Abstract
Nuclear-encoded pentatricopeptide repeat (PPR) proteins are site-specific factors for C-to-U RNA editing in plant organelles coevolving with their targets. Losing an editing target by C-to-T conversion allows for eventual loss of its editing factor, as recently confirmed for editing factors CLB19, CRR28, and RARE1 targeting ancient chloroplast editing sites in flowering plants. Here, we report on alternative evolutionary pathways for DOT4 addressing rpoC1eU488SL, a chloroplast editing site in the RNA polymerase β′ subunit mRNA. Upon loss of rpoC1eU488SL by C-to-T conversion, DOT4 got lost multiple times independently in angiosperm evolution with intermediate states of DOT4 orthologs in various stages of degeneration. Surprisingly, we now also observe degeneration and loss of DOT4 despite retention of a C in the editing position (in Carica, Coffea, Vicia, and Spirodela). We find that the cytidine remains unedited, proving that DOT4 was not replaced by another editing factor. Yet another pathway of DOT4 evolution is observed among the Poaceae. Although the rpoC1eU488SL edit has been lost through C-to-T conversion, DOT4 orthologs not only remain conserved but also have their array of PPRs extended by six additional repeats. Here, the loss of the ancient target has likely allowed DOT4 to adapt for a new function. We suggest rps3 antisense transcripts as previously demonstrated in barley (Hordeum vulgare) arising from promotor sequences newly emerging in the rpl16 intron of Poaceae as a new candidate target for the extended PPR stretch of DOT4. Altogether, DOT4 and its target show more flexible pathways for evolution than the previously explored editing factors CLB19, CRR28, and RARE1. Certain plant clades (e.g., Amaranthus, Vaccinium, Carica, the Poaceae, Fabales, and Caryophyllales) show pronounced dynamics in the evolution of editing sites and corresponding factors.
Keywords: RNA-binding PPR proteins, protein-RNA recognition, PPR-RNA code, plant mitochondria and chloroplasts, angiosperm evolution
Introduction
Plant organelle C-to-U RNA editing remains a mystery with regard to the reasons for its existence in the first place, even three decades after its discovery (Covello and Gray 1989; Gualberto et al. 1989; Hiesel et al. 1989). Hundreds, and in some species even thousands, of RNA editing sites in the transcriptomes of chloroplasts and mitochondria mainly reconstitute amino acid codon identities that could be correctly encoded in the organelle DNAs (Hecht et al. 2011; Oldenkott et al. 2014). This fact is well reflected by the (likely ancestral) absence of RNA editing in algae (Cahoon et al. 2017) or its loss in the marchantiid liverworts (Steinhauser et al. 1999; Rüdinger et al. 2012) where conserved organelle genes have no need for correction at the transcript level.
Variability in RNA editing efficiencies across different plant tissues, in different developmental stages or under different environmental conditions, is occasionally discussed as having possible regulatory roles (Bock et al. 1993; Karcher and Bock 1998, 2002a, 2002b; Miyata and Sugita 2004; Kahlau and Bock 2008). However, RNA editing varies widely even among closely related taxa with C-to-U editing sites present in one species but with a “pre-edited” thymidine at genomic level making editing obsolete in a sister taxon and vice versa. Essentially, the same can be said about “reverse” U-to-C RNA editing accompanying C-to-U editing in hornworts, lycophytes, and ferns, even when it exceeds the latter (Guo et al. 2015; Knie et al. 2016).
The reasons for the existence of RNA editing become yet more puzzling when the complex molecular apparatus to perform RNA editing is considered (Takenaka 2014; Schallenberg-Rüdinger and Knoop 2016; Sun et al. 2016; Gutmann et al. 2017). At the core of the C-to-U editing machinery are RNA-binding pentatricopeptide repeat (PPR) proteins targeting individual or multiple editing sites. Accordingly, the numbers of members in the gene families encoding PPR proteins run into the hundreds in land plant nuclear genomes (O’Toole et al. 2008; Barkan and Small 2014). PPR proteins acting as editing factors are of a characteristic “PLS”-type with P-, L-, and S-type PPR variants making up the PPR array for RNA target recognition. Additionally, the PPR proteins acting as RNA editing factors feature highly conserved carboxyterminal protein domains E1, E2, and DYW directly behind their PLS-type PPRs (Cheng et al. 2016). The DYW domain, so named after the conserved tripeptide motif at its very end, is of particular interest given its evident similarity to characterized cytidine deaminases, suggesting it to carry the enzymatic activity for cytidine-to-uridine conversion by deamination (Salone et al. 2007; Iyer et al. 2011; Hayes et al. 2013; Boussardon et al. 2014; Hayes et al. 2015; Wagoner et al. 2015).
Loss of an RNA editing site by C-to-T conversion in the organelle genome can be expected to be a prerequisite for subsequent loss of its corresponding editing factor in the nucleus. This scenario has indeed been well confirmed. We have recently found that chloroplast editing factors CLB19, CRR28, and RARE1 are highly conserved in flowering plant nuclear genomes as long as RNA editing at their respective chloroplast target sites is required (Hein et al. 2016; Hein and Knoop 2018). The editing factors may disappear, however, once their target sites are converted into thymidines, making RNA editing unnecessary. A single-target editing factor like RARE 1 (addressing editing site accDeU794SL) proved to be lost more frequently during flowering plant evolution than the dually targeted editing factors CRR28 (addressing edits ndhBeU467PL and ndhDeU878SL) or CLB19, which targets RNA editing sites rpoAeU200SF and clpPeU559HY. Expected intermediate stages of evolution with loss of the editing site(s) under retention of the editing factor were identified for RARE1 and CLB19 with more flowering plant species included in the taxon sampling (Hein and Knoop 2018).
Here, we have investigated DOT4, which has been characterized as the editing factor addressing the chloroplast rpoC1eU488SL editing site in Arabidopsis thaliana (Hayes et al. 2013). The DOT4 target editing site was found conserved in the early-branching angiosperm Amborella trichopoda (Hein et al. 2016) and we now report on an ancient and phylogenetically wide conservation of DOT4 and its target among flowering plants. We find that DOT4 shows evidence for functional disintegration, or is lost altogether, several times independently in our sampling of 121 angiosperms following C-to-T conversions at the chloroplast rpoC1eU488SL target, making editing of a serine into a leucine codon obsolete.
However, we now also observe surprising alternative evolutionary pathways for evolution of DOT4 and its target. Losses of DOT4 may also occur despite retention of its editing target rpoC1eU488SL in Carica, Vicia, Coffea, and Spirodela where editing would still be necessary to reconstitute the evolutionarily conserved leucine residue in rpoC1. Checking upon these cases, we find that the unedited cytidine is retained at cDNA level, evidently indicating that no other editing factor is replacing the lost DOT4 functionality. Accordingly, the lack of editing causing the exchange of the otherwise highly conserved leucine into serine in the RNA polymerase β′ subunit is tolerated, possibly compensated by other changes in the chloroplast RNA polymerase holoenzyme. Conversely, the loss of edit rpoC1eU488SL by C-to-T conversion in Poaceae is only accompanied by degeneration of the carboxyterminal DYW domain of DOT4, whereas its upstream stretch of PPRs for RNA recognition is retained and even extended by six additional PPRs. We suggest that C-to-T conversion at its primordial editing site has allowed DOT4 in Poaceae to adapt to a new function.
Materials and Methods
Identifying DOT4 Orthologs and Phylogenetic Analyses
Arabidopsis thaliana editing factor DOT4 (AT4G18750, Hayes et al. 2013) was used as protein query in BlastP and TBlastN searches (Altschul et al. 1990) against the angiosperm (magnoliophyte) data in the NCBI protein database and in the Transcribed Shotgun Assemblies (TSA) and Whole Genome Shotgun sequences data, respectively (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The previous sampling of angiosperms with high-quality genomic and/or transcriptomic data (Hein and Knoop 2018) was extended by Anthurium amnicola, Persea americana, Helianthus annuus, and Kalanchoe fedtschenkoi to now comprise 121 flowering plant species. Determination of genome and/or transcriptome data quality was done as described previously (Hein and Knoop 2018). The MEGA alignment explorer (Tamura et al. 2013) was used for sequence alignment and processing. Gaps and missing or inaccurate C- and N-terminal sequences in evidently erroneous protein models could be corrected in most cases by TBlastN searches against respective nucleotide databases and rechecking nucleotide sequences for possible sequence errors. Camptotheca acuminata included in our previous taxon sampling (Hein and Knoop 2018) was here retained although its DOT4 homolog remains incomplete at present owing to a small assembly gap of about 60 amino acids in the TSA data.
Care was taken to exclude paralogous PPR proteins. All sequences were used as queries to check whether DOT4 was consistently identified as the closest Arabidopsis homolog and the growing sequence collection was sequentially rechecked with phylogenetic tree constructions to avoid inclusion of paralogs outside the DOT4 ingroup including the Amborella DOT4 ortholog. The ultimate DOT4 alignment is available from the authors upon request. “WebLogos” were created at http://weblogo.threeplusone.com (Crooks et al. 2004).
Phylogenetic Tree Construction
Final alignments were used for calculation of maximum likelihood phylogenetic trees using the IQ-tree webserver (Trifinopoulos et al. 2016) at http://iqtree.cibiv.univie.ac.at. The JTT+F+R5 model of sequence evolution was used as the best-fitting model identified with the implemented ModelFinder (Kalyaanamoorthy et al. 2017). Node reliability was determined from 1,000 bootstrap replicates with ultrafast bootstrap approximation “UFBoot” (Hoang et al. 2018).
Collection of Chloroplast Sequences and RNA Editing Predictions
Wherever available, chloroplast data were collected from completely determined chloroplast genome sequences matching the species of the nuclear genome taxon sampling. In a few cases, cp data were alternatively obtained from Whole Genome Shotgun sequences data, from closely related sister species (indicated by asterisks in fig. 2) or newly determined during this study (e.g., for Metrosideros, Aquilegia, Anthurium, Eichhornia, and Rauvolfia).
Fig. 2.
—Cladograms of 121 angiosperms with reliable nuclear genome or transcriptome data. Asterids, Caryophyllales and Liliopsida have been collapsed in the left panel and Rosids in the right panel, respectively. The cladograms shown follow a modern understanding of flowering plant (Magnoliophyta) phylogeny, as, for example, reflected in the “Open Tree of Life” under https://tree.opentreeoflife.org (Hinchliff et al. 2015). DOT4 orthologs could be detected in all taxa except the ones marked by downward-pointing triangles. The upward-pointing triangles indicate DOT4 orthologs with partially deleted DYW domains. Chloroplast DNA data for rpoC1 were mostly taken from GenBank at the NCBI (www.ncbi.nlm.nih.gov) or obtained in this study. Asterisks indicate species with missing cpDNA information, which were retained given information from closely related taxa lacking nuclear gene assemblies, for example, own data for Metrosideros carminea instead of M. polymorpha, Aquilegia chrysantha instead of A. coerula. Branches colored in blue indicate a thymidine at genomic level, making RNA editing obsolete. Red branches indicate taxa (Carica, Vicia, Cicer, Medicago, Trifolium, Catharanthus, Coffea, Vaccinium, and Spirodela) where an unedited cytidine was found to be retained in the cDNA (see table 1). Species are shaded as indicated according to the number of deviations from 18 most highly conserved amino acid identities in crucial DYW domain motifs (the “PG box,” “HxEx[n]CxxC,” and “Hx[n]CSCxDYW” at the C-terminus underlined in the following: PGCSWIEIKGRVNIFVAGDSSNPETENIEAFLRKVRARMIEEGYSPLTKYALIDAEEMEKEEALCGHSEKLAMALGIISSGHGKIIRVTKNLRVCGDCHEMAKFMSKLTRREIVLRDSNRFHQFKDGHCSCRG/DFW (see supplementary figs. 3 and 4, Supplementary Material online). Camptotheca acuminata (unshaded) was retained in the DOT4 collection although its homolog remains incomplete at present owing to a small assembly gap (of ca. 60 aa) in the TSA data. Stippled boxes highlight the Alismatales investigated for rpoC1eU488SL editing with a widely increased taxon sampling (see fig. 3) and the Fabales with two coexisting DOT4 paralogs in some taxa, here indicated with “(2)” (see supplementary figs. 1 and 4, Supplementary Material online).
Plant Material and Molecular Work
Plant material for Am.trichopoda, Illicium oligandrum, Spirodela polyrhiza, An.amnicola, Agave americana, Phoenix dactylifera, Eichhornia paniculata, Ananas comosus, Aquilegia chrysantha, Banksia serrata, Camptotheca acuminata, Vaccinium macrocarpon, Coffea canephora, Rauvolfia serpentina, Glycine max, Phaseolus vulgaris, Vigna angularis, Cicer arietinum, Medicago truncatula, Trifolium pratense, Vicia faba, Metrosideros carminea, Citrus sinensis, Eutrema salsugineum, Capsella rubella, Symplocarpus foetidus, Lemna minor, Wolffia columbiana, Spathiphyllum wallisi, Dieffenbachia seguine, Amorphophallus titanum, Alocasia odora, and Colocasia esculenta as well as seeds for Lupinus angustifolius, V.angularis, Vic.faba, and C.arietinum were obtained from the Bonn University Botanic Garden. Seeds were kept on humid filter paper until germination and then transferred to soil for several days before nucleic acid preparation. Cocos nucifera, Epipremnum aureum, and Persea americana were obtained from local stores. Total plant nucleic acids were isolated using CTAB-based (Cetyltrimethylammonium bromide) protocols (Doyle and Doyle 1990; Liao et al. 2004). RNA preparations were alternatively done with the TRI reagent protocol (Sigma Aldrich) and different kits of the Macherey-Nagel NucleoSpin series. Random hexamers or gene-specific primers were used for cDNA synthesis with the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific/Fermentas). Gene-specific primers (supplementary table 1, Supplementary Material online) were used to amplify rpoC1 and dot4 gene regions. Polymerase chain reaction (PCR) products were isolated from agarose gels using the NucleoSpin Extract II Kit (Macherey-Nagel) and 2–3 replicates were sequenced directly. Commercial Dideoxy (Sanger) sequencing was done at Macrogen Europe (Amsterdam, the Netherlands). Where RNA editing could not be detected at the rpoC1eU488SL site, procedures were repeated.
An RNA-linker-adaptor strategy was used to detect specific antisense RNA following and adapting published procedures (Bensing et al. 1996; Georg et al. 2010). RNA of Hordeum vulgare was prepared using a modified CTAB protocol (Liao et al. 2004). Following DNase treatment with DNase I (Thermo Fisher Scientific), RNA-5′-polyphosphatase (Epicentre) was used instead of tobacco acid pyrophosphatase to remove the terminal triphosphates at transcription initiation sites. A ribo-oligonucleotide (5′-GAUAUGCGCGAAUUCCUGUAGAACGAACACUAGAAGAAA-3′, Integrated DNA Technologies) was ligated to RNA 5′-monophosphate ends using T4-RNA ligase (Thermo Fisher Scientific). Purification steps performed with NucleoSpin RNA Plant (Macherey-Nagel) were applied after the respective enzymatic treatments. Random hexamers were used for cDNA synthesis prior to Reverse transcription polymerase chain reaction (RT-PCR) with an adaptor primer matching the 5′-region of the ligated ribonucleotide (5′-ATATGCGCGAATTCCTGTAGAACGAACA-3′) and a gene-specific primer in the upstream rps3 coding region (5′-TYGGTTTCAGACTTGGTACAACCC-3′). PCR products were isolated from agarose gels using the NucleoSpin Extract II Kit (Macherey-Nagel) and cloned into the pGEMT-easy vector (Promega). Dideoxy sequencing was done at Macrogen Europe (Amsterdam, the Netherlands).
Results
Conservation of Chloroplast Editing Site rpoC1eU488SL and Its Corresponding Editing Factor DOT4
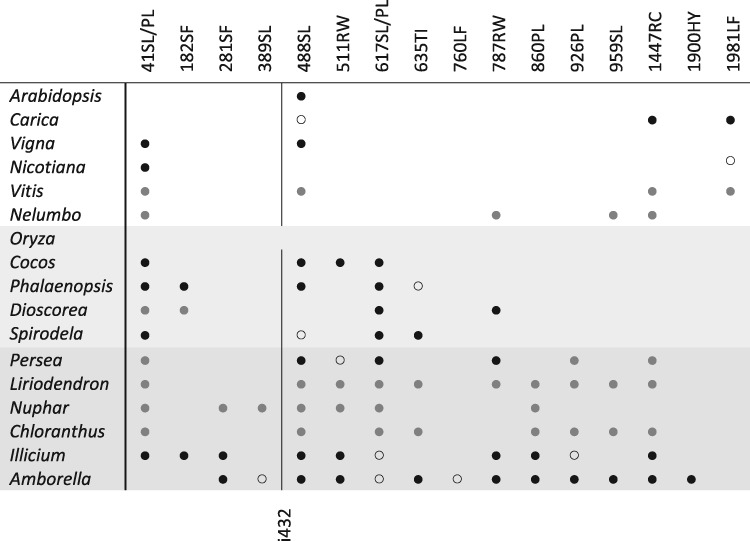
DOT4 (“Defectively Organized Tributaries,” A.thaliana locus At4g18750) is a PPR protein with 18 PLS-type PPRs and a terminal DYW domain that has been characterized as the site-specific RNA editing factor for editing site rpoC1eU488SL converting codon 163 from serine into leucine in the chloroplast rpoC1 mRNA (Hayes et al. 2013). This is the only documented, and expected, RNA editing event in the Arabidopsis rpoC1 mRNA. It is shared with the early-branching flowering plant Am.trichopoda (Hein et al. 2016). Inspecting other early angiosperm lineages, we now document altogether 13 ancient C-to-U RNA editing sites in the rpoC1 mRNA occurring in different patterns (fig. 1). Other than rpoC1eU488SL only rpoC1eU41SL appears to be more widely conserved between eudicots and monocots. We inspected the now available, high-quality genomic and transcriptomic data of 121 flowering plants for the presence of DOT4 orthologs.
Fig. 1.
—Overview on RNA editing in rpoC1 transcripts of selected flowering plants distinguishing early-branching, monocots and eudicot taxa. A total of altogether 13 editing sites was identified in the early-branching angiosperms Amborella, Illicium (star anise), and Persea (avocado). Black dots indicate presence of a C-to-U editing site indicated on top. The slash (SL/PL) indicates that editing to create a leucine codon may alternatively emanate from a proline instead of a serine codon in some species. Gray dots indicate RNA editing predicted at homologous positions in other taxa using PREPACT with default settings (Lenz et al. 2018). Open circles indicate expected editing events, which were not confirmed in cDNA analyses. The vertical line indicates conserved group II intron rpoC1i432g2, which is lost in Oryza, possibly as a consequence of recombination with cDNA from mature mRNA simultaneously erasing editing events as previously documented (e.g., Grewe et al. 2011).
Unequivocal DOT4 orthologs could be clearly identified in (nearly) all angiosperms where RNA editing rpoC1eU488SL remains necessary to re-establish the evolutionarily conserved leucine codon in the chloroplast rpoC1 gene (fig. 2 and supplementary fig. S1, Supplementary Material online). When DOT4 orthologs were identified, they feature full suites of 18 PLS-type PPRs like in the case of the Arabidopsis DOT4 protein with conserved positions 5 and “Last” (L) known to be important for RNA sequence recognition (Barkan et al. 2012) and with carboxyterminal DYW domains including the conserved cytidine deaminase signatures in most cases. However, we also identified several noteworthy exceptions that document a highly dynamic evolution of DOT4 and its target, which we will discuss in the following.
Frequent and Independent Degeneration and Loss of DOT4 after Loss of rpoC1eU488SL
DOT4 orthologs are missing altogether in many taxa where RNA editing rpoC1eU488SL has become obsolete after a cytidine-to-thymidine conversion in the chloroplast DNA (cpDNA) (fig. 2). This is the case for Lamiales, Myrtales, Solanales, and Zingiberales suggesting early losses in these orders. Indeed, checking upon all available cpDNAs in these orders, we find that the rpoC1eU488SL editing site is absent in all Myrtales and Solanales for which sequence information is available. In contrast, the simultaneous absence of rpoC1eU488SL and DOT4 in Allium, Arachis, and Macadamia (fig. 2) suggests more recent losses within the Asparagales, Fabales, and Proteales, respectively. Moreover, the DOT4 case now reveals an interesting evolutionary spectrum for the stepwise degeneration of an RNA editing factor upon loss of its editing target. Despite becoming unnecessary after C-to-T conversion at the previous rpoC1eU488SL editing site, DOT4 shows no evident signs of degeneration in Camelina (Brassicales), in Aquilaria (Malvales), in Aquilegia (Ranunculales), and in Dioscorea and only minor sequence deviations in otherwise highly conserved positions in Agave and in Eichhornia, likely indicating only recent conversion of the editing site (fig. 2).
In other cases, however, the degeneration of DOT4 after loss of its target becomes more pronounced. This is reflected not only by further amino acid exchanges in otherwise highly conserved positions but also by partial or complete deletions of the terminal DYW domain in Daucus (Apiales), Diospyros (Ericales), Helianthus (Asterales), Kalanchoe (Saxifragales), and Nelumbo (Proteales). The Caryophyllales display a particularly wide spectrum of DOT4 sequence degeneration after an early, partial DYW domain deletion, ultimately resulting in complete loss of DOT4 in Amaranthus hypochondriacus, whereas a degenerated DOT4 ortholog is still present in the sister species A. tricolor (fig. 2).
Losses of DOT4 without Previous Loss of the rpoC1eU488SL Editing Target
The above cases show that DOT4 orthologs are mostly retained, while the RNA editing target rpoC1eU488SL is present in the chloroplast genome and that DOT4 becomes stepwise degenerated and ultimately lost upon C-to-T conversions at the previous editing site. However, we now also identified cases where no DOT4 ortholog could be identified in the nuclear genomes although RNA editing in position rpoC1eU488SL was still expected: in Carica papaya, in Vic.faba, and in S.polyrhiza (fig. 2). It could be imagined that another editing factor paralog would act as a substitute for DOT4 in these cases. However, our cDNA studies showed that the genomic C indeed remained unedited in the rpoC1 transcripts (table 1) and that accordingly a serine would be retained instead of a leucine in this otherwise highly conserved position. Notably, in extending our cDNA sampling, we observed “partial” editing at the rpoC1eU488SL site in many cases, hence resulting in a mix of unedited and edited mRNAs, as also observed in independent studies (table 1). It remains to be seen whether differences in reported editing frequencies, like in the case of A.thaliana (table 1), may be due to variability among biological isolates, tissues, or developmental stages.
Table 1.
Extent of RNA Editing at the rpoC1eU488SL Site for Selected Angiosperm Species
| Species | Order | Tissue | Editing (%) | Source |
|---|---|---|---|---|
| Arabidopsis thaliana | Brassicales | Leaves, Col_0 | 40 | This study |
| Mature leaves | 50 | Hayes et al. (2013) | ||
| Leaves | 24 | Bentolila et al. (2013) | ||
| Col_0 | 15 | Ruwe et al. (2013) | ||
| Capsella rubella | Brassicales | Leaves | 60 | This study |
| Brassica rapa | Brassicales | Mature leaves | 100 | Hayes et al. (2013) |
| Eutrema salsugineum | Brassicales | Leaves | 30 | This study |
| Carica papaya | Brassicales | Leaves | 0 | This study |
| Citrus sinensis | Sapindales | Leaves | 45 | This study |
| Mangifera indica | Sapindales | Fruit skin | 70 | This study |
| Betula nana | Fagales | Leaves | 60 | This study |
| Vigna radiata | Fabales | Seedling leaves | 67 | Lin et al. (2015) |
| Vigna angularis | Fabales | Seedling leaves | 30 | This study |
| Phaseolus vulgaris | Fabales | Leaves | 33 | This study |
| Glycine max | Fabales | Leaves | 25 | This study |
| Lupinus angustifolius | Fabales | Seedling leaves | 100 | This study |
| Cicer arietinum | Fabales | Seedling leaves | 0 | This study |
| Medicago truncatula | Fabales | Leaves and flowers | 0 | This study |
| Trifolium pratense | Fabales | Leaves and flowers | 0 | This study |
| Vicia faba | Fabales | Seedling leaves | 0 | This study |
| Vaccinium macrocarpon | Ericales | Leaves | 0 (TI) | This study |
| Catharanthus roseus | Gentianales | Leaves | 0 | This study |
| Coffea canephora | Gentianales | Leaves | 0 | This study |
| Camptotheca acuminata | Cornales | Leaves | 30 | This study |
| Banksia serrata | Proteales | Leaves | 60 | This study |
| Spirodela polyrhiza | Alismatales | ∼20 plantlets | 0 | This study |
| Anthurium amnicola | Alismatales | Leaves | 60 | This study |
| Phoenix dactylifera | Arecales | Leaves | 100 | This study |
| Cocos nucifera | Arecales | Leaves | 50 | This study |
| Ananas comosus | Poales | Leaves | 50 | This study |
| Persea americana | Laurales | Fruit skin | 40 | This study |
| Amborella trichopoda | Amborellales | Leaves | 30 | Hein et al. (2016) |
Note.—Species highlighted in red lack DOT4 homologs in their genomic data (see fig. 2). In the case of Vaccinium macrocarpon, a threonine instead of a serine codon is present in the cpDNA, which could be converted into an isoleucine codon, but this is not observed. Tissue information was kindly provided by the first author of the study involving Arabidopsis thaliana and Brassica rapa (M. Hayes, personal communication).
Because the two Alismatales taxa in the angiosperm-wide sampling (fig. 2) appeared to represent the full range between a species with an apparently intact DOT4 ortholog (An.amnicola) and a species with no recognizable DOT4 ortholog (S.polyrhiza), we chose to investigate the Alismatales more closely. The results for a sampling of 15 Alismatales species is shown in figure 3. The absence of RNA editing, and hence retention of an unedited cytidine, as initially detected in Spirodela, is shared with the other two Lemnoideae (duckweed) species in our sampling, Lemna minor and Wolffia columbiana. In the other taxa of the Araceae editing efficiencies vary in the full range from 100% in Spathiphyllum wallisi to no detectable RNA editing in Epipremnum aureum (fig. 3). Outside the Araceae, a C-to-T conversion has occurred in Potamogeton perfoliatus (pondweed) and the two Hydrocharitaceae taxa Elodea and Najas.
Fig. 3.
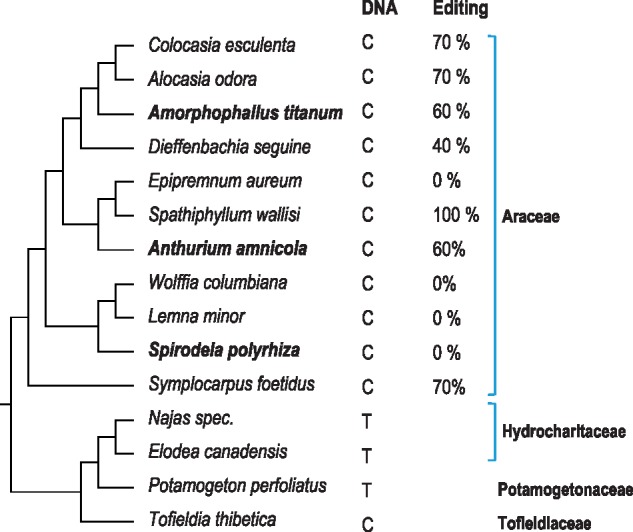
—Editing extent of rpoC1eU488SL in species of the Alismatales with a focus on the Araceae. The column labeled “DNA” indicates the nucleotide identity at genomic level. The extent of editing observed in cDNA investigation is indicated in the next column. Spirodela polyrhiza has no detectable DOT4 ortholog whereas evidently intact DOT4 orthologs could be identified in Anthurium amnicola and Amorphophallus titanum (bold).
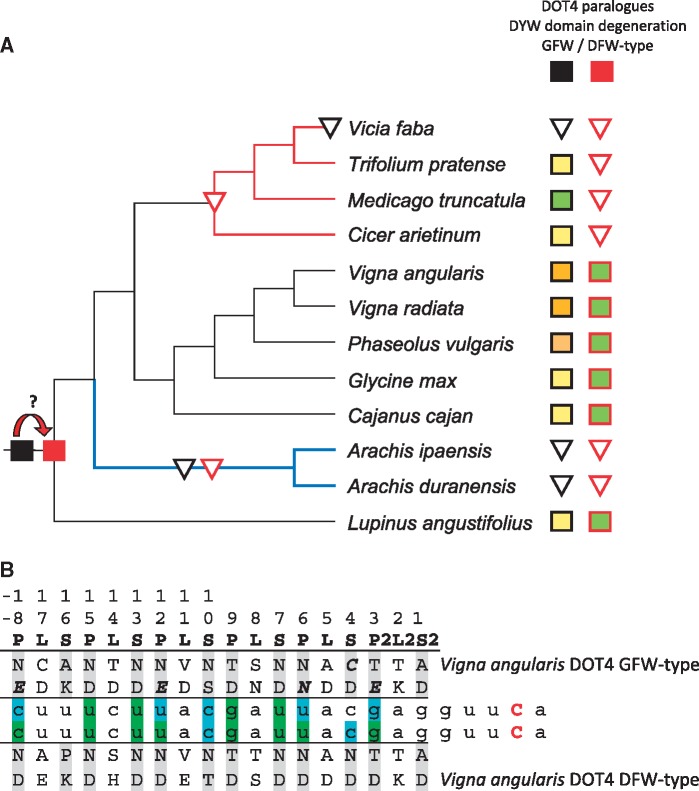
A DOT4 Duplication among Fabales
The DOT4 orthologs exist as single copies in the respective plant nuclear genomes and their phylogeny largely coincides with the phylogeny of flowering plants as currently understood, indicating common ancestry of a single ortholog (supplementary fig. S1, Supplementary Material online). One noteworthy exception are the Fabales where two DOT4 paralogs coexist in Vigna, Phaseolus, Glycine, Cajanus, and Lupinus (fig. 2 and supplementary fig. S1, Supplementary Material online). The phylogeny suggests a deep gene duplication early in Fabales creating two DOT4 copies, a (likely ancient) “GFW-type” and a (likely derived) “DFW-type,” so named after the terminal tripeptides in their DYW domains (fig. 4 and supplementary fig. S1, Supplementary Material online). The DOT4 duplicates may trace back to whole-genome duplication about 58 Myr in the history of the Papilionoideae subfamily ago (Cannon et al. 2015), to which all the species of the current Fabales taxon sampling belong. The DOT4 paralogs have been subsequently lost independently during Fabales diversification. Both copies are lost in Arachis coinciding with a C-to-T conversion at the former editing site and in Vicia where RNA editing activity has been lost (fig. 4). Evidently, this scenario needs to be tested with additional Fabales genome information and denser taxon sampling in the future, especially also outside the Papilionoideae, which show accelerated cp genome evolution (Schwarz et al. 2017).
Fig. 4.
—(A) A possible scenario for DOT4 evolution among Fabales. The likely ancestral “GFW-type” DOT4 (black square) may have given rise to a “DFW-type” paralog (red square). Both paralogs may get lost independently (inverted triangles) during further diversification of the Fabales, leading to the absence of both copies in Arachis concomitant with a C-to-T conversion at the former rpoC1eU488SL editing site (blue branch) and in Vicia, where editing activity has likely been lost earlier (red branches). When present, the (likely derived) DFW-type paralog shows better conservation of crucial residues in the DYW domain than the (likely ancestral) GFW-type copy (color coding as in fig. 2). (B) Fit of the PPR arrays of the two DOT4 paralogs to the rpoC1eU488SL target site, exemplarily shown for Vigna angularis. Color shading follows canonical rules for the PPR-RNA code of P- and S-type PPR repeats (gray shading) in positions 5 and “Last” (T/S + N: A, T/S + D: G, N + N/S: C = U, N + D: C > U). Green indicates perfect match, blue indicates match for pyrimidines by N in position 5. Positions in italics and bold highlight deviations from the likely ancestral state.
Intriguingly, the DFW-type of the duplicated DOT4 paralogs shows better sequence conservation in the DYW domains when present in parallel with the GFW-type copy (fig. 4A), especially within and directly upstream of the PG-Box where the GFW-type copies strongly deviated in most species (see supplementary fig. S3, Supplementary Material online). This is also reflected in the relevant PPR amino acid positions 5 and “Last” (L) important for RNA recognition as exemplarily shown for V.angularis (fig. 4B). The long PPR array of DOT4 generally shows a very good match to its corresponding rpoC1eU488SL target (see also fig. 5) and no evidence for any reassignment to an alternative target can be deduced for the second DOT4 copy, which rather shows a degeneration of the positions identified as relevant for RNA targeting (fig. 4B). The GFW-type DOT4 factors might have lost their ancestral RNA editing functionality early considering that RNA editing is absent in Cicer, Medicago, and Trifolium where (only) the DFW-type DOT4 is lost (figs. 2 and 4). The taxa lacking rpoC1eU488SL editing represent the galegoid or “IR-lacking” IRLC subclade of Papilionoideae, which separated from the milletoid sister subclade about 54 Ma (Schwarz et al. 2017).
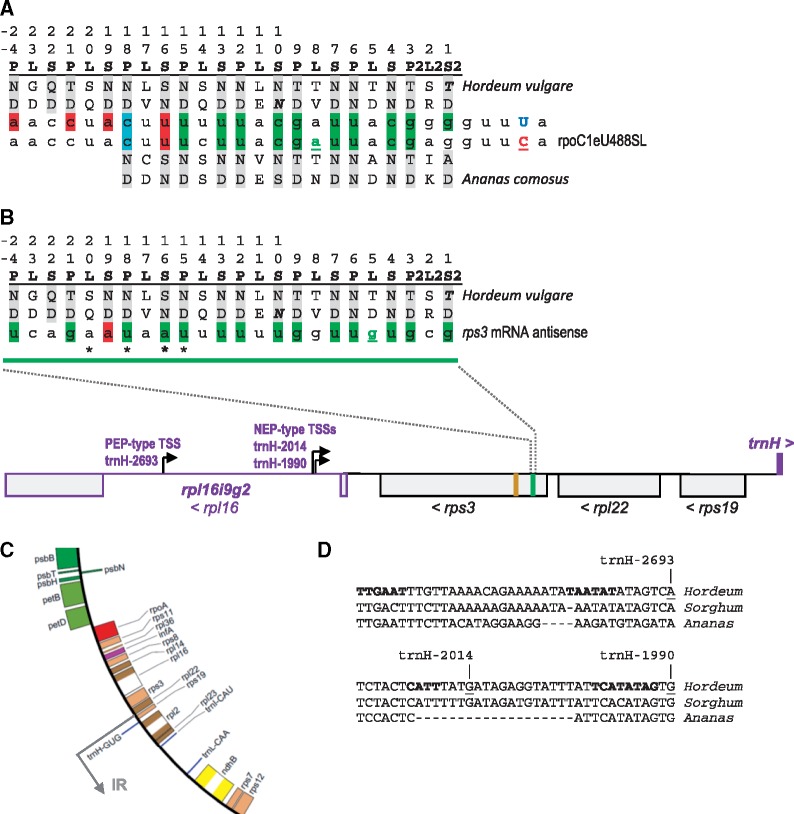
Fig. 5.
—The extended PPR array in DOT4 of Poaceae. Color shading for matches in the P- and S-type PPRs is as in figure 4 plus additional red shading for purine versus pyrimidine mismatches. (A) Ananas comosus representing Bromeliaceae among the Poales features 18 PLS-type PPRs like the Arabidopsis and most other DOT4 homologs (bottom). The Poaceae family shows an amino-terminally elongated PPR array with 6 additional PLS-type PPR repeats (P-24 to S-19), here exemplarily shown for barley, H. vulgare (top). Differences to the Ananas DOT4 homolog affecting binding to the editing target rpoC1eU488SL, lost in the Poaceae by C-to-T conversion (U in blue font), are indicated in bold and italics. (B) The extended PPR array of Poaceae matches to a sequence motif (green) in the upstream part of the chloroplast rps3 gene in antisense orientation with only one mismatch at PPR S-19. Contribution of L-type PPRs to RNA binding is not yet understood, but it is noteworthy that the TD-combination of L-5 in Hordeum matches the guanidine in the rps3 candidate site (B) and that the TN-combination of L-8 matches the adenosine (A) in the rpoC1eU488SL editing target of Ananas in panel (A) (nucleotides underlined and in green font). The rps3 gene is embedded between rpl22 and rpl16 in a large cluster of ribosomal protein genes. A comprehensive barley chloroplast transcriptome study (Zhelyazkova et al. 2012) identified one PEP-type (trnH-2693) and two NEP-type (trnH-2014 and trnH-1990) TSSs within group II intron rpl16i9g2 driving transcription in antisense orientation to the ribosomal gene cluster and in sense orientation to the trnH-GUG gene located upstream of rps19. Asterisks below the rps3 antisense sequence in positions 5, 7, 9 (dominating, bold), and 10 of the candidate DOT4 target indicate variable 5′-ends of antisense RNAs now detected by an RNA oligonucleotide ligation approach. The potential Poaceae DOT4 binding site is located 65 nucleotides apart from a 69 bp (23 amino acid) insertion in rps3 in Poaceae (ocher box). (C) Section of the H. vulgare cpDNA (accession NC_008590) at the LSC-IRB border. Poales cpDNAs are characterized by an extension of the IRs creating a second copy of trnH-GUG in antisense orientation between rpl2 and rps19 within the ancestral ribosomal protein gene cluster. (D) Sequences upstream of the PEP-type (top) and of the two NEP-type TSSs (bottom) identified in Hordeum include sequence motifs (bold) assumed to contribute to promoter activity (Zhelyazkova et al. 2012). The corresponding sequences in rpl16i9g2 are identical in Aegilops and Triticum but increasingly different in more distant taxa like Sorghum and Ananas, here included for comparison.
Loss of the rpoC1eU488SL Editing Target under Retention of DOT4 Orthologs
Whereas the earlier sections document losses of DOT4 despite retention of its editing target rpoC1eU488SL, exactly the opposite is observed among the Poales. The editing target site rpoC1eU488SL is ancestrally lost through C-to-T conversion after a split of Ananas (Bromeliaceae) in the remaining Poales (Poaceae), but unequivocal DOT4 orthologs remain conserved in their nuclear genomes, although with evident signs of degeneration in the terminal DYW domain (fig. 2 and supplementary fig. 3, Supplementary Material online). All Poaceae share a mutation of the HSE motif to HSS (supplementary fig. S3, Supplementary Material online). Because the glutamate (E) residue is of unequivocal importance for RNA editing function (Hayes et al. 2015), the DYW domain is likely unfunctional in Poaceae. Surprisingly, however, the retained DOT4 homologs in the Poaceae not only feature a conserved PPR array, but even have this extended by six additional, amino-terminal PLS-type repeats. Moreover, these six additional PLS-type PPRs feature amino acids in positions 5 and L following canonical PPR-RNA recognition rules, indicative of RNA-binding. However, these would not fit the sequence further upstream of the original editing target (fig. 5A).
We used the new TargetScan module implemented with the recent update of PREPACT (Lenz et al. 2018) to explore potential binding sites for the extended DOT4 homologs in the Poaceae. To this end we used the arbitrary weightings for purine and pyrimidine recognition as introduced before and (optionally) with additional weights of 40% for positions −3 to +2 to match the (former) rpoC1eU488SL editing site (supplementary fig. S2, Supplementary Material online). The latter was intended to account for potential, yet uncharacterized, sequence preferences imposed by the E1, E2, and (degenerating) DYW domain but no differences in top-scoring hits were seen without these extra weights (not shown). Searching for best matches upstream of RNA editing sites in all 15 angiosperm cp editome references currently available in PREPACT, the rpoC1eU488SL edit is identified with top scores despite the fact the six additional PPRs do not at all contribute to binding further upstream (supplementary fig. S2A, Supplementary Material online).
Scanning for matches in complete chloroplast genomes, we find that the extended PPR stretch of DOT4 in Poaceae would best fit to a sequence in the 5′-region of the rps3 coding sequence as exemplarily shown for the H.vulgare cpDNA, accession NC_008590 (supplementary fig. S2B, Supplementary Material online, fig. 5B). However, this sequence fit is in inverse sequence orientation suggesting binding to a potential antisense RNA, which is not immediately to be expected given the location of rps3 in the ribosomal protein cluster rps19-rpl22-rps3-rpl16. On the other hand, several publications have meanwhile reported on the presence of antisense transcripts in chloroplasts (see discussion). Most relevant to our above findings is a comprehensive study on the chloroplast transcriptome of barley (H.vulgare, Poaceae) systematically identifying and differentiating transcription start sites (TSSs) of the plastid-encoded RNA polymerase PEP and the nucleus-encoded (“phage-type”) RNA polymerase NEP (Zhelyazkova et al. 2012). This work identified altogether three TSSs (one of the PEP-type and two of the NEP-type) in antisense orientation within the rpl16 group II intron (rpl16i9g2) immediately downstream of the rps3 gene (fig. 5B). The TSSs were labeled according to their distance upstream of trnH-GUG (Zhelyazkova et al. 2012), the next gene orientated in that direction of transcription. A unique extension of the IRs among Poales cpDNAs places a second copy of trnH-GUG in antisense orientation between rpl2 and rps19 of the ancestral ribosomal protein gene cluster (fig. 5C). To independently confirm rps3 antisense transcripts and check whether they would indeed cover the candidate DOT4 binding site we treated total barley RNA with 5′-RNA-polyphosphatase prior to ligation of a ribo-oligonucleotide to the 5′-monophosphate ends of transcripts as an adaptor target. Followed by specific RT-PCR, this strategy indeed identified rps3 antisense RNAs in the proposed DOT4 binding region extending toward the 5′-end of rps3 for at least 52 nucleotides toward the binding site of a corresponding PCR primer. Intriguingly, the ribooligonucleotide adaptor strategy identified variable 5′-termini of the antisense RNAs likely indicative of ribonucleolytic processing, which clustered in the 5′-region of the proposed DOT4 target (fig. 5B). Dominating among RT-PCR clones was position +9 with five clones, followed by positions +7 and +10 with two clones each and position +5 (one clone). Only one clone revealed a larger antisense RNA with a 5′-end 48 nt. upstream of the suggested DOT4 target.
Aligning the H.vulgare sequences upstream of the antisense TSSs in rpl16i9g2 with homologous sequences in other taxa to check for conservation of the suggested promoter motifs (Zhelyazkova et al. 2012), we observe significant conservation among the two large Poaceae clades (BOP and PACMAD) but much less in Ananas or more distant taxa, most notably for the likely −10 box of the eubacterial-type PEP promoter (fig. 5D).
A persistence of DOT4 orthologs for some evolutionary time after loss of the rpoC1eU488SL editing site, especially of those with apparently intact DYW domains as, for example, in Camelina, Aquilaria, Aquilegia, or Dioscorea (see fig. 2) could be explained by a yet unidentified second editing target of DOT4. Scanning the 15 available angiosperm chloroplast editomes identifies ndhBeU611SL as a second-best match for a (likewise ancestral) chloroplast editing site (supplementary fig. S2C, Supplementary Material online). A significantly lower matching score than for rpoC1eU488SL (950 vs. 1,260) makes this site an unlikely editing target per se. Yet more importantly, edit ndhBeU611SL is also confirmed as an editing site in Solanales taxa like Nicotiana tabacum (supplementary fig. S2C, Supplementary Material online), which lack DOT4 altogether (fig. 2).
Discussion
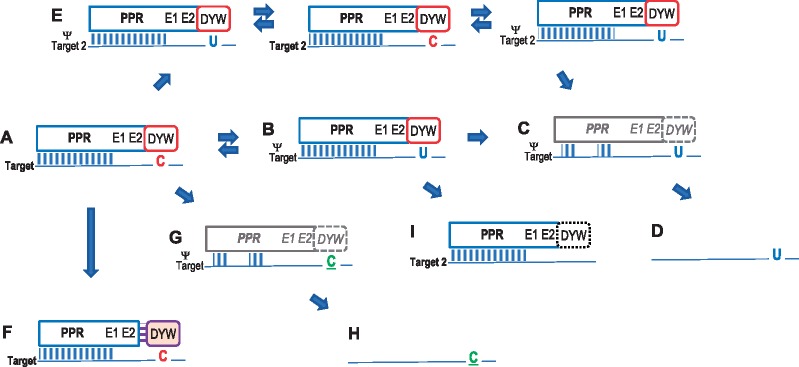
Investigating DOT4 and its RNA editing target site rpoC1eU488SL has revealed new evolutionary pathways and more overall evolutionary dynamics than previously observed for the angiosperm-wide coevolution of chloroplast RNA editing factors and their corresponding targets (fig. 6). Like the previously investigated single-target chloroplast editing factor RARE1 (Hein et al. 2016; Hein and Knoop 2018), DOT4 likewise disappears multiple times independently after C-to-T conversion of its target (fig. 6D). This has happened at least nine times during angiosperm diversification (fig. 2).
Fig. 6.
—Coevolution of PPR-type editing factors and their corresponding editing sites. An editing site (C in red font) addressed by a PPR-type editing factor (A) may get lost by C-to-T conversion (U in blue font) in the organelle genome (B), allowing for degeneration (C) and ultimate loss (D) of its corresponding editing factor. Owing to evolving sequence similarities, an editing factor may extend its activity to further targets (E) or alternative functions (I) on the respective RNA. Multiple-target editing factors need C-to-T conversion at all essential editing targets simultaneously and accordingly disintegrate and vanish more rarely. The DYW domain of editing factors may get degenerated, lost and supplemented in trans (F). The here investigated DOT4 editing factor shows alternative pathways of evolution. Degeneration of an editing factor may lead to loss of editing activity at the ancestral target and retention of the cytidine (C in green font) in the mature mRNA (G), likewise allowing for ultimate loss of the editing factor (H).
In the case of RARE1, only one single example for retention of an apparently unaffected RARE1 ortholog (in chestnut and oak among the Fagales) had been identified after loss of its editing target site accDeU794SL, likely reflecting an evolutionary intermediate state (fig. 6B). In contrast, the recently characterized chloroplast editing factor EMB2261 was found to be conserved after loss of its editing target rps14eU149PL among Fabales and Poales (Sun et al. 2018). Notably, a parallel study had found that EMB2261 (alternatively named ECD1) has a secondary effect, partially reducing editing at six other cp editing sites (Jiang et al. 2018). In contrast to RARE1 and EMB2261/ECD1, we now observed a wide spectrum of retained DOT4 orthologs in different stages of degeneration (fig. 6C) after loss of its target site rpoC1eU488SL in at least 13 independent cases, ranging from DOT4 homologs with a well-conserved terminal DYW domain to those with mutations in crucial conserved peptide motifs or with a truncated DYW domain (fig. 2). C-terminal truncations of an editing factor have previously also been identified for CRR21 after C-to-T conversions at its editing target ndhDeU383SL among Brassicaceae (Hayes et al. 2012).Yet more intriguing are the cases of DOT4 losses without a concomitant C-to-T conversion at genomic level (fig. 6H), which are hitherto unparalleled in the evolution of RARE1. Multiple independent losses not of the rpoC1eU488SL target but of RNA editing activity at the rpoC1eU488SL site (fig. 6G) are now identified in at least six independent cases (fig. 2). Again, also for this scenario, the full range from reasonably conserved (in Catharanthus) over degenerated (in Coffea and Vaccinium) to completely vanished DOT4 orthologs in Carica, Vicia, and Spirodela is observed (fig. 2). In contrast, we found no evidence that DOT4 activity could be substituted by another editing factor, that is, for losses of DOT4 under retention of editing activity at the rpoC1eU488SL target. Most importantly, no such examples for a tolerated loss of editing activity had been found in the previous investigations of CLB19, CRR28, and RARE1 and their chloroplast editing targets (Hein et al. 2016; Hein and Knoop 2018). On a much smaller taxonomic scale, however, the loss of RNA editing at the ndhBeU830SL site in the Chiifu-401 cultivar of Brassica rapa, coinciding with a nonsense-mutation in the corresponding editing factor ELI1 (Hayes et al. 2013), is another example for tolerated loss of editing activity (fig. 6G).
The intriguing discrepancy between evolution of DOT4 and RARE1 has likely to be explained through the functions of the proteins encoded by the affected mRNAs—an Acetyl-CoA decarboxylase subunit in the case of RARE1 and an RNA polymerase subunit in the case of DOT4. Firstly, it must be remembered that in addition to the RNA polymerase (PEP) encoded by the plastid rpo genes, a nuclear-encoded “phage-type” RNA polymerase (NEP) is simultaneously present in plant chloroplasts (Yu et al. 2014; Börner et al. 2015; Liebers et al. 2017). The two different chloroplast RNA polymerases are known to be active at different promoters and at different developmental stages. It could certainly be speculated that NEP activity is extended in taxa showing deficiencies in rpoC1 editing, should this affect PEP activity. More likely, however, a lack of rpoC1eU488SL editing could be compensated for by compensatory amino acid changes in the PEP/RPO holoenzyme. Protein modeling with I-TASSER (Yang and Zhang 2015) or PHYRE2 (Kelley et al. 2015), respectively, suggests the affected amino acid to be located in, or close to, the sigma factor binding pocket (not shown). It is very unlikely that an S-to-L exchange in the RNA polymerase β′ subunit is functionally irrelevant, or even evolutionary neutral per se, given the high conservation of either the rpoC1eU488SL editing event or the leucine residue at genomic level and the significant phenotype of the DOT4 mutants with erratic leaf development in A.thaliana (Petricka et al. 2008; Hayes et al. 2013). On the other hand, it is intriguing to see that 1) other expected RNA editing events in rpoC1 could not be detected in cDNA studies of certain taxa (fig. 1) and 2) that editing at the rpoC1eU488SL site is only partially detected in many studies, as we here show on a phylogenetic wider scale (table 1) and for our extended sampling focusing on Alismatales in particular (fig. 3).
The above considerations likely reflect a complex scenario. Observed differences in RNA editing frequencies may reflect cell type specific or developmentally regulated editing and/or different contributions of NEP and PEP to chloroplast transcription in different species. Alternatively, missing RNA editing at one site may be compensated by a permanent compensatory mutation elsewhere or by a compensatory editing site in the rpo subunit transcripts (A, B, C1, and C2). For example, S.polyrhiza lacking the rpoC1eU488SL editing event shows a unique (“orphan”) editing event rpoC2eU2378PL in the rpoC2 mRNA for the RNA polymerase β″ subunit that is not shared by other taxa, which retain a proline at this position (Lenz et al. 2018).
Exactly opposite to the above scenarios, the rpoC1eU488SL target is lost owing to a C-to-T conversion at genomic level in the Poaceae. In that case, however, conserved DOT4 orthologs are retained, albeit with degenerating DYW domains. Intriguingly, the retained DOT4 orthologs in the Poaceae are not only highly conserved in sequence but even have their PPR arrays extended by six PLS-type PPRs added aminoterminally (fig. 5). Except for the high degree of conservation, evidence for transcription of the deviant DOT4 homologs in Poaceae, for example, by Triticum TSA data (not shown) supports their functional role. This situation reminds of the recently explored scenario for CLB19, which is likewise retained among the Poales despite loss of its two known editing targets owing to C-T conversions in the cpDNA (Hein and Knoop 2018).
We identify a sequence located in the rps3 CDS in antisense orientation as a candidate target for the extended DOT4 PPR array of the Poaceae (fig. 5B). Several publications have meanwhile reported on the existence of antisense transcripts for individual loci in chloroplasts (Haley and Bogorad 1990; Vera et al. 1992; Nishimura et al. 2004; Zghidi et al. 2007; Marqués et al. 2008; Georg et al. 2010; Hotto et al. 2010; Sharwood, Hotto, et al. 2011; Zghidi-Abouzid et al. 2011; Chevalier et al. 2015; Castandet et al. 2016; Cavaiuolo et al. 2017; Qu et al. 2018) and plastome-wide studies have identified dozens of antisense RNAs in chloroplast transcriptomes (Hotto et al. 2011; Zhelyazkova et al. 2012; Chen et al. 2014; Michel et al. 2018). In support for our speculations, the comprehensive study of the barley chloroplast transcriptome identified three start sites for the transcription of antisense RNA within the rpl16 group II intron downstream of rps3 (Zhelyazkova et al. 2012). The corresponding PEP and NEP promoter sequences driving transcription of antisense RNAs toward the rps3 gene are highly conserved among but not beyond the Poaceae (fig. 5D). Interestingly, this region of the otherwise well-conserved angiosperm cpDNAs is characterized not only by significant sequence variation in group II intron rpl16i9g2 and in the rps3 coding sequence (fig. 5C and D) but also by variable extensions of the flanking IR, which create a second copy of trnH-GUG in inverted orientation between rpl2 and rps19 of the large ribosomal protein cluster whereas it is ancestrally present only as a single-copy gene downstream of psbA at the other end of the LSC. We identified rps3 antisense RNA with variable 5′-termini within the 5′-region of the proposed DOT4 binding site, possibly as a result from 5′-exonuclease activity. Notably, RNaseJ has previously been shown to play a key role in surveillance of chloroplast antisense RNA (Sharwood, Halpert, et al. 2011) and to participate in 5′-end maturation of chloroplast RNAs determined by binding of PPR proteins (Luro et al. 2013).
The retention of DOT4 upon loss of the editing target, including homologs with degenerating DYW domains particularly among the Caryophyllales and Asterales (fig. 2), could certainly also indicate alternative “moonlighting” functions in taxa of these orders. Exemplarily checking on candidate binding sites for the degenerating DOT4 proteins in selected genera (Helianthus, Silene, and Dianthus) with TargetScan (not shown) did not reveal similarly interesting candidate sites in the corresponding chloroplast genome sequences as in the above Poaceae case, however. Reinvestigations are certainly advisable, however, once more details about the intricacies of RNA recognition through PPR arrays are known, such as the differential contribution of different P- and S-type PPRs, the role of mismatches to native targets that are frequently observed or the relevance of L-type PPRs and of the E1 and E2 domains.
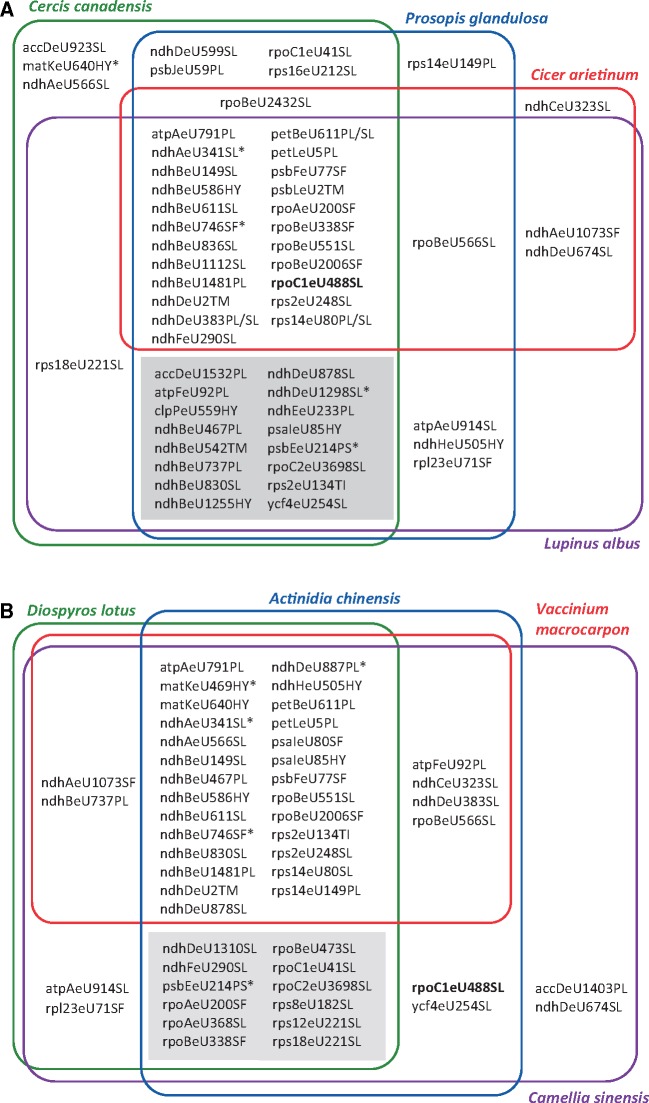
Adding the new insights for the highly dynamic evolution of DOT4 to the previously obtained data for CLB19, CRR28, and RARE1 and their targets suggests that certain angiosperm taxa could experience a particularly dynamic evolution of RNA editing. Other than the Poaceae discussed above this also holds true for the Fabaceae. The unique duplication of an editing factor here found for DOT4 is paralleled by the dynamic evolution of the two editing targets of CRR28 (ndhBeU467PL, ndhDeU878SL), resulting in the hitherto only observed case of CRR28 loss in chickpea (C.arietinum), which here turned out to have lost rpoC1eU488SL editing activity (figs. 2 and 4). Predicting chloroplast RNA editing for representatives of all four different subfamilies of the Fabaceae, we observe that C.arietinum representing the Papilionoideae indeed shows the most dramatic change in its editome having lost 16 likely ancient RNA editing sites (fig. 7A). A similar case are the Ericales where CLB19 is lost after C-to-T conversions at both of its editing targets (clpPeU559HY, rpoAeU200SF) among the Ericaceae including Vaccinium (Hein and Knoop 2018), which at the same time also has lost rpoC1eU488SL editing activity as now found (fig. 2). Comparing the chloroplast editomes of Ericales in our sampling, we again observe a significant loss of ancient RNA editing positions in Vaccinium macrocarpon (fig. 7B).
Fig. 7.
—Venn diagrams of editomes predicted for chloroplast genomes of (A) selected representative taxa of the four Fabaceae clades “Caesalpinioideae” (Cercis canadensis: KF856619), “Mimosoideae” (Prosopis glandulosa: KJ468101), “IR-containing Papilionoideae” (Lupinus albus: KJ468099), and “IR-lacking Papilionoideae” (Cicer arietinum: NC_011163), as previously defined (Schwarz et al. 2017) and (B) the Ericales taxa in the current angiosperm taxon sampling (Vaccinium macrocarpon: NC_019616, Actinidia chinensis: NC_026690, Camellia sinensis: NC_020019, and Diospyros lotus: NC_030786). RNA editing was predicted with PREPACT3.0 (Lenz et al. 2018) using a strict prediction threshold from minimally 70% from all 15 available angiosperm chloroplast editome references and minimally one case of confirmed editing at a given site. The majority of sites likely represent editing events of ancient origin because they are shared with Amborella trichopoda or at least one monocot reference. Rare exceptions possibly originating only after the monocot–eudicot split are labeled with asterisks. Accordingly, differential occurrences of edits likely reflect losses rather than gains. Editing site rpoC1eU488SL, here of interest here as a target of DOT4, is highlighted in bold. Significant numbers of edits have been lost in C. arietinum (16 sites) representing the IR-loss clade among Papilionoideae and in V. macrocarpon (12 sites) among the Ericales (gray shadings).
As yet, Amaranthus hypochondriacus represents the most intriguing case of fast evolution on genus level, having lost CLB19 and DOT4 while both editing factors can still be identified in the sister species A. tricolor. Dramatic changes in structure and mutation rates in plant mitochondrial genomes, for which the genus Silene is a paradigm (Sloan et al. 2012), are associated with massive changes in RNA editing patterns. Although plant chloroplast genomes evolve much more conservatively, the two Silene species included in our sampling were interestingly shown to have exactly opposite patterns for the presence of the two RNA editing targets of CLB19 (Hein and Knoop 2018).
As could be expected, organelle RNA editing sites and their nuclear-encoded specificity factors prove to be a vast field of molecular coevolution between the different genetic systems in a plant cell. Editing factors may frequently degenerate and ultimately get lost upon becoming obsolete after C-to-T conversions at their ancestral targets (fig. 6A–D). The now investigated case of DOT4 confirms that a single-target editing factor experiences more independent events of degeneration and loss like previously seen for RARE1 in comparison to multiple-target editing factors like CLB19 and CRR28 (fig. 6E). A novel scenario here observed for DOT4 is degeneration and loss of an editing factor under retention of a cytidine in the mRNA, which can evidently be tolerated, likely because of compensating protein function through accompanying changes at other sites. Finally, other than extending its functionality to other editing targets (fig. 6E), a former editing factor with a degenerating DYW domain may change toward a different functionality (fig. 6I). As such, it may become restricted to RNA binding to block or stabilize transcripts or RNA secondary structures, as we here speculate for DOT4 (fig. 5). The previously reported CRR2 protein may be a similar case along those lines (Hashimoto et al. 2003) and Physcomitrella patens PPR43 likely is another prime example for such an evolutionary pathway. It features a carboxyterminal DYW domain, which shows degeneration of conserved sequence motifs including the Zn2+-binding sites like we here observed for DOT4 in many cases. PpPPR43 was shown to act as an intron splicing factor instead of an RNA editing factor (Ichinose et al. 2012).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The work reported here was financed by basic funding of the University of Bonn and the state North-Rhine-Westphalia, respectively. We gratefully acknowledge discussions and comments on the manuscripts from Dr Mareike Schallenberg-Rüdinger, the helpful remarks for improvement of text and content by two anonymous reviewers, the help of our colleagues at the Bonn University Botanic Garden in supplying plant material (foremost Bernd Reinken, Felix Eisenhuth, Josef Manner, and Conny Löhne) and the experimental contributions of Ms Flavia Pavan during an internship in our group.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Barkan A, Small I.. 2014. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 65:415–442. [DOI] [PubMed] [Google Scholar]
- Barkan A, et al. 2012. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8(8):e1002910.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Meyer BJ, Dunny GM.. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci U S A. 93(15):7794–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Oh J, Hanson MRMR, Bukowski R.. 2013. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9(6):e1003584.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R, Hagemann R, Kössel H, Kudla J.. 1993. Tissue- and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids—a new regulatory mechanism? Mol Gen Genet. 240(2):238–244. [DOI] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV.. 2015. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim Biophys Acta Bioenerg. 1847(9):761–769. [DOI] [PubMed] [Google Scholar]
- Boussardon C, et al. 2014. The cytidine deaminase signature HxE(x)nCxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phytol. 203(4):1090–1095. [DOI] [PubMed] [Google Scholar]
- Cahoon AB, Nauss JA, Stanley CD, Qureshi A.. 2017. Deep transcriptome sequencing of two green algae, Chara vulgaris and Chlamydomonas reinhardtii, provides no evidence of organellar RNA editing. Genes (Basel) 8:80. [Google Scholar]
- Cannon SB, et al. 2015. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol. 32(1):193–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castandet B, Hotto AM, Strickler SR, Stern DB.. 2016. ChloroSeq, an optimized chloroplast RNA-Seq bioinformatic pipeline, reveals remodeling of the organellar transcriptome under heat stress. G3 (Bethesda) 6:2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaiuolo M, Kuras R, Wollman FA, Choquet Y, Vallon O.. 2017. Small RNA profiling in chlamydomonas: insights into chloroplast RNA metabolism. Nucleic Acids Res. 45(18):10783–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang J, Yuan G, Liu C.. 2014. Complex interplay among DNA modification, noncoding RNA expression and protein-coding RNA expression in Salvia miltiorrhiza chloroplast genome. PLoS One 9(6):e99314.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, et al. 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85(4):532–547. [DOI] [PubMed] [Google Scholar]
- Chevalier F, et al. 2015. Characterization of the psbH precursor RNAs reveals a precise endoribonuclease cleavage site in the psbT/psbH intergenic region that is dependent on psbN gene expression. Plant Mol Biol. 88(4–5):357–367. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW.. 1989. RNA editing in plant mitochondria. Nature 341:662–666. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 14(6):1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JLJJ, Doyle JLJJ.. 1990. Isolation of plant DNA from fresh tissue. Focus (Madison) 12:13–15. [Google Scholar]
- Georg J, Honsel A, Voss B, Rennenberg H, Hess WR.. 2010. A long antisense RNA in plant chloroplasts. New Phytol. 186(3):615–622. [DOI] [PubMed] [Google Scholar]
- Grewe F, et al. 2011. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 39(7):2890–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto JM, Lamattina L, Bonnard G, Weil JH, Grienenberger JM.. 1989. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341(6243):660–662. [DOI] [PubMed] [Google Scholar]
- Guo W, Grewe F, Mower JP.. 2015. Variable frequency of plastid RNA editing among ferns and repeated loss of uridine-to-cytidine editing from vascular plants. PLoS One 10(1):e0117075.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann B, Royan S, Small I.. 2017. Protein complexes implicated in RNA editing in plant organelles. Mol Plant 10(10):1255–1257. [DOI] [PubMed] [Google Scholar]
- Haley J, Bogorad L.. 1990. Alternative promoters are used for genes within maize chloroplast polycistronic transcription units. Plant Cell 2(4):323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T.. 2003. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J. 36(4):541–549. [DOI] [PubMed] [Google Scholar]
- Hayes ML, Dang KN, Diaz MF, Mulligan RM.. 2015. A conserved glutamate residue in the C-terminal deaminase domain of pentatricopeptide repeat proteins is required for RNA editing activity. J Biol Chem. 290(16):10136–101342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes ML, Giang K, Berhane B, Mulligan RM.. 2013. Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. J Biol Chem. 288(51):36519–36529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes ML, Giang K, Mulligan RM.. 2012. Molecular evolution of pentatricopeptide repeat genes reveals truncation in species lacking an editing target and structural domains under distinct selective pressures. BMC Evol Biol. 12(1):66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J, Grewe F, Knoop V.. 2011. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: the root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 3:344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein A, Knoop V.. 2018. Expected and unexpected evolution of plant RNA editing factors CLB19, CRR28 and RARE1: retention of CLB19 despite a phylogenetically deep loss of its two known editing targets in Poaceae. BMC Evol Biol. 18:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein A, Polsakiewicz M, Knoop V.. 2016. Frequent chloroplast RNA editing in early-branching flowering plants: pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors. BMC Evol Biol. 16:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R, Wissinger B, Schuster W, Brennicke A.. 1989. RNA editing in plant mitochondria. Science 246(4937):1632–1634. [DOI] [PubMed] [Google Scholar]
- Hinchliff CE, et al. 2015. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc Natl Acad Sci U S A. 112:201423041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Huston ZE, Stern DB.. 2010. Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol. 10:213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Schmitz RJ, Fei Z, Ecker JR, Stern DB.. 2011. Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the Arabidopsis transcriptome. G3 (Bethesda) 1:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M, Tasaki E, Sugita C, Sugita M.. 2012. A PPR-DYW protein is required for splicing of a group II intron of cox1 pre-mRNA in Physcomitrella patens. Plant J Cell Mol Biol. 70(2):271–278. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, Rogozin IB, Aravind L.. 2011. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 39(22):9473–9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, et al. 2018. ECD1 functions as an RNA-editing trans-factor of rps14-149 in plastids and is required for early chloroplast development in seedlings. J Exp Bot. 69(12):3037–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Bock R.. 2008. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20(4):856–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D, Bock R.. 1998. Site-selective inhibition of plastid RNA editing by heat shock and antibiotics: a role for plastid translation in RNA editing. Nucleic Acids Res. 26(5):1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D, Bock R.. 2002a. The amino acid sequence of a plastid protein is developmentally regulated by RNA editing. J Biol Chem. 277(7):5570–5574. [DOI] [PubMed] [Google Scholar]
- Karcher D, Bock R.. 2002b. Temperature sensitivity of RNA editing and intron splicing reactions in the plastid ndhB transcript. Curr Genet. 41(1):48–52. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE.. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10(6):845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knie N, Grewe F, Fischer S, Knoop V.. 2016. Reverse U-to-C editing exceeds C-to-U RNA editing in some ferns—a monilophyte-wide comparison of chloroplast and mitochondrial RNA editing suggests independent evolution of the two processes in both organelles. BMC Evol Biol. 16:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz H, Hein A, Knoop V.. 2018. Plant organelle RNA editing and its specificity factors: enhancements of analyses and new database features in PREPACT 3.0. BMC Bioinformatics 19:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z, et al. 2004. Rapid isolation of high-quality total RNA from taxus and ginkgo. Prep Biochem Biotechnol. 34(3):209–214. [DOI] [PubMed] [Google Scholar]
- Liebers M, et al. 2017. Regulatory shifts in plastid transcription play a key role in morphological conversions of plastids during plant development. Front Plant Sci. 8:23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-P, et al. 2015. Transcriptional slippage and RNA editing increase the diversity of transcripts in chloroplasts: insight from deep sequencing of Vigna radiata genome and transcriptome. PLoS One 10(6):e0129396.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luro S, Germain A, Sharwood RE, Stern DB.. 2013. RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res. 41(19):9141–9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqués J, et al. 2008. A set of novel RNAs transcribed from the chloroplast genome accumulates in date palm leaflets affected by brittle leaf disease. Phytopathology 98(3):337–344. [DOI] [PubMed] [Google Scholar]
- Michel EJS, Hotto AM, Strickler SR, Stern DB, Castandet B.. 2018. A guide to the chloroplast transcriptome analysis using RNA-Seq. New York: Humana Press; p. 295–313. [DOI] [PubMed] [Google Scholar]
- Miyata Y, Sugita M.. 2004. Tissue- and stage-specific RNA editing of rps14 transcripts in moss (Physcomitrella patens) chloroplasts. J Plant Physiol. 161(1):113–115. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Kikis EA, Zimmer SL, Komine Y, Stern DB.. 2004. Antisense transcript and RNA processing alterations suppress instability of polyadenylated mRNA in Chlamydomonas chloroplasts. Plant Cell 16(11):2849–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, Knie N, Knoop V.. 2014. Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20(10):1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole N, et al. 2008. On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol. 25:1120–1128. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Clay NK, Nelson TM.. 2008. Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 56(2):251–263. [DOI] [PubMed] [Google Scholar]
- Qu Y, et al. 2018. Ectopic transplastomic expression of a synthetic MatK gene leads to cotyledon-specific leaf variegation. Front Plant Sci. 9:1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdinger M, Volkmar U, Lenz H, Groth-Malonek M, Knoop V.. 2012. Nuclear DYW-type PPR gene families diversify with increasing RNA editing frequencies in liverwort and moss mitochondria. J Mol Evol. 74(1–2):37–51. [DOI] [PubMed] [Google Scholar]
- Ruwe H, Castandet B, Schmitz-Linneweber C, Stern DB.. 2013. Arabidopsis chloroplast quantitative editotype. FEBS Lett. 587(9):1429–1433. [DOI] [PubMed] [Google Scholar]
- Salone V, et al. 2007. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 581(22):4132–4138. [DOI] [PubMed] [Google Scholar]
- Schallenberg-Rüdinger M, Knoop V.. 2016. Coevolution of organelle RNA editing and nuclear specificity factors in early land plants In: Rensing SA, editor. Genomes and evolution of charophytes, bryophytes and ferns. Advances in Botanical Research. Vol. 78 Amsterdam: Elsevier, B.V. p. 37–93. [Google Scholar]
- Schwarz EN, et al. 2017. Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across legume subfamilies. J Mol Evol. 84(4):187–203. [DOI] [PubMed] [Google Scholar]
- Sharwood RE, Halpert M, Luro S, Schuster G, Stern DB.. 2011. Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA 17(12):2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, Hotto AM, Bollenbach TJ, Stern DB.. 2011. Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 17(2):230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10(1):e1001241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauser S, Beckert S, Capesius I, Malek O, Knoop V.. 1999. Plant mitochondrial RNA editing: extreme in hornworts and dividing the liverworts? J Mol Evol. 48(3):303–312. [DOI] [PubMed] [Google Scholar]
- Sun T, Bentolila S, Hanson MR.. 2016. The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 21(11):962–973. [DOI] [PubMed] [Google Scholar]
- Sun YK, Gutmann B, Yap A, Kindgren P, Small I.. 2018. Editing of chloroplast rps14 by PPR editing factor EMB2261 is essential for Arabidopsis development. Front Plant Sci. 9:841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M. 2014. How complex are the editosomes in plant organelles? Mol. Plant 7(4):582–585. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ.. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera A, Matsubayashi T, Sugiura M.. 1992. Active transcription from a promoter positioned within the coding region of a divergently oriented gene: the tobacco chloroplast rpl32 gene. Mol Gen Genet. 233(1–2):151–156. [DOI] [PubMed] [Google Scholar]
- Wagoner JA, Sun T, Lin L, Hanson MR.. 2015. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J Biol Chem. 290(5):2957–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y.. 2015. Protein structure and function prediction using I-TASSER In: Current protocols in bioinformatics. Vol. 52 Hoboken (NJ: ): John Wiley & Sons, Inc; p. 5.8.1–5.8.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q-B, Huang C, Yang Z-N.. 2014. Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front Plant Sci. 5:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghidi W, Merendino L, Cottet A, Mache R, Lerbs-Mache S.. 2007. Nucleus-encoded plastid sigma factor SIG3 transcribes specifically the psbN gene in plastids. Nucleic Acids Res. 35(2):455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zghidi-Abouzid O, Merendino L, Buhr F, Malik Ghulam M, Lerbs-Mache S.. 2011. Characterization of plastid psb T sense and antisense RNAs. Nucleic Acids Res. 39(13):5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, et al. 2012. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24(1):123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.