1. Purpose
Image segmentation is the process of delineating the regions occupied by objects of interest in a given image. This operation is a fundamentally required first step in numerous applications of medical imagery. In the medical imaging field, this activity has a rich literature that spans over 45 years. In spite of numerous advances, including deep learning (DL) networks (DLNs) in recent years, the problem has defied a robust, fail-safe, and satisfactory solution, especially for objects that are manifest with low contrast, are spatially sparse, have variable shape among individuals, or are affected by imaging artifacts, pathology, or post-treatment change in the body. Although image processing techniques, notably DLNs, are uncanny in their ability to harness low-level intensity pattern information on objects, they fall short in the high-level task of identifying and localizing an entire object as a gestalt. This dilemma has been a fundamental unmet challenge in medical image segmentation.
In this paper, we demonstrate that by synergistically marrying the unmatched strengths of high-level human knowledge (i.e., natural intelligence (NI)) with the capabilities of DL networks (i.e., artificial intelligence (AI)) in garnering intricate details, these challenges can be significantly overcome. Focusing on the object recognition task, we formulate an anatomy-guided DL object recognition approach named Automatic Anatomy Recognition-Deep Learning (AAR-DL) which combines an advanced anatomy-modeling strategy, model-based non-DL object recognition, and DL object detection networks to achieve expert human-like performance.
2. Methods
Outline of paper and approach
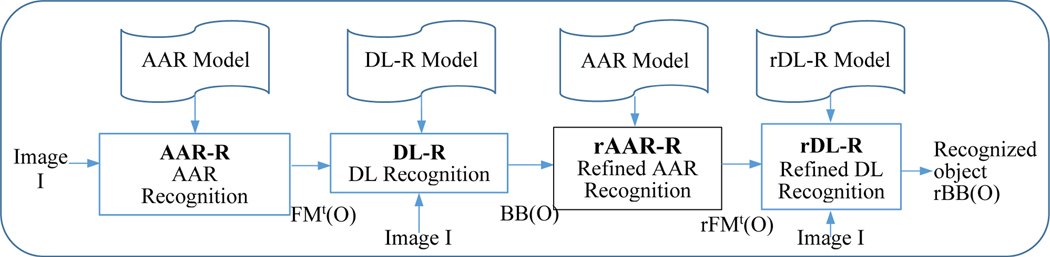
A schematic representation of the proposed approach, which we will refer to as AAR-DL, is shown in Figure 1. The AAR-DL approach consists of 4 stages which are shown by 4 modules in the figure – Automatic Anatomy Recognition-Recognition (AAR-R), Deep Learning-Recognition (DL-R), refined Automatic Anatomy Recognition-Recognition (rAAR-R), and refined Deep Learning-Recognition (rDL-R).
Figure 1.
A schematic representation of the proposed AAR-DL object recognition framework.
The first module AAR-R performs recognition of objects in a given body region B via the AAR approach [1] by making use of the already created anatomy model of the objects in B. It outputs a fuzzy model mask FMt(O), a transformed version of the original fuzzy model FM(O), for each object O included in the AAR anatomy model, which indicates where object O is likely to be in input image I. FMt(O) provides the needed region-of-interest (ROI) information in the form of a container, shaped like O, albeit fuzzy, for the second module DL-R. DL-R outputs a stack BB(O) of 2D bounding boxes (BBs) for each O by detecting the BBs in each slice within the ROI for O. In the third module rAAR-R, the AAR fuzzy model is refined by making use of the more precise localization information in BB(O) provided by DL-R. rAAR-R deforms the fuzzy model and outputs a refined recognition result (fuzzy model) rFMt(O) for each object O. Finally, the fourth module rDL-R, performs refined DL-based recognition by utilizing the refined fuzzy model rFMt(O) as an additional input channel and outputs a refined stack of 2D BBs, rBB(O), for each object O. This module has its own pre-trained network which is denoted rDL-R model.
Automatic Anatomy Recognition-Recognition (AAR-R module)
AAR is a general approach [1,9], developed before the advent of DL techniques, based on fuzzy anatomy modeling for recognizing and delineating all objects in a body region. It consists of three stages – model building, object recognition, and object delineation. Since the goal of this paper is object recognition, here we are concerned with only model building and recognition processes of AAR.
Deep Learning-Recognition (DL-R module)
DL-R uses a 3-channel image and the recognition mask FMt(O) output by AAR-R to determine the region for search for 2D BBs, and outputs a stack of 2D BBs, denoted BB(O), for each object O as described in this section. DL-R takes a slice-by-slice approach, and as such, the three channels represent the slice in question and the two neighboring slices in the superior and inferior directions.
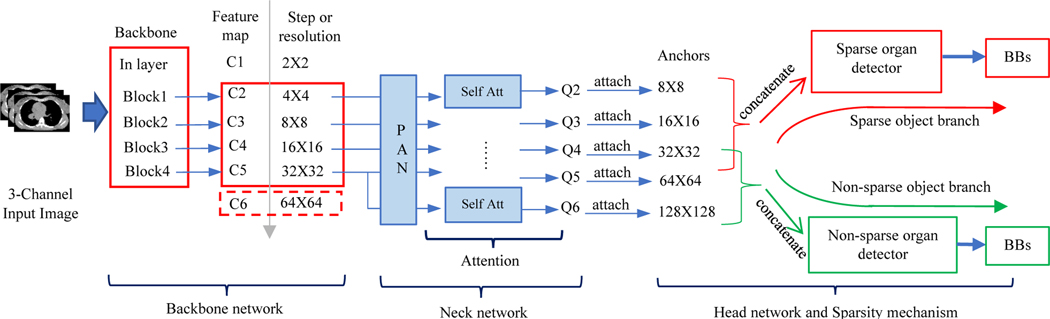
Considering accuracy, efficiency, and flexibility, our recognition network design is inspired by one-stage dense detection networks including RetinaNet [2] and YOLO [3, 4]. In order to capture richer contextual dependencies, an optimized attention mechanism is also incorporated into our recognition network. Target objects are separated into two groups: sparse and non-sparse. Spatially less compact and less filling, thin, tubular, and small objects, such as the salivary glands, thoracic esophagus, and thoracic spinal cord, form the sparse group. Large, blob-like, and spatially compact objects, such as the lungs and heart, form the non-sparse group. For non-sparse objects, DL-R cares more about semantic and abstract information from higher/deeper levels of the neural network. Conversely, the network cares more about detailed structural information from lower/shallow levels of the network for sparse objects. Thus, anatomic knowledge (NI) is infused into the network architecture at the design stage itself. The overall network architecture is depicted in Figure 2.
Figure 2.
Overall architecture of the object sparsity-specific recognition network employed in DL-R.
Although feature maps directly generated from the backbone network contain information from different levels, they are not sufficient for effective object recognition which involves classification (“what it is”) and regression (“where it is”) tasks. The neck network further integrates and refines these feature maps. A neck network is usually composed of several bottom-up paths and several top-down paths. Networks that utilize this mechanism include Feature Pyramid Network (FPN) [5], Path Aggregation Network (PAN) [6], weighted Bi-directional Feature Pyramid Network (BiFPN) [7], and Neural Architecture Search Feature Pyramid Network (NAS-FPN) [8]. To improve the performance of feature maps, FPN extracts in-network feature hierarchy, with a top-down path as well as lateral connections to propagate semantically strong features. It is known that neurons in higher layers strongly respond to entire objects while those in lower layers are more likely to be activated by local texture and patterns. To recognize a sparse object like trachea or proximal bronchial tree, not only local information but also information from deeper network layers is needed, since the locations of these structures have a close relationship to the lungs around them.
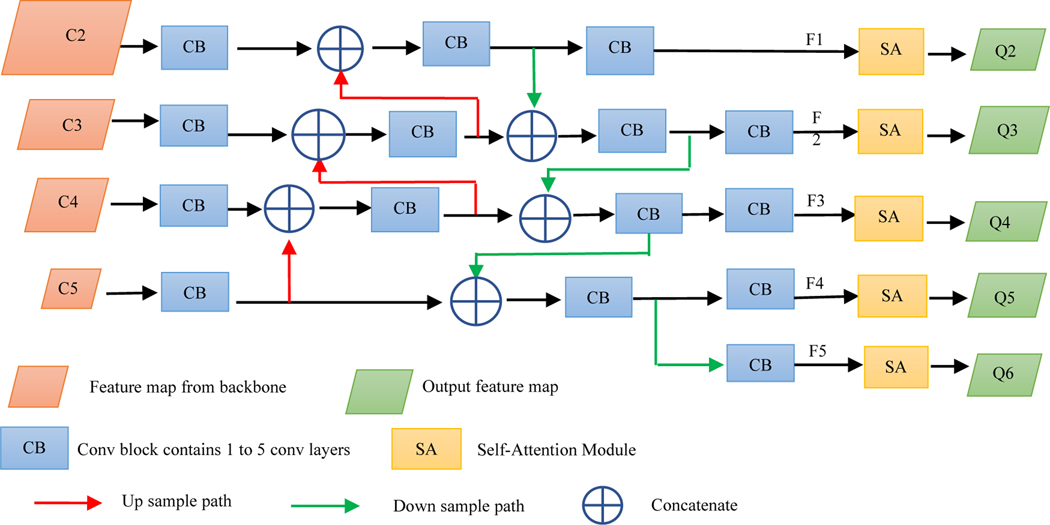
In order to further enhance the recognition capability of the entire neck network by propagating strong responses of low-level patterns, instead of the FPN and inspired by PAN, we build a path with clean lateral connections from the low level to top level. Figure 3 shows the complete structure of our neck network. This design is based on our desire that accurate recognition of non-sparse objects also requires detailed local information, especially at the object edges and corners. The lungs in 2D slices usually have sharp corners anteriorly in the superior slices. To localize lungs including such sharp angles accurately, local information from lower layers is also required. The complementary information gathered in different layers from the backbone network is fully exploited by the neck network. Each building block takes a higher resolution feature map and a coarser map through lateral connections and generates the new feature map Fi.
Figure 3.
Complete structure of the neck network.
Object recognition in medical images is often more challenging than in natural images. However, in medical images there is abundant prior information and there are reasonable assumptions that can be made to further improve recognition performance. For example, parotid glands and submandibular glands almost always occur in pairs, but are separated by a certain distance. Attention mechanisms are designed to take advantage of such information. The superiority of attention mechanisms has been demonstrated especially in complicated segmentation applications.
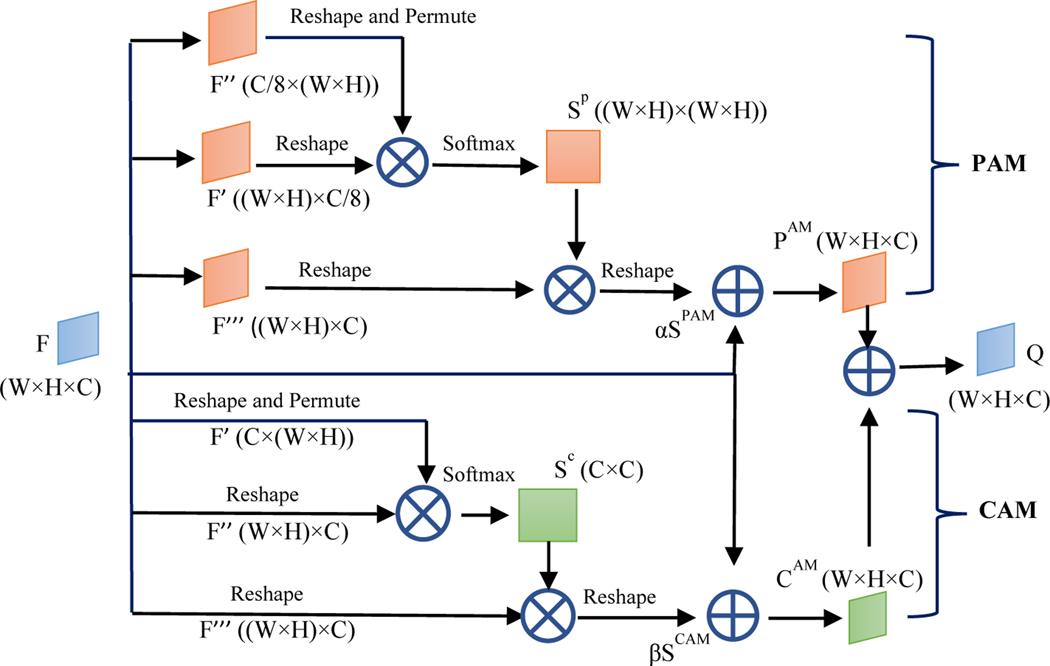
In this paper, we apply a Dual Attention Network to adaptively integrate local features with their global dependencies further for each Fi. We call this attention network Self Attention (SA) module given that it involves attention inside each feature map Fi. In the SA module, an attention mechanism is implemented in spatial and channel dimensions of each feature map. The spatial attention component selectively aggregates the feature at each location by a weighted sum of the features at all locations. Similar features have connections to each other regardless of their distances. Channel attention selectively emphasizes channel interdependence by integrating associated features between channels. At the design core of convolutional neural network (CNN), the receptive field in a convolutional operation is a local neighborhood operator. This imposes a limitation on modeling global/expansive and richer contextual representation. Owing to the complexity of human anatomy, organs are not related to just the adjacent tissues alone. The SA module enables finding any tissues that have potential connection with the target organ despite their distance. Each feature map contains spatial and channel dimension information. In order to fully process the feature maps, the SA module consists of Position Attention Module (PAM) and Channel Attention Module (CAM) as depicted in Figure 4.
Figure 4.
The structure of the SA module.
For PAM, F is an input feature map of size (W, H, C) from the previous step, denoting the width, height, and number of channels, respectively. To make Figure 4 clearer, we replace Fi by F. F is passed through a convolution layer to generate two new feature maps F’ and F’’ of size (W, H, C/8). They are reshaped to size ((W × H), C/8) and (C/8, (W × H)). Then a matrix multiplication is performed between the two feature maps and a softmax layer is attached to generate the spatial attention map Sp with size ((W × H), (W × H)). Meanwhile, F is passed through another convolution layer and reshaped into F’’’ of size ((W × H), C). Then, a matrix multiplication is performed between F’’’ and Sp to obtain a new ((W × H), C) feature map. Subsequently, a map Spam is generated with the shape as the input feature map F by a reshape operation. Finally, a trainable parameter α is attached to Spam and an element-wise sum operation with feature map F is performed. The attention feature map PAM of PAM is generated as:
where α is initialized to 0. Gradually, the network learns to increase α to enhance the importance of spatial attention. α allows the network to first rely on the cues in the local neighborhood which are easier to train, and then to gradually learn to assign more weight to the non-local cues.
For CAM, the network is similar to PAM but more simplified. Different from PAM, without the convolution operation, input feature map F is reshaped into F’ and F’’ of size (C, (W × H)) and ((W × H), C), respectively. Then a matrix multiplication is performed between them and a softmax operation is followed to generate SC which has size (C, C). Meanwhile, F is passed through another convolution layer and reshaped into F’’’ of size ((W × H), C). Then, a matrix multiplication is performed between SC and F’’’ to obtain a new feature map of size ((W × H), C). Subsequently, a map Scam is generated with the shape as the input feature map F by a reshape operation. Finally, the attention feature map CAM of CAM is generated in the same way as PAM:
Similar to α in PAM, β is a trainable parameter and initialized to 0, and gradually, the network learns to increase β. Finally, the output of the SA module is the sum of the two attention feature maps:
2.2.3. Object sparsity and head network
To detect objects in natural images, usually one has to search the whole image, since objects may appear at any position with any pose. In medical images, however, target organ characteristics such as position, size, shape, and sparsity are relatively fixed and known a priori. This information can be used as prior knowledge to improve recognition performance. AAR-DL recognizes multiple objects simultaneously by dividing target objects into two groups according to their sparsity. By observing the structure of the DL-R network shown in Figure 2, prediction maps denoted Q2-Q6 are generated by the neck network which have different resolutions and are sensitive to organs with different sizes. Since the sparsity of target organs is known, there is no need to take all feature maps to detect a specific object. More feature maps will not improve the recognition performance, but may introduce noise or even lead to wrong classification.
In order to improve the accuracy and efficiency of DL-R, organs with different sparsity take different prediction maps to perform classification. On the one hand, non-sparse organs are recognized using maps Q4, Q5, and Q6 associated with anchors with base sizes 32×32, 64×64, and 128×128, respectively. Larger receptive fields and semantically stronger information from higher level layers are crucial for recognizing non-sparse organs. On the other hand, sparse organs are recognized using maps Q2, Q3, and Q4 associated with anchors with base sizes 8×8, 16×16, and 32×32, respectively. More detailed structural and spatial characteristics are important to recognize sparse organs. In order to get a denser prediction, recognition candidate cells are then augmented with anchors with different aspect ratio and scale based on anchor base sizes. Lastly, the head network which contains only convolution layers predicts the category and location of target organs based on corresponding prediction maps and anchors.
Refined AAR-Recognition (rAAR-R module)
AAR model building and recognition are performed 3-dimensionally. AAR models are population models, and as such they may not match the detailed intensity intricacies seen in the input image I. However, the DL-R process is exceptional in matching the intricacies, when present, but runs into difficulty when the details are compromised due to artifacts, pathology, or post-treatment change. Thus, the idea of combining the information present in the AAR model FMt(O) and DL-R output BB(O) is to merge the best evidence from the two sources (NI and AI) to create the modified model mFMt(O). The rAAR-R module uses mFMt(O) as the initial model for performing the AAR recognition operation again in place of the initialization done via one-shot recognition for the AAR-R module. The output of this recognition process is a refined fuzzy model rFMt(O) that “agrees” with the DL-R output BB(O) as well as AAR modeling and recognition principles.
Refined DL-Recognition (rDL-R module)
The rDL-R module uses the same DL-R architecture as described above, with one difference. It now uses a 4-channel input – 3 gray-level channels, plus the fuzzy recognition map contained in the slices of the modified fuzzy mask produced as explained in the previous section.
3. Results
This retrospective study was conducted following approval from the Institutional Review Board at the Hospital of the University of Pennsylvania along with a Health Insurance Portability and Accountability Act waiver. Utilizing 225 neck computed tomography (CT) data sets of cancer patients undergoing routine radiation therapy planning, the recognition performance of the AAR-DL approach is evaluated 16 neck organs in comparison to pure model-based (AAR-R) and pure DL approaches without anatomy guidance. Recognition accuracy is assessed via location error/ centroid distance error, scale or size error (ratio of estimated size to true size with the ideal value being 1), and wall distance error.
We compared recognition accuracies among four different strategies listed below corresponding to the 4 modules identified in Figure 1 taken in different sequences. AAR-R: The basic AAR recognition method [9] on its own; DL-R: DL-based recognition on its own without help from AAR-R; AAR-R-DL-R: The first stage of anatomy-guided DL recognition with output BB(O); rAAR-R-DL-R: The full sequence of operations for the AAR-DL approach of anatomy-guided DL recognition, involving the first stage and the second refined stage, with output rBB(O).
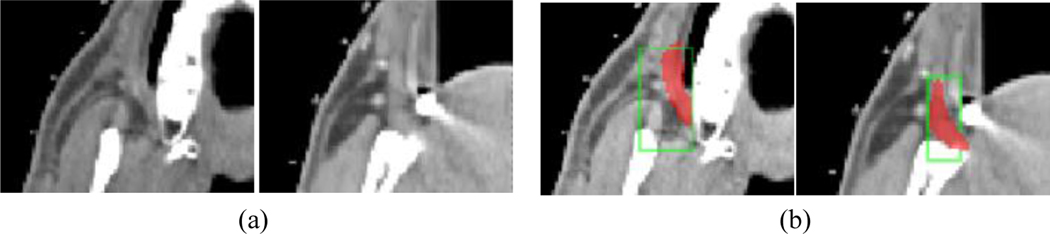
The results summarized in Table 1 demonstrate how errors are gradually and systematically reduced from the first module to the fourth module as high-level knowledge is infused via NI at various stages into the processing pipeline. This improvement is especially dramatic for sparse and artifact-prone challenging objects (where simple DL-R simply fails), achieving a location error over all objects of 4.3 mm for the neck body region. Figure 5 shows an example of extremely challenging cases where the results exhibit human-like decision making, which we firmly believe is due to the combination of NI and AI synergistically. Object abbreviations: CtSC – cervical spinal cord; CtEs – cervical esophagus; Mnd – mandible; OHPh – orohypopharynx constrictor muscle; SpGLx – supraglottic/glottic larynx; LPG & RPG – left & right parotid gland; LSmG & RSmG – left & right submandibular gland; LBMc & LBMc – left and right Buccal mucosa; TG – thyroid gland; eOC – extended oral cavity; Lp – lips; CtTr – cervical trachea; CtBrStm – cervical brainstem. Our comparison with methods from the literature shows that AAR-DL outperforms them all, and substantially for sparse, artifact-prone, and pathology-prone objects.
Table 1.
Error in recognition, location error (LE) and scale error (SE), for neck objects for the 4 different methods. Mean (1st entry) & standard deviation (2nd entry) values over the tested samples are shown. Error metrics over all objects are also listed (last column). For each method, the P-values of the paired t-test comparing the method to the other 3 methods for each object are also listed as S = significant (P ≤ 0.05) or NS = non-significant (P > 0.05). Entries “-” mean that DL-R failed and hence there was no metric or P value.
| Mnd | CtEs | CtSC | LPG | RPG | LSmG | RSmG | OHPh | SpGLx | CtBrStm | CtTr | eOC | LBMc | RBMc | LP | TG | All | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAR-R | LE (mm) | 16.3 5.0 |
13.9 7.9 |
9.9 7.5 |
18.7 13.2 |
18.7 10.4 |
11.7 10.8 |
13.9 10.5 |
18.1 9.8 |
15.7 11.0 |
13.3 6.8 |
10.7 7.0 |
12.7 3.3 |
10.5 8.0 |
11.7 7.5 |
8.9 8.1 |
15.9 10.7 |
13.7 8.6 |
| SE | 1.00 0.06 |
0.77 0.12 |
0.92 0.08 |
1.38 0.17 |
1.37 0.17 |
1.09 0.26 |
0.97 0.18 |
1.33 0.17 |
0.93 0.16 |
1.32 0.24 |
0.74 0.11 |
1.35 0.14 |
1.24 0.21 |
1.20 0.21 |
1.07 0.14 |
1.01 0.16 |
1.10 0.16 |
|
| DL-R | LE (mm) | 6.6 4.1 |
5.9 5.6 |
13.1 9.8 |
- | - | - | - | 8.9 6.8 |
5.5 3.1 |
4.8 2.5 |
6.9 6.1 |
3.9 3.0 |
- | - | 4.8 4.4 |
6.5 5.8 |
6.7 5.1 |
| SE | 0.92 0.03 |
0.87 0.17 |
0.51 0.30 |
- | - | - | - | 0.92 0.08 |
1.01 0.16 |
1.01 0.07 |
0.95 0.10 |
1.06 0.05 |
- | - | 0.96 0.07 |
0.99 0.16 |
0.92 0.12 |
|
| AAR-R-DL-R | LE (mm) | 5.4 5.2 |
4.7 5.8 |
2.3 1.3 |
5.0 2.8 |
5.0 3.1 |
4.7 4.5 |
5.4 3.5 |
5.5 7.4 |
5.6 3.00 |
2.4 1.1 |
5.5 5.9 |
4.0 3.1 |
5.4 5.9 |
6.9 4.0 |
4.6 2.4 |
6.5 5.7 |
4.9 3.9 |
| SE | 0.95 0.06 |
0.94 0.14 |
0.99 0.02 |
0.89 0.08 |
0.90 0.09 |
0.78 0.14 |
0.84 0.12 |
0.96 0.13 |
0.95 0.08 |
0.97 0.07 |
0.98 0.09 |
0.99 0.03 |
0.76 0.16 |
0.77 0.11 |
0.94 0.07 |
0.98 0.14 |
0.91 0.09 |
|
| rAAR-R-DL-R | LE (mm) | 5.4 5.7 |
5.0 3.2 |
2.2 1.5 |
3.4 3.0 |
2.9 2.6 |
3.7 5.7 |
3.5 2.4 |
5.6 7.6 |
4.6 2.9 |
2.0 1.0 |
5.3 5.4 |
4.0 3.3 |
3.2 4.6 |
5.4 6.7 |
3.6 1.8 |
8.8 8.9 |
4.3 4.1 |
| SE | 0.92 0.03 |
0.91 0.18 |
0.99 0.05 |
1.06 0.11 |
1.12 0.11 |
0.91 0.19 |
0.92 0.12 |
0.91 0.13 |
0.97 0.14 |
1.09 0.10 |
1.10 0.07 |
1.06 0.05 |
0.89 0.13 |
0.85 0.13 |
0.97 0.05 |
0.90 0.07 |
0.97 0.10 |
Figure 5.
Illustration of the recognition of an exceptionally challenging object. (a) CT slices from two patients at different locations that contain right buccal mucosa (RBMc). (b) The BBs detected by rAAR-R-DL-R and the corresponding ground truth delineations (red) for RBMc.
4. Conclusions
(i) High-level anatomy guidance improves recognition performance of DL methods. (ii) This improvement is especially noteworthy for spatially sparse, low-contrast, inconspicuous, and artifact-prone objects. (iii) Once anatomy guidance is provided, 3D objects can be detected much more accurately via 2D BBs than 3D BBs, and the 2D BBs represent object containment with much more specificity. (iv) Anatomy guidance brings stability and robustness to DL approaches for object localization. (v) The training time can be greatly reduced by making use of anatomy guidance.
Acknowledgements
This work is supported by a grant from the National Cancer Institute R42 CA199735.
5. References
- [1].Udupa JK, Odhner D, Zhao L, Tong Y, Matsumoto MMS, & Ciesielski KC, Falcao A,X, Vaideeswaran P, Ciesielski V, Saboury B, Mohammadianrasanani S, Sin S, Arens R, Torigian DA. (2014). Body-wide hierarchical fuzzy modeling, recognition, and delineation of anatomy in medical images. Medical Image Analysis, 18(5): 752–771, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lin TY, Goyal P, Girshick R, He K, & Dollár Piotr. (2017). Focal loss for dense object detection. IEEE Transactions on Pattern Analysis & Machine Intelligence, 99: 2999–3007, 2017. [DOI] [PubMed] [Google Scholar]
- [3].Redmon J, & Farhadi A (2018). Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767, 2018. [Google Scholar]
- [4].Bochkovskiy A, Wang CY, & Liao HYM (2020). YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv preprint arXiv:2004.10934, 2020. [Google Scholar]
- [5].Lin TY, Dollár P, Girshick R, He K, Hariharan B, & Belongie S, 2017. Feature pyramid networks for object detection. In Proceedings of the IEEE conference on computer vision and pattern recognition, 2117–2125. [Google Scholar]
- [6].Liu S, Qi L, Qin H, Shi J, & Jia J, 2018. Path aggregation network for instance segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition: 8759–8768. [Google Scholar]
- [7].Tan M, Pang R, & Le QV 2020. Efficientdet: Scalable and efficient object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 10781–10790, 2020. [Google Scholar]
- [8].Ghiasi G, Lin TY, & Le QV, 2019. Nas-fpn: Learning scalable feature pyramid architecture for object detection, In Proceedings of the IEEE conference on computer vision and pattern recognition: 7036–7045. [Google Scholar]
- [9].Wu X, Udupa JK, Tong Y, Odhner D, Pednekar GV, Simone II CB, McLaughlin D, Apinorasethkul C, Lukens J, Mihailidis D, Shammo G, James P, Camaratta J, Torigian DA. AAR-RT - A system for auto-contouring organs at risk on CT images for radiation therapy planning: Principles, design, and large-scale evaluation on head-and-neck and thoracic cancer cases, Medical Image Analysis, 54: 45–62, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]