| Entry |
|
|---|
| Name |
Novobiocin biosynthesis - Rhodocyclaceae bacterium Thauera-like |
|---|
| Class |
Metabolism; Biosynthesis of other secondary metabolites
|
|---|
| Pathway map |
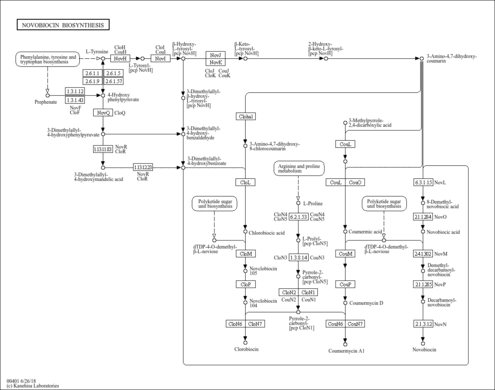
|
|---|
| Other DBs |
|
|---|
| Organism |
Rhodocyclaceae bacterium Thauera-like [GN: rbh]
|
|---|
| Gene |
|
|---|
| Compound |
| C01179 | 3-(4-Hydroxyphenyl)pyruvate |
| C12456 | 3-Dimethylallyl-4-hydroxyphenylpyruvate |
| C12457 | 3-Dimethylallyl-4-hydroxymandelic acid |
| C12458 | 3-Dimethylallyl-4-hydroxybenzoate |
| C12459 | 3-Dimethylallyl-4-hydroxybenzaldehyde |
| C12460 | 3-Dimethylallyl-beta-hydroxy-L-tyrosyl-[pcp] |
| C12462 | beta-Hydroxy-L-tyrosyl-[pcp] |
| C12463 | beta-Keto-L-tyrosyl-[pcp] |
| C12467 | 2-Hydroxy-beta-keto-L-tyrosyl-[pcp] |
| C12468 | 3-Amino-4,7-dihydroxycoumarin |
| C12469 | 3-Amino-4,7-dihydroxy-8-chlorocoumarin |
| C12475 | Demethyldecarbamoylnovobiocin |
| C12477 | 3-Methylpyrrole-2,4-dicarboxylic acid |
| C12481 | dTDP-5-dimethyl-L-lyxose |
| C12483 | Pyrrole-2-carbonyl-[pcp] |
| C20629 | 8-Demethylnovobiocic acid |
| C20637 | 5-[[(4,7-Dihydroxy-2-oxo-2H-1-benzopyran-3-yl)amino]carbonyl]-4-methyl-1H-pyrrole-3-carboxylate |
|
|---|
| Reference |
|
|---|
| Authors |
Pojer F, Li SM, Heide L. |
|---|
| Title |
Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Chen H, Walsh CT. |
|---|
| Title |
Coumarin formation in novobiocin biosynthesis: beta-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Schmutz E, Steffensky M, Schmidt J, Porzel A, Li SM, Heide L. |
|---|
| Title |
An unusual amide synthetase (CouL) from the coumermycin A1 biosynthetic gene cluster from Streptomyces rishiriensis DSM 40489. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Xu H, Kahlich R, Kammerer B, Heide L, Li SM. |
|---|
| Title |
CloN2, a novel acyltransferase involved in the attachment of the pyrrole-2-carboxyl moiety to the deoxysugar of clorobiocin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Xu H, Wang ZX, Schmidt J, Heide L, Li SM. |
|---|
| Title |
Genetic analysis of the biosynthesis of the pyrrole and carbamoyl moieties of coumermycin A1 and novobiocin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Freel Meyers CL, Oberthur M, Xu H, Heide L, Kahne D, Walsh CT |
|---|
| Title |
Characterization of NovP and NovN: completion of novobiocin biosynthesis by sequential tailoring of the noviosyl ring. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pojer F, Kahlich R, Kammerer B, Li SM, Heide L. |
|---|
| Title |
CloR, a bifunctional non-heme iron oxygenase involved in clorobiocin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Li SM, Westrich L, Schmidt J, Kuhnt C, Heide L. |
|---|
| Title |
Methyltransferase genes in Streptomyces rishiriensis: new coumermycin derivatives from gene-inactivation experiments. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pojer F, Wemakor E, Kammerer B, Chen H, Walsh CT, Li SM, Heide L. |
|---|
| Title |
CloQ, a prenyltransferase involved in clorobiocin biosynthesis. |
|---|
| Journal |
|
|---|
Related
pathway |
| rbh00330 | Arginine and proline metabolism |
| rbh00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| rbh00523 | Polyketide sugar unit biosynthesis |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|