each proposed component within our MSRCovXNet affects the final performance in the detection of COVID-19.
1). Selection of Feature Stages:
In this section, the selection of feature stages will be discussed. As suggested in the feature pyramid network [64], a single convolution layer is applied as a replacement of the single-stage feature enhancement module (SSFEM). Meanwhile, multi-stage feature enhancement module (MSFEM) is not utilized.
Experimental results are shown in Table III. By fusing the feature maps of C4 and C5, the F1 scores are increased by 0.9%, 1.3% and 0.6% on normal class, pneumonia class and COVID-19 class, respectively. However, after adding the C3, the F1 scores on normal and pneumonia are dropped by 1.2% on average. This indicates that the low-level feature maps (C2 and C3) does not bring benefits to this task. Thus, we only adopt C4 and C5 for feature fusion in the proposed MSRCovXNet.
TABLE III. Selection of Feature Stages.
| Blocks | F1 score(%) | ||
|---|---|---|---|
| Normal | Pneumonia | COVID-19 | |
| C5 | 93.5 | 93.1 | 95.3 |
| C5+C4 | 94.6 | 94.4 | 95.9 |
| C5+C4+C3 | 92.6 | 94.0 | 95.9 |
2). Number of Layers in the Single-Stage Feature Enhancement Module:
In this subsection, the number of layers in the SSFEM is discussed. According to the results shown in Table IV, as the number of layers increases, the F1 score of COVID-19 increases by 0.5% on maximum. However, the F1 scores on normal and pneumonia classes decrease. Take
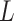 = 6 for example, the F1 scores on normal and pneumonia classes are reduced by 0.9% and 0.3%, respectively. This is mainly caused by the size of the dataset, i.e., the complexity of models, where
= 6 for example, the F1 scores on normal and pneumonia classes are reduced by 0.9% and 0.3%, respectively. This is mainly caused by the size of the dataset, i.e., the complexity of models, where
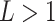 , is overfitting for normal, and pneumonia classes in COVIDx. As the training accuracies are ranged between 97% and 100% for the models in Table IV, we deduce that the performance of SSFEM with deeper layers could be further improved by adding more available samples with more variants in the future. Thus, in this paper, we select
, is overfitting for normal, and pneumonia classes in COVIDx. As the training accuracies are ranged between 97% and 100% for the models in Table IV, we deduce that the performance of SSFEM with deeper layers could be further improved by adding more available samples with more variants in the future. Thus, in this paper, we select
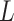 = 1 for the following experiments.
= 1 for the following experiments.
TABLE IV. F1 Scores in Terms of the SSFEM Layer Numbers.
Number of
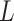
|
F1 score(%) | ||
|---|---|---|---|
| Normal | Pneumonia | COVID-19 | |
| 1 | 94.6 | 94.4 | 95.9 |
| 2 | 94.1 | 93.0 | 95.9 |
| 4 | 93.3 | 94.4 | 96.4 |
| 6 | 93.5 | 94.1 | 96.4 |
3). Effect of the Multi-Stage Feature Enhancement Module:
In this subsection, we will verify the effect of the MSFEM. Experimental results are shown in Table V. As seen, though the F1 score on normal cases decreases by 0.4%, the F1 scores on the pneumonia and COVID-19 classes increase by 1% and 0.5% respectively. Overall, the performance has been further improved with the MSFEM.
TABLE V. Ablation Study of the MSFEM Layer.
| F1 score(%) | |||
|---|---|---|---|
| Normal | Pneumonia | COVID-19 | |
| ResNet-18 +SSFEM (c=1) |
94.6 | 94.4 | 95.9 |
| ResNet-18 +SSFEM (c=1) +MSFEM |
94.2 | 95.4 | 96.4 |
C. Compared With the State-of-the-Art Methods
1). Result Comparison on the COVIDx Dataset:
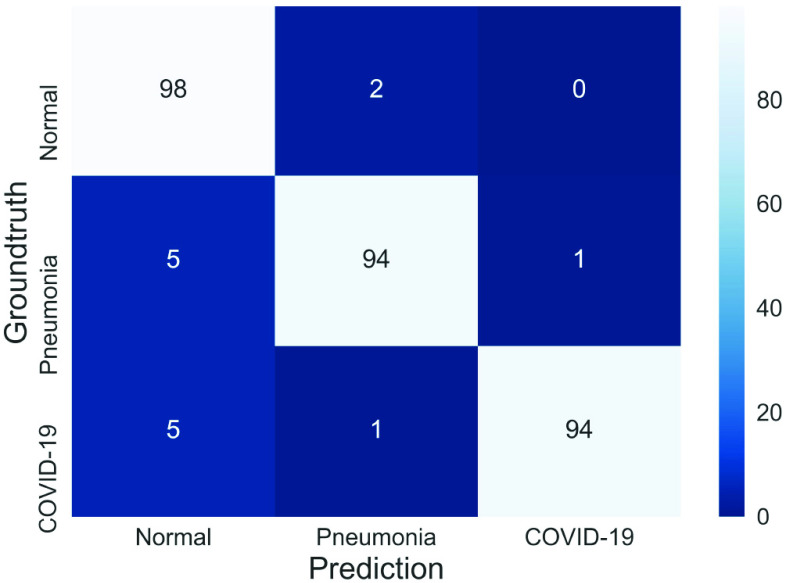
Here we compared the proposed MSRCovXNet with the state-of-the-art deep learning models. First of all, the proposed MSRCovXNet is compared with the other methods that are trained on the same COVIDx dataset. Experimental results are shown and compared in Table VI, and the confusion matrix is visualized in the Fig. 3. As seen, the proposed MSRCovXNet has achieved state-of-the-art performance in all three classes.
TABLE VI. Results Comparison in Terms of F1 Score (%) With Several State-of-the-Art Deep Learning Models on the COVIDx Test Dataset.
| Methods | F1 Score(%) | ||
|---|---|---|---|
| Normal | Pneumonia | COVID-19 | |
| ResNet-18* [22] | 93.5 | 93.1 | 95.3 |
| ResNet-50* [22] | 93.3 | 93.9 | 95.9 |
| Res2Net-50* [58] | 93.7 | 94.9 | 96.4 |
| ChexNet* [23] | 94.2 | 94.9 | 95.9 |
| COVID-Net [41] | 92.5 | 91.6 | 95.9 |
| MSRCovXNet (proposed) | 94.2 | 95.4 | 96.4 |
Trained by author with 5 runs
Fig. 3.
Confusion matrix for the proposed MSRCovXNet on the COVIDx test dataset. Precision and recall of each class is shown in Table VIII.
2). Result Comparison With Methods Trained on Other Datasets:
Due to the limited number of image samples, some of the existing methods are trained and evaluated on a subset of the COVIDx dataset [19], [23], [27], [28] or self-collected dataset [22], [24]. However, as the classification task in these datasets are the same, it is worthy to compare the performance with these methods as well. Results are shown in Table VII. Here we compared with methods that used the same classes (normal, pneumonia, and COVID-19) as our methods. Methods with different classes are not compared, e.g., the method in [26] which classified four categories: bacterial pneumonia, coronavirus, COVID-19, and normal.
TABLE VII. Results Comparison in Terms of F1 Score (%) with Methods Trained on Other Datasets.
| Methods | Number of images for training | Number of images for testing | F1 Score (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | Normal | Pneumonia | COVID-19 | |
| Karim [20] | 5647 | 6030 | 182 | 2419 | 2584 | 77 | 86.5 | 84.5 | 81.6 |
| Zhang [24] | 8154 | 2306 | 258 | 885 | 0 | 60 | n/a | n/a | 93.0 |
| Kim * [25] | 1422 | 3821 | 166 | 157 | 424 | 18 | 92.2 | 94.4 | 96.6 |
| Rahimzadeh [28] | 2000 | 1634 | 149 | 6851 | 4420 | 31 | 93.2 | 89.1 | 49.1 |
| Togacar ** [29] | 46 | 69 | 207 | 19 | 29 | 88 | 95.3 | 96.5 | 99.5 |
| MSRCovXNet (proposed) | 7966 | 5451 | 286 | 100 | 100 | 100 | 94.2 | 95.4 | 96.4 |
| MSRCovXNet-s (proposed) | 100 | 100 | 100 | 7966 | 5451 | 286 | 90.0 | 90.0 | 80.0 |
A slightly higher F1 in COVID-19 cases than ours due to a very small number of test samples in COVID-19 yet the F1 scores for the other two classes are much lower.
Relatively better performance is due to too limited training and testing samples used, where the higher number of COVID-19 cases than others seems impractical in real scenarios.
As seen, the proposed MSRCovXNet outperforms most of the methods in all the three classes, except Toğaçar et al. [28] and Misra et al. [24]. When compared with Kim, the number of COVID-19 samples is less than 20% of COVIDx. However the F1 score on COVID-19 cases only outperforms by 0.2%, along with a degradation of 2% and 3% on the normal and pneumonia cases, respectively. Therefore, it is hard to say it actually outperformed the proposed MSRCovXNet. For Toğaçar et al. [28], as the total number of test images is only 136, this is also highly imbalanced for the three classes. As a result, we deduce that the difference on performance is mainly caused by the small size and imbalanced samples of the test set.
Training on small dataset size: Specifically, we trained the proposed method on the testing set of COVIDx, and test on the training set, which is to further evaluate the efficacy and robustness of the proposed approach in distinguishing COVID-19. In this case, the number of training images are 100 for each class. Results are shown in Table VII as “MSRCovXNet-s”. As seen, when the testing set is far larger than the training set, which is similar to the real-life situation, the proposed method can still achieve comparable results to other methods. This validates the effectiveness of the MSRCovXNet on a small training set.
Evaluating on COVIDGR: Results comparison on the COVIDGR dataset from different approaches are listed in Table IX. As seen, even with many early-stage cases included, the proposed MSRCovXNet can still achieve the state-of-the-art performance, surpassing the COVID-SDNet by 1.2% on the average accuracy. This has validated the effectiveness of the proposed method on detecting the early-stage COVID-19. For comparison, although COVID-Net achieves a high F1 score on the COVIDx, the F1 score on the COVIDGR dataset is only 65.64%, which is 15% lower than the proposed method. This has again validated the robustness of the proposed methodology in detection of COVID-19 in CXR images.
TABLE VIII. Precision and Recall of the Proposed MSRCovXNet.
| Normal | Pneumonia | COVID-19 | |
|---|---|---|---|
| Precision (%) | 90.7 | 96.9 | 99.0 |
| Recall (%) | 98.0 | 94.0 | 94.0 |
TABLE IX. Result Comparison (%) to State-of-the-Art Methods on the COVIDGR 1.0 Dataset. Spec., Prec., and Sens. are the Abbreviation of Specificity, Precision and Sensitivity, Respectively.
| Class | Metric | Methods | ||||
|---|---|---|---|---|---|---|
| COVID-Net [41] | COVID-CAPS [61] | FuCiTNetz [62] | COVID-SDNet [47] | MSRCovXNet (proposed) | ||
| N | Spec. | 83.42 ± 15.39 | 65.09 ± 10.51 | 82.63 ± 6.61 | 85.20 ± 5.38 | 82.35 ± 6.49 |
| Prec. | 69.73 ± 10.34 | 71.72 ± 5.57 | 79.94 ± 4.28 | 78.88 ± 3.89 | 85.12 ± 4.07 | |
| F1 | 74.45 ± 8.86 | 67.52 ± 5.29 | 81.05 ± 3.44 | 81.75 ± 2.74 | 83.46 ± 3.13 | |
| P | Sens. | 61.82 ± 22.44 | 73.31 ± 9.74 | 78.91 ± 5.88 | 76.80 ± 6.30 | 82.01 ± 5.76 |
| Prec. | 79.50 ± 11.47 | 68.40 ± 5.13 | 82.43 ± 5.43 | 84.23 ± 4.59 | 79.73 ± 5.04 | |
| F1 | 65.64 ± 15.90 | 70.20 ± 4.31 | 80.37 ± 3.16 | 80.07 ± 0.04 | 80.60 ± 2.83 | |
| Accuracy | 72.62 ± 7.6 | 69.20 ± 3.61 | 80.77 ± 3.15 | 81.00 ± 2.87 | 82.20 ± 2.83 | |
As discussed by Tabik et al. [46], the majority of COVID-19 cases in the COVIDx dataset is at the severe level. Methods reported on this dataset have achieved quite high accuracy on detecting COVID-19, due mainly to the low detecting difficulty. This can be also observed in our experiments, see in Table IX, where an accuracy of 82.2% was achieved. However, the classification accuracy drops by 19.27% when the same method is trained and tested on the COVIDGR dataset. This has clearly indicated potential issue of data quality, which may affect the detection accuracy in this context. A lesson herein will be that it is unsuitable to only apply the easy samples for training and testing, where hard samples at less severe levels of COVID-19 would be beneficial. By training on the dataset with uniformly distributed four severity levels, the discrimination of detecting the early-stage cases can be further strengthened.
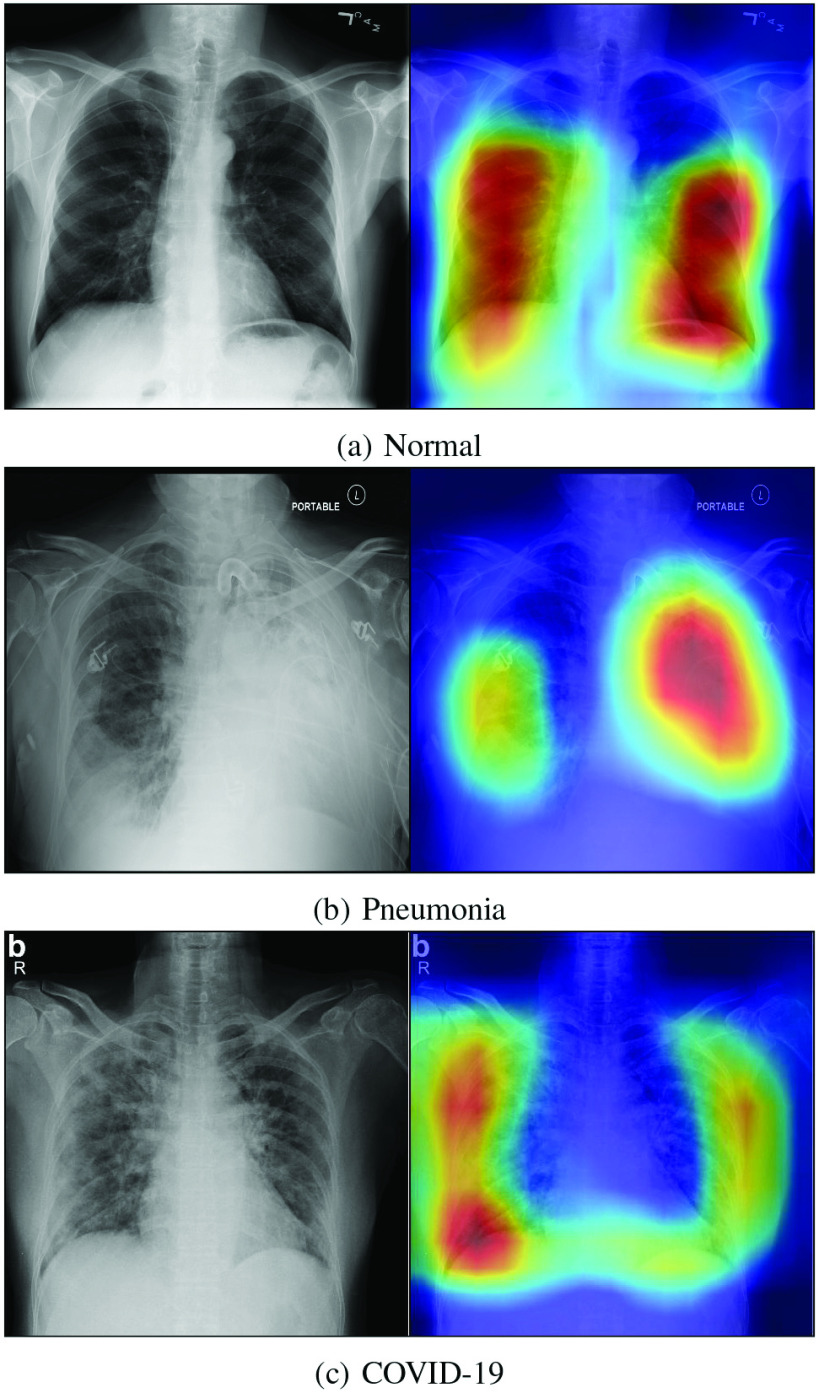
3). Visualization of the Class-Discriminating Regions:
It is important to know the decisive regions on the image where the pixels contribute most to the final decision. This is because it is able to verify the reliability of the diagnostic decision made by the CNN, which can help the clinical doctor to gain a better understanding of the proposed deep learning model. It also benefits them to find out the diseased regions on the image. In this paper, the class-discriminating regions are highlighted using gradient-guided class activation maps (Grad-CAM++) [42], which is shown in Fig. 4. As expected, the proposed method predicts based on regions with pathological features in the lungs, which also validates the high reliability of the proposed method in effective detection of COVID-19 from other cases.
Fig. 4.
Decision visualization using Grad-CAM++ with the input label: (a) Normal, (b) Pneumonia, and (c) COVID-19. Images are all from the COVIDx dataset.
V. Conclusion
In this paper, we proposed a novel COVID-19 classifier, namely MSRCovXNet, for the detection of COVID-19 from chest x-ray (CXR) imaging. To tackle the challenging problem of insufficient training samples, the ResNet-18 is used as the feature extractor. In order to improve the discriminative capability of the extracted features from this shallow network, the proposed MSRCovXNet fuses the features from multiple stages rather than adopting the feature map from the last stage for decision making. With a VGG-style subnet structure, the proposed single stage feature enhancement module (SSFEM) has effectively enhanced the feature representation of low-level features. Meanwhile, the proposed multi-stage feature enhancement module (MSFEM) has improved the performance by varying the range of the receptive fields to obtain highly discriminative features fused from multiple stages. The performance of the proposed MSRCovXNet has been validated on the COVIDx dataset, by far the largest publicly available dataset for COVID-19 detection from CXR images. Thanks to the proposed feature enhancement modules, our MSRCovXNet has demonstrated superior performance over several state-of-the-art deep learning models, under a small number of training samples and without any ensembling models. When compared with models which are trained on other datasets, the proposed MSRCovXNet still obtains a promising performance.
Future work will focus on further enhancement of the features using ResNet-style skip connections [21], [56], [65] as the VGG style subnet is suboptimal. In addition, fusion of multiple CXR images will also be utilized as the supplementary information in between can improve the discriminative capability of the learnt features. Finally, we may also explore other deep learning models and also effective methods in addressing imbalanced learning in detection of COVID-19 from CXR and other image data.
Biographies

Zhenyu Fang received the B.Eng. degree (First-Class Hons.) in electronic and electrical engineering from the University of Strathclyde Glasgow, U.K., and North China Electric Power University, Baoding, China, in 2016, and the Ph.D. degree in electronic and electrical engineering from the University of Strathclyde in July 2020. His main interests are algorithm development for image classification, object detection, and face detection.

Jinchang Ren (Senior Member, IEEE) received the B.E. degree in computer software, the M.Eng. degree in image processing, the D.Eng. degree in computer vision from Northwestern Polytechnical University, Xi’an, China, and the Ph.D. degree in electronic imaging and media communication from the University of Bradford, Bradford, U.K.
He is currently a Chair Professor of Computing Science with the National Subsea Centre, Robert Gordon University, Aberdeen, U.K. He has published over 300 peer reviewed journal/conferences papers. His research interests focus mainly on hyperspectral imaging, image processing, computer vision, big data analytics, and machine learning. He acts as an Associate Editor for several international journals, including IEEE Transactions on Geoscience and Remote Sensing and Journal of The Franklin Institute.
Calum MacLellan is currently pursuing the Ph.D. degree with the Department of Biomedical Engineering, University of Strathclyde. His research interests include biomedical image analysis and deep learning.

Huihui Li received the Ph.D. degree in computer science and engineering from the South China University of Technology, Guangzhou, China. She is currently a Lecturer with the School of Computer Science, Guangdong Polytechnic Normal University. Her research interests include image processing, machine learning, and affective computing.

Huimin Zhao was born in Shaanxi, China, in 1966. He received the B.Sc. and the M.Sc. degrees in signal processing from Northwestern Polytechnical University, Xi’an, China, in 1992 and 1997, respectively, and the Ph.D. degree in electrical engineering from the Sun Yat-sen University in 2001.
He is currently a Professor and the Dean of the School of Computer Sciences, Guangdong Polytechnic Normal University, Guangzhou, China. His research interests include image, video, and information security technology.

Amir Hussain received the B.Eng. (Highest First Class Hons. with Distinction) and Ph.D. degrees from the University of Strathclyde, Glasgow, U.K., in 1992 and 1997, respectively.
He is a Founding Director of the Centre of AI and Data Science, Edinburgh Napier University. He has led major national, EU and internationally funded projects and supervised over 30 Ph.D. students. He is currently a Chief Investigator for the COG-MHEAR programme grant funded under the EPSRC Transformative Healthcare Technologies 2050 Call. He has (co)authored three international patents and over 400 publications, including over 170 international journal papers, 20 books/monographs and over 100 book chapters. His research interests are cross-disciplinary and industry-led, aimed at developing cognitive data science and AI technologies, to engineer the smart and secure systems of tomorrow. He is a General Chair of IEEE WCCI 2020 (the world’s largest technical event in computational intelligence), a Vice-Chair of the Emergent Technologies Technical Committee of the IEEE Computational Intelligence Society (CIS), IEEE U.K. and Ireland Chapter Chair of the Industry Applications Society, and a (Founding) Vice-Chair for the IEEE CIS Task Force on Intelligence Systems for e-Health. He is the Founding Editor-in-Chief of two leading international journals: Cognitive Computation (Springer Nature), and Big Data Analytics (BMC). He is a member of the U.K. Computing Research Committee.

Giancarlo Fortino (Senior Member, IEEE) received the Ph.D. degree in computer engineering from the University of Calabria (Unical), Italy, in 2000, where he is a Full Professor of Computer Engineering with the Department of Informatics, Modeling, Electronics, and Systems. He is also a Distinguished Professor with the Wuhan University of Technology and Huazhong Agricultural University, China, a High-End Expert with HUST, China, a Senior Research Fellow with the ICAR-CNR Institute, and a CAS PIFI Visiting Scientist with SIAT, Shenzhen. He is the Director of the SPEME Lab, Unical, as well as a Co-Chair of Joint labs on IoT established between Unical and WUT and SMU and HZAU Chinese universities. He has authored over 430 papers in international journals, conferences, and books. His research interests include agent-based computing, wireless (body) sensor networks, and IoT. He is a Co-Founder and a CEO of SenSysCal S.r.l., a Unical spinoff focused on innovative IoT systems. He is a (Founding) Series Editor of IEEE Press Book Series on Human–Machine Systems and a EiC of Springer Internet of Things series and AE of many international journals, such as IEEE Transactions on Automatic Control, IEEE Transactions on Human–Machine Systems, IEEE Internet of Things Journal, IEEE Systems Journal, Information Fusion, Journal of Network and Computer Applications, Engineering Applications of Artificial Intelligence, and IEEE SMCM. He is currently member of the IEEE SMCS BoG and of the IEEE Press BoG, and a Chair of the IEEE SMCS Italian Chapter.
Funding Statement
This work was supported in part by the Dazhi Scholarship of the Guangdong Polytechnic Normal University; in part by the Key Laboratory of the Education Department of Guangdong Province under Grant 2019KSYS009; and in part by the National Natural Science Foundation of China under Grant 62072122 and Grant 62006049.
Footnotes
In this case, viral and bacterial pneumonias are grouped into a single ‘pneumonia’ class for comparing with images of the COVID-19 and healthy patients to give the three classes.
References
- [1].Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV), World Health Org., Geneva, Switzerland, 2005. [Google Scholar]
- [2].Who Director-General’s Opening Remarks at The Media Briefing on COVID-19-11 March 2020. World Health Org., Geneva, Switzerland, 2020. [Google Scholar]
- [3].Coronavirus Disease (COVID-19): Situation Report, 135, World Health Org., Geneva, Switzerland, 2020. [Google Scholar]
- [4].Mahase E., “Coronavirus covid-19 has killed more people than SARS and MERS combined, despite lower case fatality rate,” Brit. Med. J., vol. 368, p. m641, Feb. 2020. [DOI] [PubMed] [Google Scholar]
- [5].Wang C., Horby P. W., Hayden F. G., and Gao G. F., “A novel coronavirus outbreak of global health concern,” Lancet, vol. 395, no. 10223, pp. 470–473, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pan L.et al. , “Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: A descriptive, cross-sectional, multicenter study,” Amer. J. Gastroenterol., vol. 115, no. 5, pp. 766–773, May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fernandes N., “Economic effects of coronavirus outbreak (COVID-19) on the world economy,” IESE Bus. School, Barcelona, Spain, Working Paper, 2020. [Google Scholar]
- [8].Ren J.et al. , “A novel intelligent computational approach to model epidemiological trends and assess the impact of non-pharmacological interventions for COVID-19,” IEEE J. Biomed. Health Inform., vol. 24, no. 12, pp. 3551–3563, Dec. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Candace M. M. and Daniel . (2020). COVID-19. Accessed: Jun. 2020. [Online]. Available: https://radiopaedia.org/articles/covid-19-3?lang=us [Google Scholar]
- [10].Chan J. F.-W.et al. , “A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster,” Lancet, vol. 395, no. 10223, pp. 514–523, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huang C.et al. , “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yoon S. H.et al. , “Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): Analysis of nine patients treated in Korea,” Korean J. Radiol., vol. 21, no. 4, pp. 494–500, Apr. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shi F.et al. , “Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19,” IEEE Rev. Biomed. Eng., vol. 14, pp. 4–15, Apr. 2020. [DOI] [PubMed] [Google Scholar]
- [14].Chung M.et al. , “CT imaging features of 2019 novel coronavirus (2019-nCoV),” Radiology, vol. 295, no. 1, pp. 202–207, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schlegl T.et al. , “Fully automated detection and quantification of macular fluid in OCT using deep learning,” Ophthalmology, vol. 125, no. 4, pp. 549–558, 2018. [DOI] [PubMed] [Google Scholar]
- [16].Ragab D. A., Sharkas M., Marshall S., and Ren J., “Breast cancer detection using deep convolutional neural networks and support vector machines,” PeerJ, vol. 7, p. e6201, Jan. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Laukamp K. R.et al. , “Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI,” Eur. Radiol., vol. 29, no. 1, pp. 124–132, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rajpurkar P.et al. , “CheXNet: Radiologist-level pneumonia detection on chest X-rays with deep learning,” 2017. [Online]. Available: https://arXiv:1711.05225 [Google Scholar]
- [19].Karim M., Döhmen T., Rebholz-Schuhmann D., Decker S., Cochez M., and Beyan O., “DeepCOVIDExplainer: Explainable COVID-19 predictions based on chest X-ray images,” 2020. [Online]. Available: https://arXiv:2004.04582 [Google Scholar]
- [20].Deng J., Dong W., Socher R., Li L.-J., Li K., and Fei-Fei L., “ImageNet: A large-scale hierarchical image database,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Miami, FL, USA, 2009, pp. 248–255. [Google Scholar]
- [21].He K., Zhang X., Ren S., and Sun J., “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Las Vegas, NV, USA, 2016, pp. 770–778. [Google Scholar]
- [22].Bassi P. R. A. S. and Attux R., “A deep convolutional neural network for COVID-19 detection using chest X-rays,” 2020. [Online]. Available: https://arXiv:2005.01578 [Google Scholar]
- [23].Zhang Y.et al. , “COVID-DA: Deep domain adaptation from typical pneumonia to COVID-19,” 2020. [Online]. Available: https://arXiv:2005.01577 [Google Scholar]
- [24].Misra S., Jeon S., Lee S., Managuli R., and Kim C., “Multi-channel transfer learning of chest X-ray images for screening of COVID-19,” 2020. [Online]. Available: https://arXiv:2005.05576 [Google Scholar]
- [25].Yamac M., Ahishali M., Degerli A., Kiranyaz S., Chowdhury M. E. H., and Gabbouj M., “Convolutional sparse support estimator based COVID-19 recognition from X-ray images,” 2020. [Online]. Available: https://arXiv:2005.04014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].El Asnaoui K. and Chawki Y., “Using X-ray images and deep learning for automated detection of coronavirus disease,” J. Biomol. Struct. Dyn., vol. 39, no. 10, pp. 3615–3626, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rahimzadeh M. and Attar A., “A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2,” Informat. Med. Unlocked, vol. 19, May 2020, Art. no. 100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Toğaçar M., Ergen B., and Cömert Z., “COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches,” Comput. Biol. Med., vol. 121, Jun. 2020, Art. no. 103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lv D., Qi W., Li Y., Sun L., and Wang Y., “A cascade network for detecting COVID-19 using chest X-rays,” 2020. [Online]. Available: https://arXiv:2005.01468 [Google Scholar]
- [30].Gozes O.et al. , “Rapid AI development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis,” 2020. [Online]. Available: https://arXiv:2003.05037 [Google Scholar]
- [31].Sohaib A. and Yi W.. (2020). Automatic Detection of COVID-19 Using X-Ray Images With Deep Convolutional Neural Networks and Machine Learning. [Online]. Available: https://doi.org/10.1101/2020.05.01.20088211 [Google Scholar]
- [32].Wang S.et al. , “A deep learning algorithm using CT images to screen for Corona virus disease (COVID-19),” Eur. Radiol., vol. 31, no. 8, pp. 6096–6104, Aug. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shi F.et al. , “Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification,” 2020. [Online]. Available: https://arXiv:2003.09860 [DOI] [PubMed] [Google Scholar]
- [34].Ouyang X.et al. , “Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2595–2605, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [35].Kang H.et al. , “Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2606–2614, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [36].Ghoshal B. and Tucker A., “Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection,” 2020. [Online]. Available: https://arXiv:2003.10769 [Google Scholar]
- [37].Wang B.et al. , “AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system,” Appl. Soft Comput., vol. 98, pp. 106897–106897, Jan. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jin C.et al. , “Development and evaluation of an artificial intelligence system for COVID-19 diagnosis,” Nature Commun., vol. 11, no. 1, pp. 1–14, Oct. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang J., Xie Y., Li Y., Shen C., and Xia Y., “COVID-19 screening on chest X-ray images using deep learning based anomaly detection,” 2020. [Online]. Available: https://arXiv:2003.12338 [Google Scholar]
- [40].Wang L. and Wong A., “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images,” 2020. [Online]. Available: https://arXiv:2003.09871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wong A.et al. , “Towards computer-aided severity assessment via deep neural networks for geographic and opacity extent scoring of SARS-CoV-2 chest X-rays,” Sci. Rep., vol. 11, no. 1, pp. 1–8, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chattopadhay A., Sarkar A., Howlader P., and Balasubramanian V. N., “Grad-CAM++: Generalized gradient-based visual explanations for deep convolutional networks,” in Proc. IEEE Winter Conf. Appl. Comput. Vis. (WACV), Lake Tahoe, NV, USA, 2018, pp. 839–847. [Google Scholar]
- [43].Wehbe R. M.et al. , “DeepCOVID-XR: An artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large U.S. clinical data set,” Radiology, vol. 299, no. 1, pp. E167–E176, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wong H. Y. F.et al. , “Frequency and distribution of chest radiographic findings in patients positive for COVID-19,” Radiology, vol. 296, no. 2, 2020, Art. no. 201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Warren M. A.et al. , “Severity scoring of lung oedema on the chest radiograph is associated with clinical outcomes in ARDS,” Thorax, vol. 73, no. 9, pp. 840–846, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tabik S.et al. , “COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest X-ray images,” IEEE J. Biomed. Health Inform., vol. 24, no. 12, pp. 3595–3605, Dec. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li L.et al. , “Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy,” Radiology, vol. 296, no. 2, pp. E65–E71, Mar. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Javaheri T.et al. , “CovidCTNet: An open-source deep learning approach to identify COVID-19 using CT image,” 2020. [Online]. Available: https://arXiv:2005.03059 [Google Scholar]
- [49].Wang J.et al. , “Prior-attention residual learning for more discriminative COVID-19 screening in CT images,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2572–2583, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [50].Roy S.et al. , “Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2676–2687, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [51].Bhattacharya S.et al. , “Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey,” Sustain. Cities Soc., vol. 65, Feb. 2020, Art. no. 102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chi C., Zhang S., Xing J., Lei Z., Li S. Z., and Zou X., “Selective refinement network for high performance face detection,” in Proc. AAAI Conf. Artif. Intell., vol. 33, 2019, pp. 8231–8238. [Google Scholar]
- [53].Sun G.et al. , “Deep fusion of localized spectral features and multi-scale spatial features for effective classification of hyperspectral images,” Int. J. Appl. Earth Observ. Geoinf., vol. 91, Sep. 2020, Art. no. 102157. [Google Scholar]
- [54].Fang Z.et al. , “Triple loss for hard face detection,” Neurocomputing, vol. 398, pp. 20–30, Jul. 2020. [Google Scholar]
- [55].Huang G., Liu Z., Van Der Maaten L., and Weinberger K. Q., “Densely connected convolutional networks,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Honolulu, HI, USA, 2017, pp. 4700–4708. [Google Scholar]
- [56].Gao S.-H., Cheng M.-M., Zhao K., Zhang X.-Y., Yang M.-H., and Torr P. H., “Res2Net: A new multi-scale backbone architecture,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 43, no. 2, pp. 652–662, Feb. 2021. [DOI] [PubMed] [Google Scholar]
- [57].Belongie S., Malik J., and Puzicha J., “Shape matching and object recognition using shape contexts,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 24, no. 4, pp. 509–522, Apr. 2002. [DOI] [PubMed] [Google Scholar]
- [58].Lowe D. G., “Distinctive image features from scale-invariant keypoints,” Int. J. Comput. Vis., vol. 60, no. 2, pp. 91–110, 2004. [Google Scholar]
- [59].Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K. N., and Mohammadi A., “COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images,” 2020. [Online]. Available: https://arXiv:2004.02696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rey-Area M., Guirado E., Tabik S., and Ruiz-Hidalgo J., “FuCiTNet: Improving the generalization of deep learning networks by the fusion of learned class-inherent transformations,” 2020. [Online]. Available: https://arXiv:2005.08235 [Google Scholar]
- [61].Paszke A.et al. , “Automatic differentiation in PyTorch,” in Proc. 31st Conf. Neural Inf. Process. Syst., 2017, pp. 1–4. [Google Scholar]
- [62].Kingma D. P. and Ba J., “Adam: A method for stochastic optimization,” 2014. [Online]. Available: https://arXiv:1412.6980 [Google Scholar]
- [63].Loshchilov I. and Hutter F., “SGDR: Stochastic gradient descent with warm restarts,” 2016. [Online]. Available: https://arXiv:1608.03983 [Google Scholar]
- [64].Lin T.-Y., Dollár P., Girshick R., He K., Hariharan B., and Belongie S., “Feature pyramid networks for object detection,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Honolulu, HI, USA, 2017, pp. 2117–2125. [Google Scholar]
- [65].Fang Z., Ren J., Marshall S., Zhao H., Wang S., and Li X., “Topological optimization of the densenet with pretrained-weights inheritance and genetic channel selection,” Pattern Recognit., vol. 109, Jan. 2021, Art. no. 107608. [Google Scholar]