Google Cloud and Apollo24|7: Building Clinical Decision Support System (CDSS) together

Chaitanya Bharadwaj
Head of Clinical AI Products, Apollo 24|7
Sharmila Devi
AI Consultant, Google
Clinical Decision Support System (CDSS) is an important technology for the healthcare industry that analyzes data to help healthcare professionals make decisions related to patient care. The market size for the global clinical decision support system appears poised for expansion, with one study predicting a compound annual growth rate (CAGR) of 10.4%, from 2022 to 2030, to $10.7 billion.
For any health organization that wants to build a CDSS system, one key block is to locate and extract the medical entities that are present in the clinical notes, medical journals, discharge summaries, etc. Along with entity extraction, the other key components of the CDSS system are capturing the temporal relationships, subjects, and certainty assessments.
At Google Cloud, we know how critical it is for the healthcare industry to build CDSS systems, so we worked with Apollo 24|7, the largest multi-channel digital healthcare platform in India, to build the key blocks of their CDSS solution.
We helped them to parse the discharge summaries and prescriptions to extract the medical entities. These entities can then be used to build a recommendation engine that would help doctors with the “Next Best Action” recommendation for medicines, lab tests, etc.
Let’s take a sneak peek at Apollo 24|7’s entity extraction solutions, and the various Google AI technologies that were tested to form the technology stack.
Datasets Used
To perform our experiments on entity extraction, we used two types of datasets.
i2b2 Dataset - i2b2 is an open-source clinical data warehousing and analytics research platform that provides annotated deidentified patient discharge summaries made available to the community for research purposes. This dataset was primarily used for training and validation of the models.
Apollo 24|7’s Dataset - De-identified doctor’s notes from Apollo24|7 were used for testing. Doctors annotated them to label the entities and offset values.
Experimentation and choosing the right approach — Four models put to test
For entity extraction, both Google Cloud products and open-source approaches were explored. Below are the details:
1. Healthcare Natural Language API: This is a no-code approach that provides machine learning solutions for deriving insights from medical text. Using this, we parsed unstructured medical text and then generated a structured data representation of the medical knowledge entities stored in the data for downstream analysis and automation. The process includes:
Extract information about medical concepts like diseases, medications, medical devices, procedures, and their clinically relevant attributes;
Map medical concepts to standard medical vocabularies such as RxNorm, ICD-10, MeSH, and SNOMED CT (US users only);
Derive medical insights from text and integrate them with data analytics products in Google Cloud.
The advantage of using this approach is that it not only extracts a wide range of entity types like MED_DOSE, MED_DURATION, LAB_UNIT, LAB_VALUE, etc, but also captures functional features such as temporal relationships, subjects, and certainty assessments, along with the confidence scores. Since it is available on Google Cloud, this offers long-term product support. It is also the only fully-managed NLP service among all the approaches tested and hence, it requires the least effort to implement and manage.
But one thing to keep in mind is that since the Healthcare NL API offers natural language models that are pre-trained, it currently cannot be used for custom entity extraction models trained using custom annotated medical text or to extract custom entities. This has to be done via AutoML Entity Extraction for Healthcare, another Google Cloud service for custom model development. Custom model development is important for adapting the pre-trained models to new languages or region-specific natural language processing, such as medical terms whose use may be more prevalent in India than in other regions
2. Vertex AutoML Entity Extraction for Healthcare: This is a low-code approach that’s already available on Google Cloud. We used AutoML Entity Extraction to build and deploy custom machine learning models that analyzed documents, categorized them, and identified entities within them. This custom machine learning model was trained on the annotated dataset provided by the Apollo 24|7 team.
The advantage of AutoML Entity Extraction is that it gives the option to train on a new dataset. However, one of the prerequisites to keep in mind is that it needs a little pre-processing to capture the input data in the required JSONL format. Since this is an AutoML model just for Entity Extraction, it does not extract relationships, certainty assessments, etc.
3. BERT-based Models on Vertex AI: Vertex AI is Google Cloud’s fully managed unified AI platform to build, deploy, and scale ML models faster, with pre-trained and custom tooling. We experimented with multiple custom approaches based on pre-trained BERT-based models, which have shown state-of-the-art performance in many natural language tasks. To gain better contextual understanding of medical terms and procedures, these BERT-based approaches are explicitly trained on medical domain data. Our experiments were based on BioClinical BERT, BioLink BERT, Blue BERT trained on Pubmed dataset, and Blue BERT trained on Pubmed + MIMIC datasets.
The major advantage of these BERT-based models is that they can be finetuned on any Entity Recognition task with minimal efforts.
However, since this is a custom approach, it requires some technical expertise. Additionally, it does not extract relationships, certainty assessments, etc. This is one of the main limitations of using BERT-based models.
4. ScispaCy on Vertex AI: We used Vertex AI to perform experiments based on ScispaCy, which is a Python package containing spaCy models for processing biomedical, scientific or clinical text.
Along with Entity Extraction, Scispacy on Vertex AI provides additional components like Abbreviation Detector, Entity Linking, etc. However, when compared to other models, it was less precise, with too many junk phrases, like “Admission Date,” captured as entities.
“Exploring multiple approaches and understanding the pros/cons of each approach helped us to decide the one that would fit our business requirements." according to Abdussamad M, Engineering Lead at Apollo 24|7.
Evaluation Strategy
In order to match the parsed entity with the test data labels, we used extensive matching logic that comprised of the below four methods:
Exact Match - Exact match captures entities where the model output and the entities in the test dataset match. Here, the offset values of the entities have also been considered. For example, the entity “gastrointestinal infection” that is present as-is in both the model output and the test label will be considered an “Exact Match.”
Match-Score Logic - We used a scoring logic for matching the entities. For each word in the test data labels, every word in the model output is matched along with the offset. A score is calculated between the entities and based on the threshold, it is considered as a match.
Partial Match - In this matching logic, entities like “hypertension” and “hypertensive” are matched based on the Fuzzy logic.
UMLS Abbreviation Lookup - We also observed that the medical text had some abbreviations, like AP meaning abdominal pain. These were first expanded by doing a lookup on the respective UMLS (Unified Medical Language System) tables and then passed to the individual entity extraction models.
Performance Metrics
We used precision and recall metrics to compare the outcomes of different models/experiments.
Precision (also called positive predictive value) is the fraction of relevant instances among the retrieved instances, while recall (also known as sensitivity) is the fraction of relevant instances that were retrieved.
The below example shows how to calculate these metrics for a given sample.
Example sample: “Krish has fever, headache and feels uncomfortable”
Expected Entities: [“fever”, “headache”]
Model Output: [“fever”, “feels”, “uncomfortable”]


Thus,


Experimentation Results
The following table captures the results of the above experiments on Apollo24|7’s internal datasets.
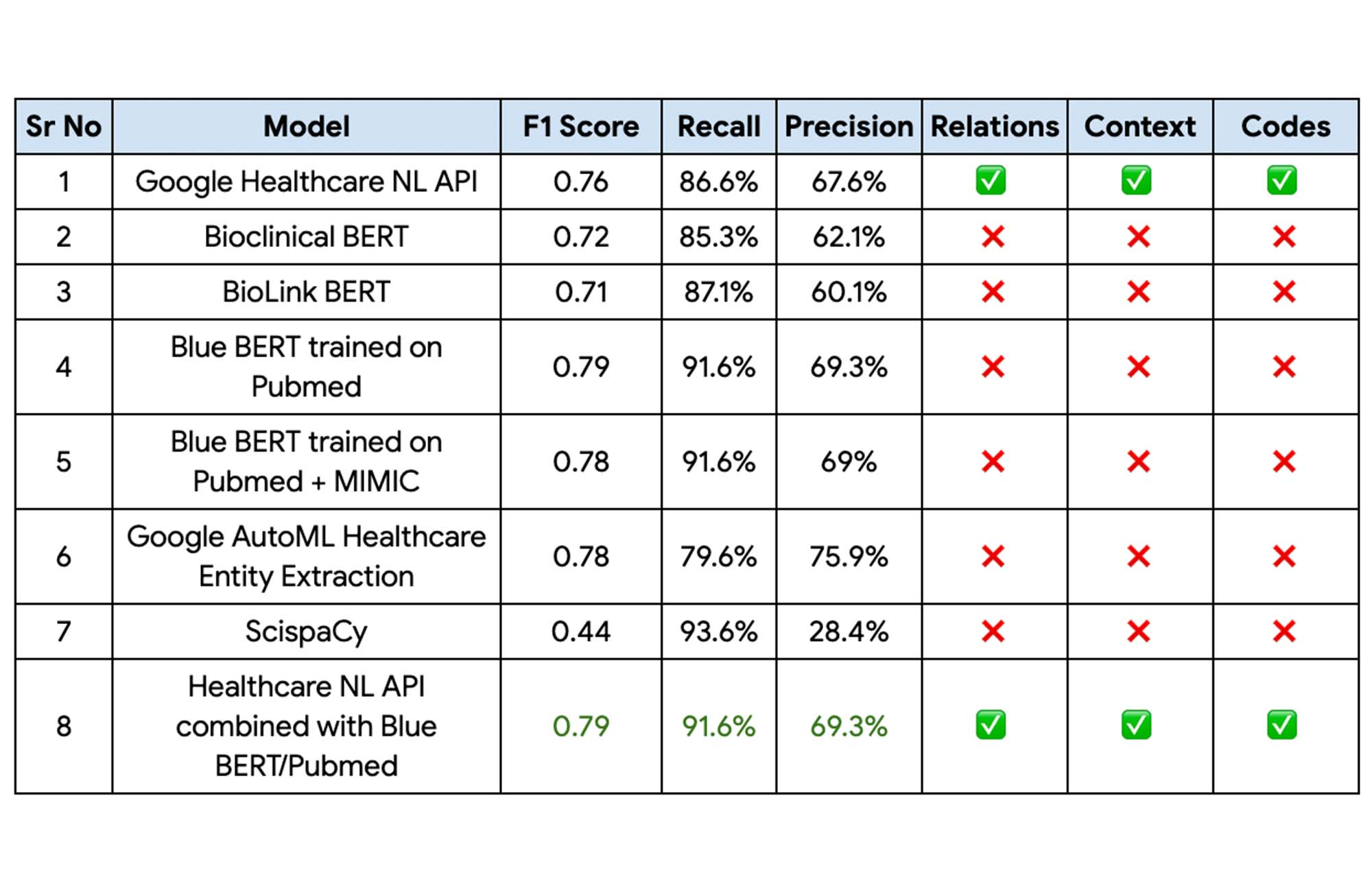

Finally, the Blue BERT model trained on the Pubmed dataset had the best performance metrics with a 81% improvement on Apollo 24|7’s baseline mode with the Healthcare Natural Language API providing the context, relationships, and codes. This performance could be further improved by implementing an ensemble of these two models.
“With the Blue BERT model giving the best performance for entity extraction on Vertex AI and the Healthcare NL API being able to extract the relationships, certainty assessments etc, we finally decided to go with an ensemble of these 2 approaches,“ Abdussamad added.
Fast track end-to-end deployment with Google Cloud AI Services (AIS)
Google AIS (Professional Services Organization) helped Apollo24|7 to build the key blocks of the CDSS system.
The partnership between Google Cloud and Apollo 24|7 is just one of the latest examples of how we’re providing AI-powered solutions to solve complex problems to help organizations drive the desired outcomes. To learn more about Google Cloud’s AI services, visit our AI & ML Products page, and to learn more about Google Cloud solutions for health care, explore our Google Cloud Healthcare Data Engine page.
Acknowledgements
We’d like to give special thanks to Nitin Aggarwal, Gopala Dhar and Kartik Chaudhary for their support and guidance throughout the project. We are also thankful to Manisha Yadav, Santosh Gadgei and Vasantha Kumar for implementing the GCP infrastructure. We are grateful to the Apollo team (Chaitanya Bharadwaj, Abdussamad GM, Lavish M, Dinesh Singamsetty, Anmol Singh and Prithwiraj) and our partner team from HCL/Wipro (Durga Tulluru and Praful Turanur) who partnered with us in delivering this successful project. Special thanks to the Cloud Healthcare NLP API team (Donny Cheung, Amirhossein Simjour, and Kalyan Pamarthy).




