Abstract
Traditionally, the tribe Sophoreae sensu lato has been considered a basal but also heterogeneous taxonomic group of the papilionoid legumes. Phylogenetic studies have placed Sophoreae sensu stricto (s.s.) as a member of the core genistoids. The recently suggested new circumscription of this tribe involved the removal of traditional members and the inclusion of Euchresteae and Thermopsideae. Nonetheless, definitions and inter- and intra-taxonomic issues of Sophoreae remain unclear. Within the field of legume systematics, the molecular characteristics of a plastid genome (plastome) have an important role in helping to define taxonomic groups. Here, we examined the plastome of Maackia fauriei, belonging to Sophoreae s.s., to elucidate the molecular characteristics of Sophoreae. Its gene contents are similar to the plastomes of other typical legumes. Putative pseudogene rps16 of Maackia and Lupinus species imply independent functional gene loss from the genistoids. Our overall examination of that loss among legumes suggests that it is common among all major clades of Papilionoideae. The M. fauriei plastome has a novel 24-kb inversion in its large single copy region, as well as previously recognized 50-kb and 36-kb inversions. The 36-kb inversion is shared by the core genistoids. The 24-kb inversion is present in the eight genera belonging to three tribes: Euchresteae, Sophoreae s.s., and Thermopsideae. The phylogenetic distribution of this 24-kb inversion strongly supports the monophyly of members of Sophoreae s.s. with Euchresteae and Thermopsideae. Hence, it can be used as a putative synapomorphic characteristic for the newly circumscribed Sophoreae, including Euchresteae and Thermopsideae. However, plastome conformation suggests a slightly narrower taxonomic group because of heterogeneous results from Bolusanthus and Dicraeopetalum. The phylogenetic analysis, based on plastome sequences from 43 legumes, represents well our understanding of legume systematics while resolving the genistoid clade as a sister group to an Old World clade. It also demonstrates the value that plastomes are powerful marker for systematic studies of basal papilionoid legumes.
Introduction
Fabaceae (legumes) is the third largest angiosperm family, with approximately 751 genera and 20,000 species [1–2]. Three subfamilies—Mimosoideae, Papilionoideae, and Caesalpinioideae—are typically recognized although the first two are generally placed within the latter, which is paraphyletic. Papilionoideae is the most diverse and includes economically important legume crops. Recent comprehensive molecular phylogenies [3–4] have provided strong support for the relationships and composition of many major Papilionoideae clades. However, a systematic understanding is lacking for “basal papilionoids” [5] or “early-branching papilionoids” [3], which include tribes Swartzieae, Sophoreae, Dipterygeae, and Dalbergieae [2]. In particular, the polyphyletic tribe Sophoreae is in a systematics state of flux.
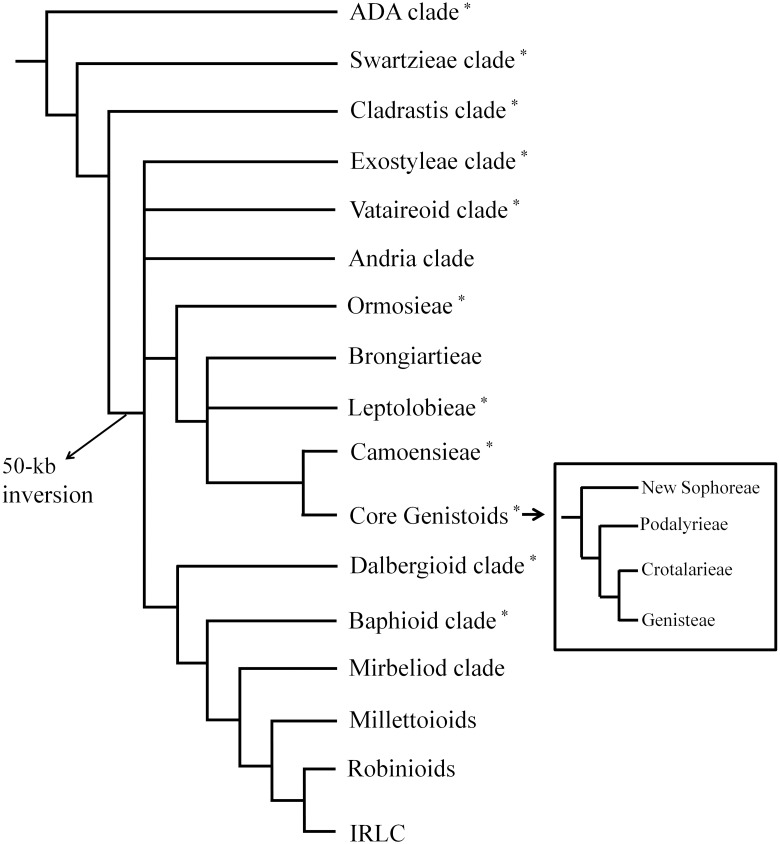
The taxonomic circumscription of tribe Sophoreae has been debated during last several decades. The traditional tribe Sophoreae sensu lato (sensu Polhill; hereafter, ‘Sophoreae s.l.’) has been circumscribed as a vast and heterogeneous assemblage of taxa that are considered basal papilionoid legumes [6–7]. This has led to taxonomic disputes about how Sophoreae relates to early-branching papilionoid tribes [8–10]. Modern molecular phylogenies [3–5, 11–13] have shown that Sophoreae s.l. is indeed a polyphyletic group comprising genera [Sophoreae pro parte (p.p.)] that are scattered throughout the phylogenetic tree of the Papilionoideae (Fig 1). Meanwhile, as established by Crisp et al. [14], Sophoreae sensu stricto (s.s.) is considered a member of the core genistoids that include the tribes of Genisteae, Crotalarieae, Euchresteae, Podalyrieae, and Thermopsideae. Recently, Cardoso et al. [4] suggested a new circumscription of Sophoreae (hereafter, ‘new Sophoreae’) that includes 122 species of 14 genera that were formerly treated as Sophoreae s.s. (Ammodendron Fisch. ex DC., Ammothamnus Bunge, Bolusanthus Harms, Dicraeopetalum Harms, Maackia Rupr. & Maxim, Platycelyphium Harms, Salweenia Baker f., and Sophora L.), Euchresteae (Euchresta Benn.), and Thermopsideae (Ammopiptanthus S.H. Cheng, Anagyris L., Baptisia Vent., Piptanthus Sweet, and Thermopsis R. Br. ex W.T. Aiton). This new circumscription involved huge rearrangement of genera of Sophoreae s.l. and inclusion of all genera belonging to Euchresteae and Thermopsideae except Pickeringia Nutt. ex Torr. & A. Gray. However, their taxon sampling did not cover all genera of new Sophoreae (nine of 14 genera were included) and other studies [15–16] demonstrated that most Thermopsideae genera could also be a monophyletic group independent from Sophoreae s.s. Moreover, new Sophoreae lacks morphological criteria that can define it in the core genistoids. Thus, its taxonomic boundary remains unclear.
Fig 1. Simplified schematic phylogeny of Papilionoideae, originally published as Figure 1 in Cardoso et al. [4].
Phylogenetic positions are shown for new Sophoreae (incl. Euchresteae and Thermopsideae). *; evolutionary group that includes some former members of Sophoreae s.l. (sensu Polhill [6–7]).
The plastid genome (plastome) is generally conserved among seed plants based on its quadripartite architecture [a pair of inverted repeats (IRs), small single copy (SSC) region, and large single copy (LSC) region], gene content, and order [17]. Plastomes are also generally free of recombinants, maternally inherited, and have a slow rate of evolution [18–19]. Thus, the sequence variations associated with a plastome are useful tools for studies that are phylogeographic [20–23], phylogenetic [4, 24–26], or phylogenomic [27–30] in nature. For example, within the legume phylogeny, IR losses (IRLC; Inverted Repeat-Lacking Clade); large inversions such as a 50-kb inversion (valid for all papilionoid tribes except some members of Swartzieae, Sophoreae s.l., and Dipterygeae); and gene and/or intron losses serve as important taxonomic characters [17, 31]. For genistoids, a 36-kb inversion which is embedded within 50-kb inversion and caused by flip-flop recombination from short repeat sequences of the trnS (GGA) and trnS (GCU) regions has been proposed as proof of molecular synapomorphy among a core genistoid clade [32].
Morphologically Sophoreae s.l. has been a "tribe of convenience" rather than a natural group that is clearly distinctive from both Caesalpinioideae and Papilionoideae [6]. Its non- or imperfect papilionaceous flower, with 10 free stamens, has been considered a taxonomically significant characteristic [6, 33]. However, taxa with more specialized floral features are now dominant element of new Sophoreae sensu Cardoso et al. [4]. Although such morphological characters can provide critical and fundamental evidence when determining taxonomic boundaries, convergent evolution of their morphology is often problematic, especially at higher taxonomic levels [34]. Therefore, it is difficult to elucidate evolutionary relationships and delimit taxonomic boundaries based only on morphology [35]. The distinct plastome characteristic has been used as a powerful marker for systematic evaluations of legumes [17, 31]. Thus, plastome studies could be promising for testing the validity of the new Sophoreae.
Here, we sequenced and analyzed the first plastome of Sophoreae s.s. from Maackia fauriei (H. Lév.) Takeda, a large tree endemic to Jeju Island, Korea. Although a single plastid genome sequence is not sufficient to answer all of the taxonomic problems surrounding Sophoreae s.l., we did identify a distinct plastome organization from M. fauriei and then screened those features in representatives of genistoid and Sophoreae s.l. We also reconstructed a legume phylogeny based on the plastome sequences found in our current study as well as those reported recently by other researchers.
Materials and methods
Ethics statement
Maackia fauriei is not an endangered or protected species in Korea. We did not collect plants from the protected areas that required permission.
Plastid genome sequencing
Leaves of Maackia fauriei were collected from Jeju Island, Korea, and preserved with silica gel. Voucher specimen (J.Y. Lee & I.S. Choi 1208046) were deposited in the Herbarium of Inha University (IUI), Incheon, Korea. Genomic DNA was extracted from the dried tissues by a protocol that utilized the DNeasy Plant Mini Kit (Qiagen, Seoul, Korea). The quantity and quality of the extracted genomic DNA were assessed by gel electrophoresis and spectroscopy with a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA). The gDNAs were then fragmented into 500-bp segments for library construction per the manufacturer’s instructions (Illumina Inc., San Diego, CA, USA), and were sequenced using the Illumina HiSeq 2000 System (Illumina Inc.) at the BML Co. in Daejeon, Korea.
Plastid genome assembly
The sequencing run produced 25,318,060 paired-end reads (101 bp each). Poor-quality sequences were removed by Trimmomatic 0.32 [36]. To isolate plastid-related reads and assemble the plastome, we largely followed the method described by Wang and Messing [37]. Briefly, the paired-end reads were mapped on the reference genome of Lupinus luteus L. (GenBank Accession Number NC_023090) using Geneious ver. 7.1.3 (Biomatters Ltd., Auckland, NZ). The mapped reads were then reassembled de novo using Geneious and the generated plastome contigs were re-aligned to the plastome of L. luteus to identify gaps among the contigs. This generated four contigs (two for LSC and two each for the SSC and IR regions) with a total length of approximately 154 kb. To fill those gaps, we used purified DNA and performed polymerase chain reactions (PCRs) with primers designed via Primer3 [38].
Gene annotation
The complete plastome for M. fauriei was annotated using DOGMA [39] and Geneious. The tRNAs were confirmed by tRNAscan-SE [40]. Other protein-coding regions were checked based on data from the NCBI (http://blast.ncbi.nlm.nih.gov/), and manual corrections were made for the start and stop codons. Particular gene features of the plastome were illustrated with the Web-based tool OGDraw [41].
Investigation of rps16 among legume plastomes
The loss of rps16 was investigated in complete plastomes from 33 legume species (Table 1). Each rps16 gene was extracted from the available sequenced genomes and sequences were aligned by MUSCLE 3.8.31 [42] using default parameters. We re-analyzed and categorized this gene in four ways, as in Kim et al. [43]: 1) intact gene (full-length and in-frame), 2) putative pseudogene (mutation in start or stop codon or frame shift-inducing indels), 3) truncated gene (significant deletion), and 4) complete deletion.
Table 1. Comparison of features from legume plastomes selected for this study.
| Subfamily | Tribe | Species | Accession number | rps16 | Total genome size (bp) |
|---|---|---|---|---|---|
| Caesalpinioideae | Cercideae | Cercis canadensis | KF856619 | intact gene | 158,995 |
| Caesalpinioideae | Detarieae | Tamarindus indica | KJ468103 | intact gene | 159,551 |
| Caesalpinioideae | Caesalpinieae | Ceratonia siliqua | KJ468096 | intact gene | 156,367 |
| Caesalpinioideae | Cassieae | Chamaecrista fasciculata | KP126855 | partial genome | |
| Caesalpinioideae | Caesalpinieae | Caesalpinia coriaria | KJ468095 | intact gene | 158,045 |
| Caesalpinioideae | Caesalpinieae | Haematoxylum brasiletto | KJ468097 | intact gene | 157,728 |
| Mimosoideae | Mimoseae | Prosopis glandulosa | KJ468101 | intact gene | 163,042 |
| Mimosoideae | Mimoseae | Leucaena trichandra | NC_028733 | intact gene | 164,692 |
| Mimosoideae | Mimoseae | Desmanthus illinoensis | KP126868 | partial genome | |
| Mimosoideae | Ingeae | Inga leiocalycina | NC_028732 | intact gene | 175,489 |
| Mimosoideae | Acacieae | Acacia ligulata | NC_026134 | intact gene | 174,233 |
| Papilionoideae | Amorpheae | Amorpha canescens | KP126852 | partial genome | |
| Papilionoideae | Dalbergieae | Arachis hypogaea | KJ468094 | complete deletion | 156,395 |
| Papilionoideae | Genisteae | Lupinus albus | KJ468099 | putative pseudogene | 154,140 |
| Papilionoideae | Genisteae | Lupinus luteus | NC_023090 | putative pseudogene | 151,894 |
| Papilionoideae | Sophoreae | Maackia fauriei | KX388160 | putative pseudogene | 154,541 |
| Papilionoideae | Thermopsideae | Baptisia alba | KP126860 | partial genome | |
| Papilionoideae | Thermopsideae | Baptisia bracteata | KP126854 | partial genome | |
| Papilionoideae | Indigofereae | Indigofera tinctoria | KJ468098 | intact gene | 158,367 |
| Papilionoideae | Millettieae | Millettia pinnata | NC_016708 | intact gene | 152,968 |
| Papilionoideae | Phaseoleae | Apios americana | KF856618 | complete deletion | 152,828 |
| Papilionoideae | Phaseoleae | Pachyrhizus erosus | KJ468100 | intact gene | 151,947 |
| Papilionoideae | Phaseoleae | Glycine max | NC_007942 | intact gene | 152,218 |
| Papilionoideae | Psoraleeae | Pediomelum argophyllum | KP126866 | partial genome | |
| Papilionoideae | Psoraleeae | Psoralidium tenuiflorum | KP126859 | partial genome | |
| Papilionoideae | Phaseoleae | Phaseolus vulgaris | NC_009259 | truncated gene | 150,285 |
| Papilionoideae | Phaseoleae | Strophostyles leiosperma | KP126853 | partial genome | |
| Papilionoideae | Phaseoleae | Vigna unguiculata | KJ468104 | intact gene | 151,866 |
| Papilionoideae | Robinieae | Robinia pseudoacacia | KJ468102 | truncated gene | 154,835 |
| Papilionoideae | Loteae | Lotus japonicus | NC_002694 | intact gene | 150,519 |
| Papilionoideae | Millettieae | Wisteria floribunda | NC_027677 | putative pseudogene | 130,960 |
| Papilionoideae | Galegeae | Glycyrrhiza glabra | NC_024038 | truncated gene | 127,943 |
| Papilionoideae | Galegeae | Astragalus mongholicus var. nakaianus | NC_028171 | truncated gene | 123,633 |
| Papilionoideae | Galegeae | Oxytropis lambertii | KP126858 | partial genome | |
| Papilionoideae | Cicereae | Cicer arietinum | NC_011163 | truncated gene | 125,319 |
| Papilionoideae | Trifolieae | Melilotus albus | KP126850 | partial genome | |
| Papilionoideae | Trifolieae | Medicago truncatula | NC_003119 | complete deletion | 124,033 |
| Papilionoideae | Trifolieae | Trifolium aureum | NC_024035 | truncated gene | 126,970 |
| Papilionoideae | Trifolieae | Trifolium subterraneum | NC_011828 | truncated gene | 144,763 |
| Papilionoideae | Fabeae | Pisum sativum | NC_014057 | complete deletion | 122,169 |
| Papilionoideae | Fabeae | Lathyrus sativus | NC_014063 | complete deletion | 121,020 |
| Papilionoideae | Fabeae | Vicia faba | KF042344 | truncated gene | 123,722 |
| Papilionoideae | Fabeae | Lens culinaris | NC_027152 | complete deletion | 123,096 |
Whole-genome alignments
Other plastome sequences for relevant legumes (Table 1) were obtained from GenBank. Single IR copies were manually removed. Afterward, the complete plastome of M. fauriei and other sequences [Tamarindus indica L. (KJ468103), Arachis hypogaea L. (KJ468094), and Lupinus luteus (NC_023090)] were aligned using genome alignment software Mauve 2.3.1 [44] to check for inversion events.
Survey of inversion events among genistoids and Sophoreae s.l.
To verify the distribution of 36-kb and 24-kb inversions on the phylogenetic tree, we used methods based on PCR amplifications. In all, 16 representative species were selected that belong to five tribes and 15 genera (Table 2). They roughly cover the genistoids and Sophoreae s.l. group. Those plant materials had been collected from East Asia (China, Korea, and Japan) and deposited at the IUI herbarium. Other DNA samples were obtained from the DNA Banks of the Royal Botanic Gardens, Kew (http://apps.kew.org/dnabank/homepage.html) and the University of Johannesburg (UJ), South Africa (http://acdb.co.za/index.php/dna-bank/introduction-2.html). The remaining materials needed for this study were also collected through KNRRC (Medicinal Plants Resources Bank NRF-2010-0005790), supported by the Korea Research Foundation and the Ministry of Education, Science and Technology in 2014. Vouchers were deposited at the herbarium of Gachon University (GCU). The gDNAs were extracted from silica gel-dried leaves, as described above. All PCRs were conducted with a GeneAmp® PCR System 2700 Thermal Cycler (Applied Biosystems, Foster City, CA, USA) according to a program of initial denaturation that was followed by 30 cycles of 10 s at 98°C, 7 s at 58°C, and 2 min at 72°C; and then a final extension for 5 min at 72°C. Each 50-μL reaction mixture included 1 μL of genomic DNA (~ 20 ng), 1 μL each of forward and reverse primers (10 pMol), and 25 μL of PrimeSTAR HS Premix (TaKaRa, Seoul, Korea). The existence of the 36-kb inversion was tested using primer pairs designed by Martin et al. [32]. The pair of rps4-bef-F and ycf3-bef-R was used for determining its absence while ycf3-inv-F and psbI-int-R were used to detect its presence. Three primers were designed to test for the 24-kb inversion: the pair of FGA-ndhJ-F and FGA-trnF-R for its absence and the pair of FGA-ndhJ-F and FGA-trnC-R for its presence (Table 3). The PCR products were visualized on 2% agarose gels, purified by PCR quick-spin TM (iNtRON Biotechnology, Seongnam, Korea), and sequenced with an ABI 3100 Genetic Analyzer and an ABI BigDyeTM Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) at the Macrogen, Seoul, Korea.
Table 2. Taxa sampled to screen for inversions.
| Taxon | Classification | Inversion events | Voucher information | |||
|---|---|---|---|---|---|---|
| Polhill (1981 and 1994) [6–7] | Lewis et al. (2005) [1] | Cardoso et al. (2013) [4] | 36-kb | 24-kb | ||
| Cladrastis wilsonii | Sophoreae | Sophoreae p.p. | Cladrastis clade | - | - | I.S. Choi & D.P. Jin 1209001 (IUI) |
| Styphnolobium japonicum | Sophoreae | Sophoreae p.p. | Cladrastis clade | - | - | B.H. Choi 1317 (IUI) |
| Camoensia brevicalyx | Sophoreae | Sophoreae s.s. | Camoensieae | - | - | Champluvier 5205 (K)(Kew 33888) |
| Crotalaria capensis | Crotalarieae | Crotalarieae | Crotalarieae | + | - | O. Maurin et al. OM2692 (JRAU)(UJ 05004) |
| Lupinus luteus | Genisteae | Genisteae | Genisteae | + | - | ABH 31123 (ABH)(Kew 15870) |
| Bolusanthus speciosus | Sophoreae | Sophoreae s.s. | Sophoreae | + | - | O. Maurin et al. OM240 (JRAU)(UJ 00040) |
| Dicraeopetalum mahafaliense | Sophoreae | Sophoreae s.s. | Sophoreae | + | - | D. Puy et al. 1029 (Kew 13059) |
| Anagyris foetida | Thermopsideae | Thermopsideae | Sophoreae | + | + | Crespo & E. galante 38544 (ABH)(Kew 15865) |
| Baptisia australis | Thermopsideae | Thermopsideae | Sophoreae | + | + | I.S. Choi 1405001 (IUI) |
| Piptanthus nepalensis | Thermopsideae | Thermopsideae | Sophoreae | + | + | H. Sun s.n. (GCU)(MPRBP 01013) |
| Thermopsis fabacea | Thermopsideae | Thermopsideae | Sophoreae | + | + | B.H. Choi & I.S. Choi 1406001 (IUI) |
| Maackia fauriei | Sophoreae | Sophoreae s.s. | Sophoreae | + | + | J.Y. Lee & I.S. Choi 1208046 (IUI) |
| Salweenia bouffordiana | Sophoreae | Sophoreae s.s. | Sophoreae | + | + | H. Sun s.n. (GCU)(MPRBP 01012) |
| Sophora koreensis | Sophoreae | Sophoreae s.s. | Sophoreae | + | + | J.Y. Lee & B.H. Choi s.n. (IUI) |
| Sophora flavescens | Sophoreae | Sophoreae s.s. | Sophoreae | + | + | S.G. Jung s.n. (IUI) |
| Euchresta japonica | Euchresteae | Euchresteae | Sophoreae | + | + | B.H. Choi 99078 (IUI) |
For inversion events: + indicates presence of inversion; –, absence. DNA bank numbers are in parentheses after voucher information.
Table 3. PCR primers used for screening of inversion events.
| Primer | Sequence | Source |
|---|---|---|
| rps4-bef-F | 5′-CAATCAAATAATAGATAGTAAATGGGTTG-3′ | Martin et al. [32] |
| ycf3-bef-R | 5′-GGAATTATTCGTAATAATATATTGGCTAC-3′ | Martin et al. [32] |
| ycf3-inv-F | 5′-CGTAATAAGATATTGGCTAC-3′ | Martin et al. [32] |
| psbI-int-R | 5′-CTCTTTTCATCTTCGGATTC-3′ | Martin et al. [32] |
| FGA-ndhJ-F | 5′-CGTTCCCAATGTGCCTAT-3′ | This study |
| FGA-trnF-R | 5′-TGGTAGAGCAGAGGACTG-3′ | This study |
| FGA-trnC-R | 5′-CAAATCCTTTTTCCCCAGTT-3′ | This study |
Phylogenetic analysis
A phylogenetic tree was constructed using 43 complete or partial plastomes for legumes, as collected from the GenBank database (Table 1). The outgroup included Morus mongolica (Bureau) C.K. Schneid. (KM491711), Fragaria vesca L. (NC_015206), Castanea mollissima Blume (NC_014674), and Cucumis sativus L. (DQ119058), as described by Schwarz et al. [45]. The final data set from these 47 plastomes comprised 71 conserved plastid protein-coding genes: atpA, B, E, F, H, and I; ccsA; cemA; clpP; matK; ndhA, B, C, D, E, F, G, H, I, J, and K; petA, B, D, G, L, and N; psaA, B, C, I, and J; psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, and Z; rbcL; rpl2, 14, 16, 20, 23, 32, 33, and 36; rpoA, B, C1, and C2; rps2, 3, 4, 7, 8, 11, 12, 14, 15, and 19; and ycf3. The alignments were made with MAFFT (v. 7.017) and default parameters. Poorly aligned regions were either refined or deleted using Geneious. A maximum likelihood (ML) analysis of the complete, final alignment of all taxa was conducted with RAxML Blackbox [46], using the gamma model of rate heterogeneity and an ML search.
Results
Plastome sequence of Maackia fauriei and variation in rps16 loss among legumes
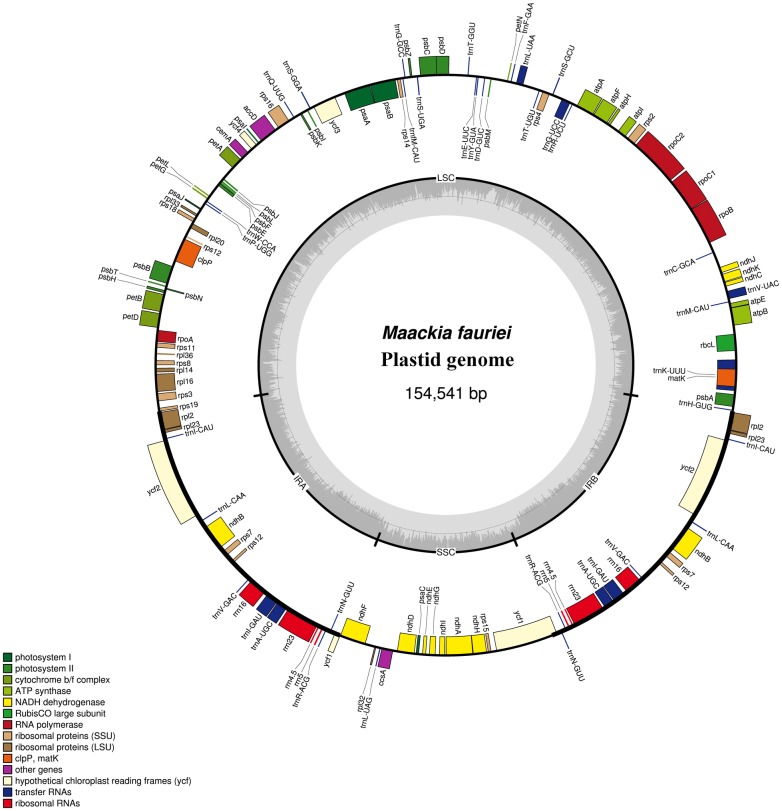
The complete plastome of Maackia fauriei (GenBank Accession No. KX388160) is 154,541 bp long and has two IR regions (25,494 bp each) that are separated by LSC and SSC regions of 85,140 bp and 18,413 bp, respectively (Fig 2). Approximately 57.9% of the sequence is composed of protein-coding regions while the remaining 42.1% contains non-coding sequences that include introns and intergenic spacers (IGS). The AT and GC contents are 63.5% and 36.5%, respectively. Among the 135 recognized genic features are four unique rRNAs, 31 tRNAs, and 76 protein-coding genes. The rpl22 loss, which is typical legume plastomes, is shared by Maackia. The rps16 is a putative pseudogene caused by “AAAC” duplication in exon2 that results in a frame shift mutation and internal stop codon.
Fig 2. Map of Maackia fauriei plastome.
Genes on outside of outer circle are transcribed in clockwise direction; those on inside of outer circle are transcribed in counterclockwise direction. Functional categories of genes are color-coded. Dashed area in inner circle indicates GC content of plastid genome.
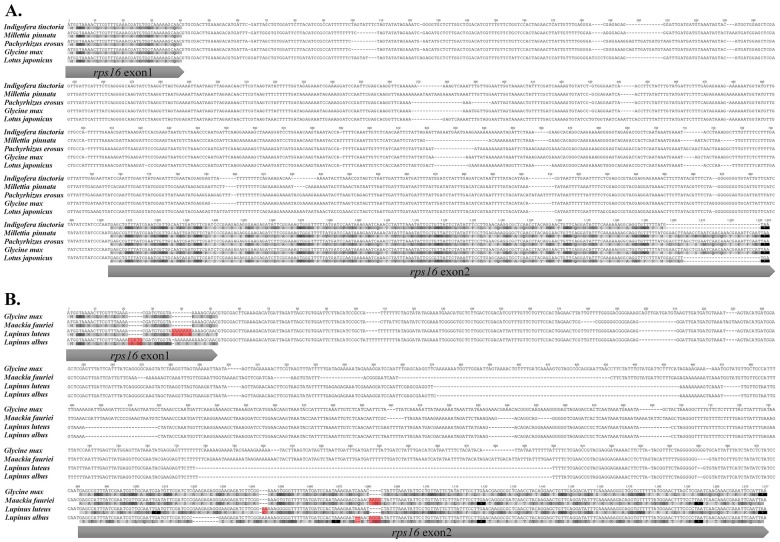
We also investigated the loss of rps16 among 33 complete legume plastomes (Table 1) and found that the gene was intact in 15 species, a putative pseudogene in four species, truncated pseudogene in eight species, and deleted in six species. All of the Caesalpinioideae and Mimosoideae samples contained intact rps16. The types of losses varied for rps16 and were restricted to Papilionoideae. We considered the rps16s from Lupinus L. species as putative pseudogenes similar to that of M. fauriei due to several indel events on the exons and an approximately 200-bp deletion of intron sequences (Fig 3).
Fig 3. Sequence alignment of intact genes and putative pseudogenes of rps16.
A, intact rps16 in papilionoids; B, putative pseudogene rps16 in genistoids, with comparison to Glycine max (L.) Merr. Frameshift-inducing indels are shaded in red.
The 36-kb and 24-kb inversion events and their phylogenetic distribution among genistoids and Sophoreae s.l.
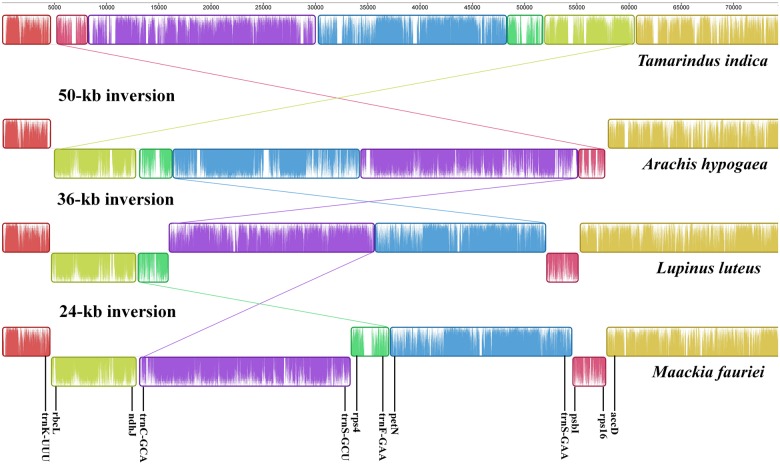
The legume species used for our comparisons revealed only subtle differences in their gene content. However, the gene order for Maackia fauriei did not resemble that of any other species. To trace these evolutionary changes, we selected four legume plastomes based on the results of phylogenetic analysis. Our Mauve alignment of the LSC region among M. fauriei and other legumes generated seven locally collinear blocks (LCBs) that represented a homologous region without rearrangement (Fig 4). These LCBs indicated that M. fauriei experienced at least three inversion events: 1) a 50-kb inversion between IGSs near accD and trnK (UUU), which is the major landmark in most papilionoid legume plastomes; 2) a 36-kb inversion situated between the 29-bp identical sequences of trnS (GGA) and trnS (GCU), a feature shared by Lupinus among core genistoids and Robinia L. in the robinioids; and 3) a 24-kb inversion embedded in the 50-kb inversion region, an event that is newly discovered from the M. fauriei plastome and which occurs between IGSs near trnC (GCA) and trnF (GAA).
Fig 4. Plastome alignment of LSC regions from Maackia fauriei and other legume species.
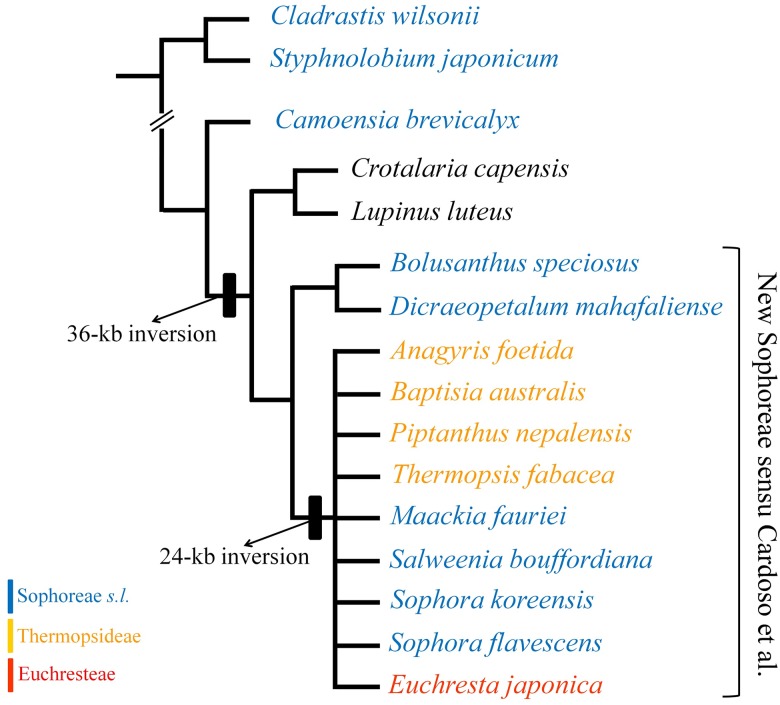
To verify the distribution of these 36-kb and 24-kb inversions on our phylogenetic tree, we used PCR-screening with 16 representative species belonging to five tribes—14 genera of genistoids and the Sophoreae s.l. group. All sequences were deposited in GenBank (Accessions KX430180 through KX430211). This strategy demonstrated that three genera—Cladrastis Raf., Styphnolobium Schott, and Camoensia Welw. ex Benth.–did not have those inversions (Table 2 and Fig 5). However, the 36-kb inversion was distributed within the five tribes of core genistoids while the 24-kb inversion was shared by eight genera in three genistoid tribes—Euchresteae, Sophoreae s.s., and Thermopsideae. Two other genera from Sophoreae s.s.—Bolusanthus and Dicraeopetalum—did not share the 24-kb inversion and had only 36-kb inversions in their plastomes.
Fig 5. Phylogenetic distribution of 36-kb and 24-kb inversion events in plastomes, as surveyed in our study.
All taxa and their classifications are detailed in Table 2. Arrangement of taxa is based on current understanding about phylogenies of these taxa [3–4, 14–16, 25, 47].
Phylogenetic analysis
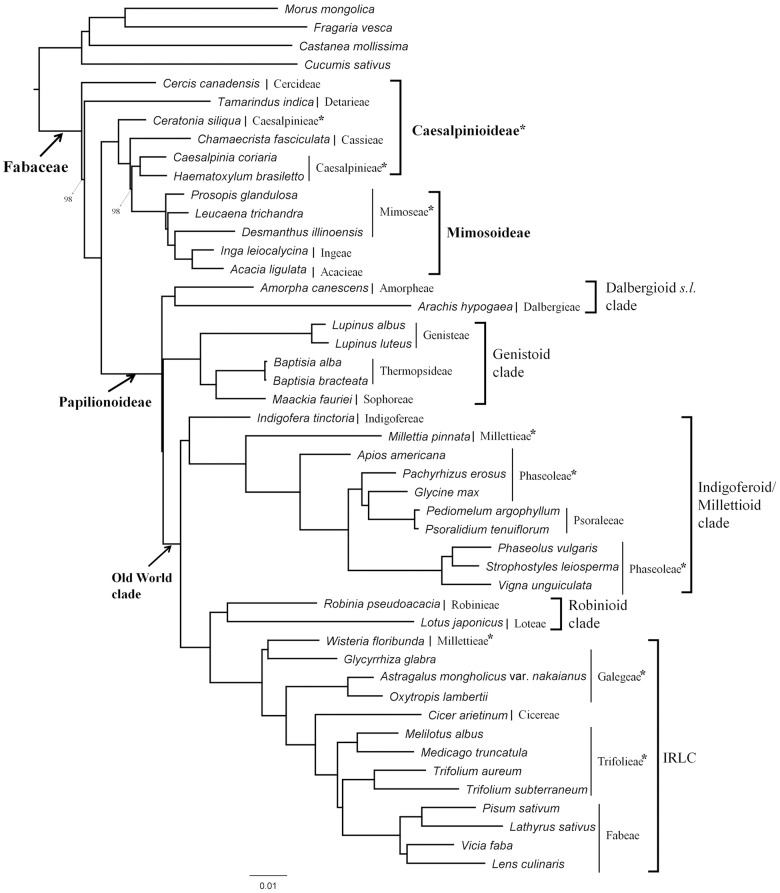
Our data set for the phylogenetic analysis contained 71 protein-coding genes from 47 taxa, including 43 legumes and four outgroups. This accounted for 53,307 nucleotide positions. The ML analysis of those 47 taxa resulted in a single best-scoring tree with—lnL = 358979.245. Bootstrap analyses indicated that, except for two nodes outside of Papilionoideae, all nodes were supported by values of 100%. The ML phylogeny indicated that Caesalpinioideae was basal and paraphyletic and that Mimosoideae and Papilionoideae formed monophyletic groups nested within the Caesalpinioideae (Fig 6). We also recognized five sub-clades of papilionoids, i.e., dalbergioid s.l., genistoid, indigoferoid/millettioid, robinioid, and IRLC. The dalbergioid s.l. was the first diverging clade, followed by genistoids. Branch lengths were very short between nodes of dalbergioid s.l. and the genistoids. Maackia fauriei was nested within the genistoids with Thermopsideae (Baptisia) and Genisteae (Lupinus). The remaining three sub-clades formed the Old World clade, with indigoferoid/millettioid being the first to branch, followed by robinioid and IRLC. Among the 22 tribes of legumes examined here, six (Caesalpinieae, Mimoseae, Millettieae, Phaseoleae, Galegeae, and Trifolieae) were deemed non-monophyletic. Overall, the relationships within legumes were in agreement with recently described phylogenies [2–4]. The exception was the genistoids, which were resolved as a sister group to the Old World clade.
Fig 6. Maximum likelihood phylogeny tree of legume species inferred by RAxML Blackbox.
Phylogenetic relationships among taxa were generated from concatenated matrix of 71 protein-coding genes with combined aligned length of 53,307 characters. Values at nodes are shown when bootstrap support values are not 100%. Asterisks mark taxa that are not monophyletic.
Discussion
Characteristics of the Maackia fauriei plastome and independent rps16 loss in genistoids
The complete plastome of Maackia fauriei includes four rRNA, 31 tRNA, and 76 protein-coding gene species. As with the other legume plastomes [45], this genome lacks rpl22, which is transferred to the nucleus [48]. Moreover, this genome has sequences consistent with the presence of a putative pseudogenized rps16. Using slot blot hybridization with various legume species, Doyle et al. [49] found multiple and independent rps16 losses among papilionoids. Schwarz et al. [45] suggested that these losses have independently occurred at least five times in Papilionoideae, but not in genistoids. However, our analysis of rps16 loss patterns demonstrated that the plastomes of genistoids also experienced independent loss events. We categorized rps16 as an intact gene, putative pseudogene, truncated gene, or complete deletion (Table 2). The plastomes of Caesalpinioideae, Mimosoideae, and some of the Papilionoideae (i.e., Indigofera L., Millettia Wight & Arn., Pachyrhizus Rich. ex DC., Glycine L., and Lotus L.) have intact rps16s that have no internal indels in the exons except at the ends. Truncations occur sporadically in papilionoid legumes and are largely attributed to the severe deletion of exon2. Exon1 of rps16 is relatively conserved in papilionoid legumes, except for complete deletions, as observed with Arachis L., Apios Fabr., and most members of Fabeae. Putative pseudogenes are relatively rare and only observed within Wisteria Nutt. and genistoid species (Fig 3). The rps16 of Lupinus species has previously been annotated as in-frame and is regarded as an intact gene [32, 45]. However, it does not seem to be functional because of severe frame-shifts and deletions in the intron revealed by our investigation. Overall, rps16 losses exist for the five major clades—dalbergioid s.l., genistoid, indigoferoid/millettioid, robinioid, and IRLC—and all feature 50-kb inversions in their plastomes.
Systematic implications of plastome inversions in genistoids and Sophoreae
The genistoid is an informal taxonomic group of legumes that are characterized by quinolizidine alkaloids and a base chromosome number of n = 9 [5, 12, 50]. The tribes Genisteae, Crotalarieae, Euchresteae, Podalyrieae, Sophoreae s.s., and Thermopsideae are consistently resolved as a monophyletic group within genistoids, and are considered the core genistoids [3–4, 12, 14]. We found evidence here that the plastome of Maackia fauriei, belonging to Sophoreae s.s., has undergone three inversion events (50-kb, 36-kb, and 24-kb) based on our Mauve alignment with other legume species (Fig 4). Martin et al. [32] examined the distribution of the 36-kb inversion and their taxon sampling included two species outside of the genistoids [i.e., Cladrastis lutea (Michx.) K. Koch and “Sophora japonica”], plus nine genera of core genistoids belonging to one genus within Thermopsideae (Thermopsis) and eight genera within Genisteae (Argyrolobium Eckl. & Zeyh., Lupinus, Chamaecytisus Link, Laburnum Fabr., Retama Raf., Ulex L., Echinospartum Fourr., and Genista L.). They have argued that the 36-kb inversion is potentially a molecular synapomorphy for core genistoids. In the current study, we examined 16 representative species (Table 2) that included five core genistoid tribes (Crotalarieae, Euchresteae, Genisteae, Sophoreae s.s., and Thermopsideae), and also Sophoreae s.l., that are outside of core genistoid [Cladrastis wilsonii Takeda, Styphnolobium japonicum (L.) Schott (= Sophora japonica L.), and Camoensia brevicalyx Benth.]. The 36-kb inversion is absent from species outside of the core genistoid, including those in its putative sister group, Camoensia. The genus Camoensia was once treated as a core genistoid [3] because it was thought to be a member of Sophoreae s.s. [35]. However, Cardoso et al. [4] resurrected Camoensia as being in the monotypic tribe Camoensieae and treated it as part of a sister group instead. Our findings that differ from those of Martin et al. [32] are the plastome conformation and classification for Styphnolobium japonicum (= Sophora japonica) which is clearly not in the core genistoid group [5, 50–53]. Instead, our research indicates that S. japonicum lacks the 36-kb inversion, similar to its close relative Cladrastis. Three genera—Cladrastis, Pickeringia, and Styphnolobium—are currently known as being free from plastome rearrangements and forming a sister clade to vast papilionoid legume taxa marked by 50-kb inversion [2, 4]. Thus, this contrast with the report by Martin et al. [32] might be an outcome of mistakes made during the earlier experimental process rather than being an independent parallel-inversion event at the intra-species level. Unlike the case of Styphnolobium, the 36-kb inversion has occurred independently for the genus Robinia, which is thought to be distantly related evolutionarily but includes 50-kb inversion in its plastome [45]. Thus, the possible existence of a parallel 36-kb inversion from evolutionarily close taxa means that its value in molecular synapomorphy is questionable among genistoids. Additional plastome sequences from other early-diverging papilionoid legumes are warranted. Nevertheless, the absence of such an inversion in Camoensia but its presence in all core genistoid tribes tested here is distinct evidence that supports the recent research on core genistoids [4].
The Sophoreae s.l. have largely included the taxa with least specialized flowers that are similar to those within Caesalpinioideae [6–7] even though its type genus, Sophora, has more specialized and papilionaceous floral characteristics than other basal papilionoid legumes [33]. Recent molecular phylogenetic studies [3–5, 11–13] have also demonstrated that Sophoreae s.s. is more closely related to tribes of Euchresteae and Thermopsideae, which share specialized papilionaceous flowers. Likewise, our study demonstrated that the Sophoreae s.s. has a more specialized plastome organization (Fig 4) that has resulted from the combination of three inversion events (i.e. 50-kb, 36-kb, and 24-kb). This characteristic is also shared with Euchresteae and Thermopsideae. The new Sophoreae sensu Cardoso et al. [4] includes the merger of Euchresteae and Thermopsideae into Sophoreae and removal of a large number of genera that were traditionally considered members of Sophoreae. The formal taxonomic revision for new Sophoreae remains to be done and a synapomorphic characteristic for this group was lacking. Hence, the distinct plastome organization revealed from our study could be a putative molecular synapomorphy for new Sophoreae. In contrast, it is also conceivable that a 24-kb inversion occurred at distantly related lineages, as was the case for 36-kb parallel inversions [45]. However, the plastome conformation of Sophoreae s.s. is not likely to be a homoplasious characteristic. Researchers have assumed that the 36-kb inversion (39-kb inversion for Robinia) was mediated by flip-flop recombination between the conserved 29-bp repeats of two trnS genes [32, 45]. By comparison, the breakpoints of 24-kb inversion are IGSs near trnC (GCA) and trnF (GAA), which are not conserved sequences. Furthermore, the plastome conformation of Sophoreae s.s. was not formed by a single inversion but, combinationally and sequentially, through three independent inversion events. Hence, a 24-kb inversion seems to be unique to a monophyletic group that comprises Sophoreae s.s. and its closely related tribes Euchresteae and Thermopsideae.
Two genera, Bolusanthus and Dicraeopetalum, that are distributed in the Afro-Madagascan region, have long been included in Sophoreae (whether sensu stricto or lato) [4, 6–7, 35]. However, we found a heterogeneous plastome conformation (i.e., lack of a 24-kb inversion) when those two were compared with other Northern Hemisphere genera (Anagyris, Baptisia, Piptanthus, Thermopsis, Maackia, Salweenia, Sophora, and Euchresta) (Table 2 and Fig 5). The presence of a 36-kb inversion in Bolusanthus and Dicraeopetalum and recent phylogenetic evidence [3–4] indicate that they are members of the core genistoid clade. However, inclusion of either into the new Sophoreae (incl. Euchresteae and Thermopsideae) needs more careful consideration. The ITS phylogeny suggests that inclusion of Bolusanthus and Dicraeopetalum into new Sophoreae could make this tribe polyphyletic because those genera reside within a clade only distantly related to other Sophoreae s.s., and they are grouped with other Afro-Madagascan genera, Neoharmsia R. Vig. and Platycelyphium [54]. Furthermore, some morphologically disparate characteristics of Dicraeopetalum, e.g., radially symmetrical flowers, could make new Sophoreae too complicated to be a natural group. Hence, extensive analyses of morphology and molecular phylogeny based on nuclear as well as plastid data are needed to accomplish formalization of new Sophoreae.
Legume phylogeny with a “plastome-scale data set”
Recent progress in the sequencing of legume plastomes has provided a great deal of valuable data [30, 32, 45, 55–59] that can be used to make phylogenetic inferences based on a “plastome-scale data set” [29]. Those approaches have become more common since the development of next-generation DNA sequencing [28, 30, 60]. Moreover, the usefulness of plastomes in phylogenetic analyses has proven to show good resolution among evolutionarily puzzling taxa [17]. In our study, we examined 43 legumes belonging to 22 tribes, based on 71 conserved protein-coding genes in their plastomes (Fig 6). Previously, the plastome-scale phylogeny that featured the most comprehensive sampling for legumes had been made by Schwarz et al. [45]. They included 32 plastomes belonging to 16 tribes. When one considers the wide diversity inherent to legumes, our sample size was still small. Most tribes (14 of 22) have not been sufficiently sampled to test monophyly. Nevertheless, our ML analysis produced a phylogenetic tree that is very representative of the current understanding about legume phylogeny among higher taxa [2]. This is certainly true for the monophyly of informal groups and the non-monophyly of some legume tribes. For example, large informal taxonomic groups of Papilionoideae (dalbergioid s.l., genistoid, indigoferoid/millettioid, robinioid, and IRLC) are resolved as a monophyletic group with 100% bootstrap support. However, the subfamily Caesalpinioideae and six of the 22 legume tribes are not monophyletic (i.e., Caesalpinieae, Mimoseae, Millettieae Phaseoleae, Galegeae, and Trifolieae). In that sense, we believe it is notable that our phylogenetic analysis supports genistoids as sister to the Old World clade and dalbergioid as an earlier diverging lineage. Although the individual groupings of dalbergioid and genistoids are well-established, their phylogenetic positions and interrelationship are still ambiguous. The topology we describe here may have resulted from taxon sampling bias, due to a lack of enough basal papilionoids. However, the high bootstrap values and overall concordance in topology of Old World clades reported previously in other published studies at least suggest that additional plastome sequences for basal papilionoids will be promising data for investigating the higher-level systematics of legumes.
Conclusion
The first plastome from a member of the tribe Sophoreae s.s. (Maackia fauriei) that we have presented here illustrates the additional, independent loss of rps16 genes from genistoids. Along with the recent discovery of a 36-kb inversion [32], the novel 24-kb inversion described here is critical evidence of the systematics of genistoids and new Sophoreae sensu Cardoso et al. [4]. Our plastome phylogeny demonstrates its potential usefulness when investigating early-diverging groups of Papilionoideae. Thus, sequence variations and the structural rearrangement of these plastomes will serve as a powerful marker when making formal taxonomic treatments for currently known non-monophyletic tribes of legumes and, most importantly, the new Sophoreae of genistoids.
Acknowledgments
The first author thanks Dr. Takeshi Itoh and his colleagues at the National Institute of Agrobiological Sciences, Japan for bioinformatics training. Both authors are grateful to Prof. Joo-Hwan Kim at Gachon University for his suggestions on an early version of this paper and for providing plant materials. Some of the DNA samples were kindly provided by DNA Banks of the Royal Botanic Gardens, Kew and the University of Johannesburg, South Africa. The authors also would like to acknowledge all colleagues—W.B. Cho, J.S. Park, D.H. Lee, and D.P. Jin—at the Plant Systematics Laboratory of Inha University for commenting on this manuscript.
Data Availability
The newly sequenced plastome and related data are available in GenBank (accession numbers KX388160, KX430180-KX430211).
Funding Statement
This research was supported by Inha University grant.
References
- 1.Lewis GP, Schrire BD, Mackinder B, Lock JM, editors. Legumes of the World. Richmond: Royal Botanic Gardens, Kew; 2005. [Google Scholar]
- 2.The Legume Phylogeny Working Group. Legume phylogeny and classification in the 21st century: Progress, prospects and lessons for other species-rich clades. Taxon. 2013;62: 217–248. [Google Scholar]
- 3.Cardoso D, de Queiroz LP, Pennington RT, de Lima HC, Fonty É, Wojciechowski MF, et al. Revisiting the phylogeny of papilionoid legumes: New insights from comprehensively sampled early-branching lineages. Am J Bot. 2012;99: 1991–2013. 10.3732/ajb.1200380 [DOI] [PubMed] [Google Scholar]
- 4.Cardoso D, Pennington RT, de Queiroz LP, Boatwright JS, van Wyk B-E, Wojciechowski MF, et al. Reconstructing the deep-branching relationships of the papilionoid legumes. S Afr J Bot. 2013;89: 58–75. [Google Scholar]
- 5.Pennington RT, Lavin M, Ireland H, Klitgaard B, Preston J, Hu J-M. Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplast trnL intron. Syst Bot. 2001;26: 537–556. [Google Scholar]
- 6.Polhill RM. Sophoreae Sprengel (1818) In: Polhill RM, Raven PH, editors. Advances in legume systematics, Part 1. Richmond: Royal Botanic Gardens, Kew; 1981. pp. 213–230. [Google Scholar]
- 7.Polhill RM. Complete synopsis of legume genera In: Bisby FA, Buckingham J, Harborne JB, editors. Phytochemical dictionary of the Leguminosae, Volume 1 London: Chapman and Hall; 1994. pp. xlix–liv. [Google Scholar]
- 8.Chappill JA. Cladistic analysis of the Leguminosae: the development of an explicit phylogenetic hypothesis In: Crisp MD, Doyle JJ, editors. Advances in legume systematics, Part 7, phylogeny. Richmond: Royal Botanic Gardens, Kew; 1995. pp.1–9. [Google Scholar]
- 9.Herendeen PS. Phylogenetic relationships of the tribe Swartzieae In: Crisp MD, Doyle JJ, editors. Advances in legume systematics, Part 7, phylogeny. Richmond: Royal Botanic Gardens, Kew; 1995. pp.123–132. [Google Scholar]
- 10.Doyle JJ, Doyle JL, Ballenger JA, Palmer JD. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol Phylogenet Evol. 1996;5: 429–438. 10.1006/mpev.1996.0038 [DOI] [PubMed] [Google Scholar]
- 11.Doyle JJ, Chappill JA, Bailey CD, Kajita T. Towards a comprehensive phylogeny of legumes: evidence from rbcL sequences and non-molecular data In: Herendeen PS, Bruneau A, editors. Advances in legume systematics, Part 9. Richmond: Royal Botanic Gardens, Kew; 2000. pp. 1–20. [Google Scholar]
- 12.Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 2004;91: 1846–1862. 10.3732/ajb.91.11.1846 [DOI] [PubMed] [Google Scholar]
- 13.Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol. 2005;54: 575–594. 10.1080/10635150590947131 [DOI] [PubMed] [Google Scholar]
- 14.Crisp MD, Gilmore S, van Wyk BE. Molecular phylogeny of the genistoid tribes of papilionoid legumes In: Herendeen PS, Bruneau A, editors. Advances in legume systematics, Part 9. Richmond: Royal Botanic Gardens, Kew; 2000. pp. 249–276. [Google Scholar]
- 15.Wang HC, Sun H, Compton JA, Yang JB. A phylogeny of Thermopsideae (Leguminosae: Papilionoideae) inferred from nuclear ribosomal internal transcribed spacer (ITS) sequences. Bot J Linn Soc. 2006;151: 365–373. [Google Scholar]
- 16.Zhang M-L, Huang J-F, Sanderson SC, Yan P, Wu Y-H, Pan B-R. Molecular Biogeography of Tribe Thermopsideae (Leguminosae): A Madrean-Tethyan Disjunction Pattern with an African Origin of Core Genistoides. Biomed Res Int. 2015;Article ID 864804, 13 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen RK, Ruhlman TA. Plastid genomes of seed plants In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Dordrecht: Springer Netherlands; 2012. pp. 103–126. [Google Scholar]
- 18.Wolfe KH, Li W-H, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987;84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am J Bot. 1988;75: 1443–1458. [Google Scholar]
- 20.Petit RJ, Brewer S, Bordács S, Burg K, Cheddadi R, Coart E, et al. Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For Ecol Manag. 2002;156: 49–74. [Google Scholar]
- 21.Aoki K, Matsumura T, Hattori T, Murakami N. Chloroplast DNA phylogeography of Photinia glabra (Rosaceae) in Japan. Am J Bot. 2006;93: 1852–1858. 10.3732/ajb.93.12.1852 [DOI] [PubMed] [Google Scholar]
- 22.Sugahara K, Kaneko Y, Ito S, Yamanaka K, Sakio H, Hoshizaki K, et al. Phylogeography of Japanese horse chestnut (Aesculus turbinata) in the Japanese Archipelago based on chloroplast DNA haplotypes. J Plant Res. 2011;124: 75–83. 10.1007/s10265-010-0356-z [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, Lee DH, Choi BH. Phylogeography and genetic diversity of East Asian Neolitsea sericea (Lauraceae) based on variations in chloroplast DNA sequences. J Plant Res. 2013;126: 193–202. 10.1007/s10265-012-0519-1 [DOI] [PubMed] [Google Scholar]
- 24.Doyle JJ, Doyle JL, Ballenger JA, Dickson EE, Kajita T, Ohashi H. A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am J Bot. 1997;84: 541–554. [PubMed] [Google Scholar]
- 25.Kajita T, Ohashi H, Tateishi Y, Bailey CD, Doyle JJ. rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Syst Bot. 2001;26: 515–536. [Google Scholar]
- 26.Han JE, Chung KH, Nemoto T, Choi BH. Phylogenetic analysis of eastern Asian and eastern North American disjunct Lespedeza (Fabaceae) inferred from nuclear ribosomal ITS and plastid region sequences. Bot J Linn Soc. 2010;164: 221–235. [Google Scholar]
- 27.Jansen RK, Kaittanis C, Saski C, Lee S-B, Tomkins J, Alverson AJ, et al. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 2006;6: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104: 19369–19374. 10.1073/pnas.0709121104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stull GW, de Stefano RD, Soltis DE, Soltis PS. Resolving basal lamiid phylogeny and the circumscription of Icacinaceae with a plastome-scale data set. Am J Bot. 2015;102: 1794–1813. 10.3732/ajb.1500298 [DOI] [PubMed] [Google Scholar]
- 30.Williams AV, Miller JT, Small I, Nevill PG, Boykin LM. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol Phylogenet Evol. 2016;96: 1–8. 10.1016/j.ympev.2015.11.021 [DOI] [PubMed] [Google Scholar]
- 31.Jansen RK, Wojciechowski MF, Sanniyasi E, Lee S-B, Daniell H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol Phylogenet Evol. 2008;48: 1204–1217. 10.1016/j.ympev.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin GE, Rousseau-Gueutin M, Cordonnier S, Lima O, Michon-Coudouel S, Naquin D, et al. The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann Bot. 2014;113: 1197–1210. 10.1093/aob/mcu050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tucker SC. Floral ontogeny in Sophoreae (Leguminosae: Papilionoideae): II. Sophora sensu lato (Sophora group). Am J Bot. 1994;81: 368–380. [Google Scholar]
- 34.Fougère-Danezan M, Herendeen PS, Maumont S, Bruneau A. Morphological evolution in the variable resin-producing Detarieae (Fabaceae): do morphological characters retain a phylogenetic signal?. Ann Bot. 2010;105: 311–325. 10.1093/aob/mcp280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennington R, Stirton C, Schrire B. Tribe Sophoreae In: Lewis GP, Schrire BD, Mackinder B, Lock JM, editors. Legumes of the World. Richmond: Royal Botanic Gardens, Kew; 2005. pp. 227–249. [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Messing J. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS ONE. 2011;6: e24670 10.1371/journal.pone.0024670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40: e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20: 3252–3255. 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 40.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33: W686–W689. 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41: W575–W581. 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HT, Kim JS, Moore MJ, Neubig KM, Williams NH, Whitten WM, et al. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS ONE. 2015;10: e0142215 10.1371/journal.pone.0142215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5: e11147 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz EN, Ruhlman TA, Sabir JSM, Hajrah NH, Alharbi NS, Al-Malki AL, et al. Plastid genome sequences of legumes reveal parallel inversions and multiple losses of rps16 in papilionoids. J Syst Evol. 2015;53: 458–468. [Google Scholar]
- 46.Stamatakis A, Hoover P, Rougemont J, Renner S. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 47.Lee WK, Tokuoka T, Heo K. Molecular evidence for the inclusion of the Korean endemic genus “Echinosophora” in Sophora (Fabaceae), and embryological features of the genus. J Plant Res. 2004;117: 209–219. 10.1007/s10265-004-0150-x [DOI] [PubMed] [Google Scholar]
- 48.Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991;10: 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle JJ, Doyle JL, Palmer JD. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot. 1995;20: 272–294. [Google Scholar]
- 50.Kite GC, Pennington RT. Quinolizidine alkaloid status of Styphnolobium and Cladrastis (Leguminosae). Biochem Syst Ecol. 2003;31: 1409–1416. [Google Scholar]
- 51.Sousa M, Rudd VE. Revision del genero Styphnolobium (Leguminosae: Papilionoideae: Sophoreae). Ann Mo Bot Gard. 1993;80: 270–283. Spanish. [Google Scholar]
- 52.Palomino G, Martinez P, Bernal C, Sousa M. Diferencias cromosomicas entre algunas especies de los generos Sophora L. y Styphnolobium Schott. Ann Mo Bot Gard. 1993;80: 284–290. Spanish. [Google Scholar]
- 53.Lee ST, Cook D, Molyneux RJ, Davis TZ, Gardner DR. Alkaloid profiles of Dermatophyllum arizonicum, Dermatophyllum gypsophilum, Dermatophyllum secundiflorum, Styphnolobium affine, and Styphnolobium japonicum previously classified as Sophora species. Biochem Syst Ecol. 2013;49: 87–93. [Google Scholar]
- 54.Edwards D, Hawkins JA. Are Cape floral clades the same age? Contemporaneous origins of two lineages in the genistoids s.l. (Fabaceae). Mol Phylogenet Evol. 2007;45: 952–970. 10.1016/j.ympev.2007.09.014 [DOI] [PubMed] [Google Scholar]
- 55.Sabir J, Schwarz E, Ellison N, Zhang J, Baeshen NA, Mutwakil M, et al. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 2014;12: 743–754. 10.1111/pbi.12179 [DOI] [PubMed] [Google Scholar]
- 56.Dugas DV, Hernandez D, Koenen EJ, Schwarz E, Straub S, Hughes CE, et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci Rep. 2015;5: e16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kellar PRS, Ahrendsen DL, Aust SK, Jones AR, Pires JC. Biodiversity comparison among phylogenetic diversity metrics and between three North American prairies. Appl Plant Sci. 2015;3: e1400108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams AV, Boykin LM, Howell KA, Nevill PG, Small I. The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. PLoS ONE. 2015;10: e0125768 10.1371/journal.pone.0125768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi IS, Kim JH, Choi BH. Complete plastid genome of Astragalus mongholicus var. nakaianus (Fabaceae). Mitochondrial DNA Part A. 2016;27: 2838–2839. [DOI] [PubMed] [Google Scholar]
- 60.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci U S A. 2010;107: 4623–4628. 10.1073/pnas.0907801107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The newly sequenced plastome and related data are available in GenBank (accession numbers KX388160, KX430180-KX430211).