Abstract
We generated the PEDE (Pig EST Data Explorer; http://pede.dna.affrc.go.jp/) database using se quences assembled from porcine 5′ ESTs from oligo-capped full-length cDNA libraries. Thus far we have performed EST analysis of various organs (thymus, spleen, uterus, lung, liver, ovary and peripheral blood mononuclear cells) and assembled 68 076 high-quality sequences into 5546 contigs and 28 461 singlets. PEDE provides a search interface for getting results of homology searches and enables users to obtain information on sequence data and cDNA clones of interest. Single-nucleotide polymorphisms detected through comparison of the EST sequences are classified by origin (western and oriental breeds) and are searchable in the database. This database system can accelerate analyses of livestock traits and yields information that can lead to new applications in pigs as model systems for medical research.
INTRODUCTION
To improve our understanding of livestock productivity and enhance livestock quality, genes expressed in pigs have been investigated intensely. Moreover, porcine genomic information is gaining in importance because the pig is considered a promising animal model in regenerative medicine and a producer of useful agents in light of recent progress in cloning techniques (1). These researches in pigs require extensive knowledge of the genes in the porcine genome. Because expressed sequence tag (EST) analysis is an effective method for collecting expressed genes, several groups have been accumulating porcine ESTs, and nearly 140 000 sequences had been registered in the DDBJ/EMBL/GenBank database by the end of July 2003. However, expressed mRNA sequences that encode full-length coding sequences (CDSs) can rarely be obtained by the usual cDNA synthesis methods, and assembled EST sequences have revealed relatively few full-length protein sequences expressed in pigs. In contrast, cDNA clones containing full-length CDSs are very beneficial for analyses using protein products translated from clones, such as the preparation of antibodies against porcine antigens. To catalogue the full-length mRNA sequences expressed in pigs, we constructed libraries enriched for full-length cDNA sequences from various porcine tissues and used these libraries in EST analyses. We stored the ESTs obtained and the results of homology searches with them in a database with a web interface.
CONSTRUCTION OF PORCINE FULL-LENGTH CDNA LIBRARIES AND EST ANALYSIS
Tissues for the construction of the cDNA libraries were prepared from crossbred [(Landrace × Large White) × Duroc] pigs, which are representative of those in the Japanese pork market, and Meishan animals, a breed representative of those in China, that were housed at the National Institute of Livestock and Grassland Science (Tsukuba, Ibaraki, Japan). The cDNA libraries were constructed according to the oligo-capped method (2) and cloned unidirectionally into the pCMVFL3 vector (Invitrogen, Carlsbad, CA). The average estimated length of the inserts in the libraries is 1.5 kb. Thus far, cDNA libraries of thymus, spleen, peripheral blood mononuclear cells, uterus, ovary, liver and lung derived from crossbred pigs and those of liver and ovary from Meishan pigs have been subjected to single-pass sequencing from the 5′-ends. The EST reads obtained (83 564 by the end of May 2003) have undergone base calling using Phred (3,4), and the vector sequences have been screened by using the cross-match program in the Phrap package (P. Green, unpublished). The average length of valid data from the reads (excluding vector sequences) with Phred QV > 20 was 627 bases. Repetitive sequences and low-complexity regions [e.g. poly(A) tracts] in the reads were screened by using RepeatMasker (A. Smit, unpublished) and in-house-generated Perl scripts. Clustering and assembling of sequences were performed using the TGICL package (5) with the CAP3 (6) option ‘-f 30’ to separate the reads of alternatively spliced products from contigs to increase the accuracy of detecting single-nucleotide polymorphisms (SNPs). The sequences of 68 076 high-quality reads were clustered and assembled into 5546 contigs and 28 461 singlets, and details of the sequencing status and assembly are shown in Supplementary table 1 (http://pede.dna.affrc.go.jp/supplement/suppl_table1.php).
CHARACTERISTICS OF THE cDNA LIBRARIES AND ESTS
We subjected the contigs and singlets that were not included in the contigs obtained by assembly of the EST reads (designated PEDE assemblies) to homology searches using BLAST (7) with the UniGene clusters of humans, mice, cattle and pigs in GenBank. The human and mouse deduced protein sequences in RefSeq (8) were also used in the BLAST searches. The PEDE assemblies that possessed high similarity (BLAST score ≥ 50) with sequences including putative CDSs of the UniGene clusters in the correct direction were 4370 contigs and 11 079 singlets (Tables 1 and 2). Assemblies with sequences longer than the corresponding CDS of the UniGene clusters upstream of the aligned regions with high similarity are considered to include full-length CDSs. According to this standard, we estimate that 3579 contigs and 7920 singlets in the PEDE assemblies possess the full-length CDS described in the UniGene clusters and represent at least 5856 different genes.
Table 1. PEDE assemblies estimated to contain full-length CDSs in light of the results of BLAST homology searches of UniGene clusters.
| BLAST score ≥ 50 | BLAST score ≥ 100 | |
|---|---|---|
| No. containing entire CDS in UniGene | ||
| contigs | 3579 | 3440 |
| singlets | 7920 | 6515 |
| No. that hit all or part of CDS in UniGene | ||
| contigs | 4370 | 4163 |
| singlets | 11079 | 9219 |
| No. that hit PEDE assemblies | ||
| human | 7531 | 6658 |
| all | 9426 | 7976 |
| No. whose entire CDS are encoded in PEDE assemblies | ||
| human | 5033 | 4689 |
| all | 5856 | 5363 |
Table 2. PEDE assemblies estimated to contain full-length CDSs in light of results of BLAST homology searches of RefSeq sequences.
| BLAST score ≥50 | BLAST score ≥100 | |
|---|---|---|
| No. containing entire CDS in RefSeq | ||
| contig | 3590 | 3344 |
| singlet | 7011 | 4863 |
| No. that hit all or part of CDS in RefSeq | ||
| contig | 4082 | 3830 |
| singlet | 9471 | 6869 |
| No. that hit PEDE assemblies | ||
| human | 6356 | 5724 |
| mouse | 5644 | 5058 |
| all | 7814 | 6898 |
| No. whose entire CDS are encoded in PEDE assemblies | ||
| human | 5082 | 4539 |
| mouse | 4677 | 4111 |
| all | 6235 | 5471 |
CDS, coding sequences.
Several collections of porcine EST clusters are available, including UniGene in GenBank, which we used in our BLAST searches, and SsGI, which is available in the TIGR gene indices (9). However, all assemblies in PEDE are linked to the cDNA clones that are ready to use for analyses like expression in mammalian cells because of the high likelihood of these clones containing full-length CDS. Features of several sequences of porcine genes newly identified in the PEDE assemblies are shown in Supplementary table 2 (http://pede.dna.affrc.go.jp/supplement/suppl_table2.html).
SNPs in cDNA sequences (cSNPs) are useful for dense and reliable gene mapping and are indispensable for analyses of correlation between genes and phenotypes. Further, oriental pig breeds such as Meishan have the potential to improve various traits, including reproductive ability, in representative commercial pig breeds. Information regarding SNPs specific to oriental or western breeds is useful for gene mapping using western–oriental swine experimental families constructed at various livestock experimental stations and for exploring candidate genes responsible for economic traits in pigs. However, the number of SNPs mined from EST data may be an overestimate because of the relatively high error rate of reverse transcription (10). In the PEDE database, SNPs in the assemblies were detected by PolyPhred (11), and mutations that appeared on more than one read were considered as valid alleles according to a filtering strategy similar to that used in a previous study (12). We used regions of porcine sequences containing SNPs causing alteration of coding amino acids in BLAST searches of human or mouse RefSeq protein sequences highly similar (i.e. BLAST score ≥ 50) to the porcine sequence. Detected SNPs are classified and shown on the pig cSNP database page (http://pede.dna.affrc.go.jp/csnp/csnp_main.php). Investigation of the distribution in various breeds of several SNPs extracted randomly from the putative SNPs in the database suggests that the SNPs detected between western and oriental breeds will be useful for gene mapping in swine resource families crossbred from these breeds (Supplementary table 3; http://pede.dna.affrc.go.jp/supplement/suppl_table3.html).
DATABASE AND SEARCH INTERFACE OF THE PEDE ASSEMBLIES
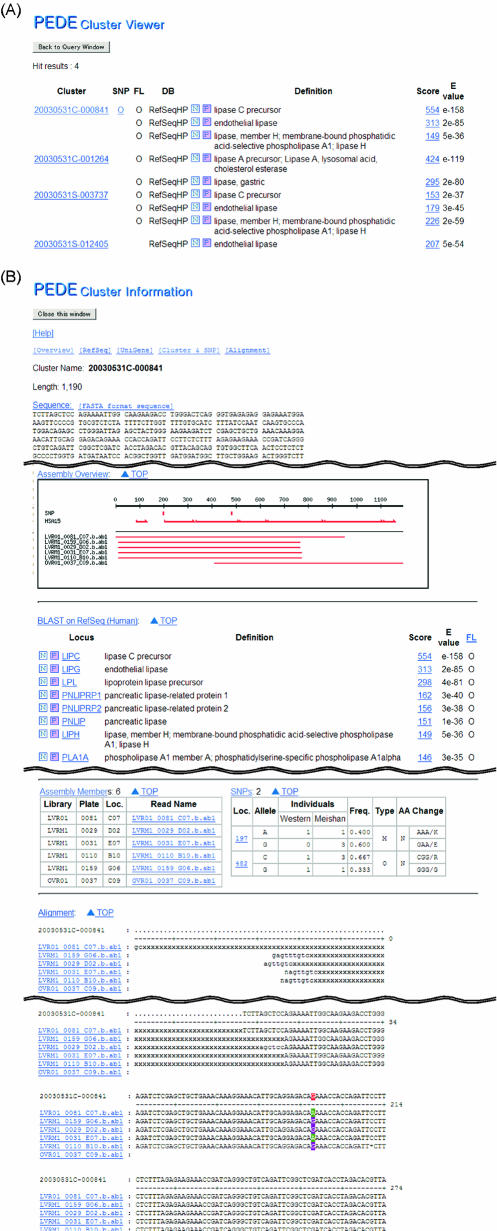
The PEDE database was constructed to store sequences and similarity data of the PEDE assemblies and to make this information available to users. The cluster viewer page of the PEDE database (http://pede.dna.affrc.go.jp/cluster/cluster_viewer.php) provides the interface for searches by keyword, locus name, database accession number and corresponding human chromosome according to the BLAST result and enables users to obtain sequence data and names for clones of interest. Search results can be limited by the existence of SNPs specific to particular breeds. The existence of any SNPs and an estimate of the amount of sequence needed to encode the full-length CDS are presented in the result view (Fig. 1A). Details of each assembly and the output of the BLAST searches can be inspected by using the links. Details of each assembly, including its nucleotide sequence, a summary of the BLAST results and the SNPs identified, are summarized in a page linked to the search result page. The page for each assembly provides the alignment of reads contributing to the contiguous sequence. To design primers for the detection of SNPs through PCR using swine genomic DNA, we used a BLAST search to compare spans of porcine sequences encoded within single exons with the draft sequence of the human genome (Fig. 1B). In addition, a sequence of interest, through another interface for BLASTN and TBLASTN (http://pede.dna.affrc.go.jp/pedeblast/pedeblast_main.html), can be used in searches against the PEDE assemblies.
Figure 1.
A representative result of queries of the PEDE assemblies. (A) List of assemblies matching the query. (B) Detailed information on each assembly is shown in an individual window. The sequence of the assembly, results of BLAST searches of RefSeq sequences and UniGene clusters, locations of the putative SNPs, reads comprising the assembly and their alignment to generate the assembled sequence are indicated. The likelihood of a putative SNP resulting from a synonymous/non-synonymous mutation is estimated for regions with high similarity to corresponding human or mouse known genes.
Another useful aspect of the PEDE database concerns the analysis of artiodactyl-specific antigens such as swine leukocyte antigen [SLA; swine major histocompatibility complex (MHC)] molecules. T cell receptors of humans and other primates can directly recognize SLA molecules, leading to acute vascular rejection of porcine grafts which, after the hyperacute rejection due to several porcine antigens that primates lack (e.g. galactose-α1,3-galactose), is one of the greatest problems in xenotransplantation (13). The genomic sequence of the porcine SLA locus is partly clarified within the region encoding representative classical and non-classical class I molecules (14,15). The PEDE assemblies include 295 assemblies and 1905 reads that have noteworthy similarity to known SLA genes. The MHC region is a hotspot for gene conversion and rearrangement, and the number of genes in any particular locus may vary even in the same species (16). The number of groups of loci, especially of class I molecules, is highly variable among species. The PEDE database facilitates the investigation of novel SLA loci and possible polymorphism(s) in each molecule.
IMPLEMENTATION
The PEDE database was developed on the PostgreSQL relational database system, and its web interface was constructed using PHP script language. The structure and tables of the database are described in Supplementary figure 1 (http://pede.dna.affrc.go.jp/supplement/suppl_figure1.html). The PEDE database is provided as one of the resources in the Animal Genome Database (http://animal.dna.affrc.go.jp/) (17) and is accessible directly at http://pede.dna.affrc.go.jp/.
CONCLUSIONS AND PERSPECTIVES
The PEDE database provides a catalogue of porcine expressed genes and cDNA clones that are estimated to include full-length CDSs and promotes functional analyses of porcine genes. Furthermore, this sequence information and recent developments in the design of primers for screening porcine BAC clones (18) including genes of interest will aid the development of transgenic and gene knockout pigs, which have been gaining in popularity because of rapid advances in cloning technology. Although sequence information regarding large regions of the porcine genome has been limited to date (14,15,19), further progress in this area and the use of the PEDE assemblies will help to clarify the precise structure of porcine genes and regulation of their expression.
We have selected a representative clone from each contig in the PEDE assemblies and are determining the complete sequence of the insert. We will add data from full-length cDNA libraries derived from other tissues and cell populations to our database to facilitate the identification of rarely expressed porcine genes. Moreover, mapping of the sequences derived from the PEDE assemblies on our porcine radiation hybrid (SSRH) map (http://ssrh.gene.staff.or.jp) (20) and linkage map (ToNMaP) (http://agp.gene.staff.or.jp/agp/db/linkage/linkage.html) (21) will augment available comparative information between human and pig.
In conclusion, the PEDE database of porcine nucleic acid sequences and cDNA clones will help users to explore genes that may be responsible for traits like disease susceptibility. This database also offers information regarding major and minor porcine-specific antigens, which should be investigated in the use of pigs for medical applications.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Yasumichi Sakai and Kazuyoshi Makino for their assistance in the computational analysis and Takako Suzuki for technical assistance. We are indebted to Tomiji Akita for preparation of the porcine tissues. We also thank Takuji Sasaki and Yoshihiro Muneta for useful suggestions. This work was supported by the Animal Genome Research Project and the Food Traceability Research Project of the Ministry of Agriculture, Forestry and Fisheries of Japan and by a Grant-in-Aid of the Japan Racing Association.
DDBJ/EMBL/GenBank accession nos+ AB116555–AB116564, BP137499–BP173623 and BP433030–BP464980
REFERENCES
- 1.Onishi A., Iwamoto,M., Akita,T., Mikawa,S., Takeda,K., Awata,T., Hanada,H. and Perry,A.C. (2000) Pig cloning by microinjection of fetal fibroblast nuclei. Science, 289, 1188–1190. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki Y., Yoshitomo-Nakagawa,K., Maruyama,K., Suyama,A. and Sugano,S. (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene, 200, 149–156. [DOI] [PubMed] [Google Scholar]
- 3.Ewing B., Hillier,L., Wendl,M.C. and Green,P. (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res., 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 4.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 5.Pertea G., Huang,X., Liang,F., Antonescu,V., Sultana,R., Karamycheva,S., Lee,Y., White,J., Cheung,F., Parvizi,B. et al. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics, 19, 651–652. [DOI] [PubMed] [Google Scholar]
- 6.Huang X. and Madan,A. (1999) CAP3: A DNA sequence assembly program. Genome Res., 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quackenbush J., Cho,J., Lee,D., Liang,F., Holt,I., Karamycheva,S., Parvizi,B., Pertea,G., Sultana,R. and White,J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res., 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerard G.F., Fox,D.K., Nathan,M. and D’Alessio,J.M. (1997) Reverse transcriptase. The use of cloned Moloney murine leukemia virus reverse transcriptase to synthesize DNA from RNA. Mol. Biotechnol., 8, 61–77. [DOI] [PubMed] [Google Scholar]
- 11.Nickerson D.A., Tobe,V.O. and Taylor,S.L. (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res., 25, 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picoult-Newberg L., Ideker,T.E., Pohl,M.G., Taylor,S.L., Donaldson,M.A., Nickerson,D.A. and Boyce-Jacino,M. (1999) Mining SNPs from EST databases. Genome Res., 9, 167–174. [PMC free article] [PubMed] [Google Scholar]
- 13.Cascalho M. and Platt,J.L. (2001) The immunological barrier to xenotransplantation. Immunity, 14, 437–446. [DOI] [PubMed] [Google Scholar]
- 14.Renard C., Vaiman,M., Chiannilkulchai,N., Cattolico,L., Robert,C. and Chardon,P. (2001) Sequence of the pig major histocompatibility region containing the classical class I genes. Immunogenetics, 53, 490–500. [DOI] [PubMed] [Google Scholar]
- 15.Chardon P., Rogel-Gaillard,C., Cattolico,L., Duprat,S., Vaiman,M. and Renard,C. (2001) Sequence of the swine major histocompatibility complex region containing all non-classical class I genes. Tissue Antigens, 57, 55–65. [DOI] [PubMed] [Google Scholar]
- 16.Kumnovics A., Takada,T. and Lindahl,K.F. (2003) Genomic organization of the mammalian MHC. Annu. Rev. Immunol., 21, 629–657. [DOI] [PubMed] [Google Scholar]
- 17.Wada Y. and Yasue,H. (1996) Development of an animal genome database and its search system. Comput. Appl. Biosci., 12, 231–235. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki K., Asakawa,S., Iida,M., Shimanuki,S., Fujishima,N., Hiraiwa,H., Murakami,Y., Shimizu,N. and Yasue,H. (2000) Construction and evaluation of a porcine bacterial artificial chromosome library. Anim. Genet., 31, 8–12. [DOI] [PubMed] [Google Scholar]
- 19.Uenishi H., Hiraiwa,H., Yamamoto,R., Yasue,H., Takagaki,Y., Shiina,T., Kikkawa,E., Inoko,H. and Awata,T. (2003) Genomic structure around joining segments and constant regions of swine T-cell receptor α/δ (TRA/TRD) locus. Immunology, 109, 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamasima N., Suzuki,H., Mikawa,A., Morozumi,T., Plastow,G. and Mitsuhashi,T. (2003) Construction of a new porcine whole-genome framework map using a radiation hybrid panel. Anim. Genet., 34, 216–220. [DOI] [PubMed] [Google Scholar]
- 21.Mikawa S., Akita,T., Hisamatsu,N., Inage,Y., Ito,Y., Kobayashi,E., Kusumoto,H., Matsumoto,T., Mikami,H., Minezawa,M. et al. (1999) A linkage map of 243 DNA markers in an intercross of Göttingen miniature and Meishan pigs. Anim. Genet., 30, 407–417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.