Abstract
Free full text

Cyclic AMP-Dependent Protein Kinase Regulates Pseudohyphal Differentiation in Saccharomyces cerevisiae
Abstract
In response to nitrogen starvation, diploid cells of the yeast Saccharomyces cerevisiae differentiate to a filamentous growth form known as pseudohyphal differentiation. Filamentous growth is regulated by elements of the pheromone mitogen-activated protein (MAP) kinase cascade and a second signaling cascade involving the receptor Gpr1, the Gα protein Gpa2, Ras2, and cyclic AMP (cAMP). We show here that the Gpr1-Gpa2-cAMP pathway signals via the cAMP-dependent protein kinase, protein kinase A (PKA), to regulate pseudohyphal differentiation. Activation of PKA by mutation of the regulatory subunit Bcy1 enhances filamentous growth. Mutation and overexpression of the PKA catalytic subunits reveal that the Tpk2 catalytic subunit activates filamentous growth, whereas the Tpk1 and Tpk3 catalytic subunits inhibit filamentous growth. The PKA pathway regulates unipolar budding and agar invasion, whereas the MAP kinase cascade regulates cell elongation and invasion. Epistasis analysis supports a model in which PKA functions downstream of the Gpr1 receptor and the Gpa2 and Ras2 G proteins. Activation of filamentous growth by PKA does not require the transcription factors Ste12 and Tec1 of the MAP kinase cascade, Phd1, or the PKA targets Msn2 and Msn4. PKA signals pseudohyphal growth, in part, by regulating Flo8-dependent expression of the cell surface flocculin Flo11. In summary, the cAMP-dependent protein kinase plays an intimate positive and negative role in regulating filamentous growth, and these findings may provide insight into the roles of PKA in mating, morphogenesis, and virulence in other yeasts and pathogenic fungi.
In response to nitrogen limitation, diploid cells of Saccharomyces cerevisiae undergo a dimorphic transition to a filamentous growth form referred to as pseudohyphal differentiation (14, 19). This filamentous growth form represents a dramatic change in the cellular program in which the cells elongate, adopt a unipolar budding pattern, remain physically connected in chains, and invade the agar (14, 23). This alternative growth form may enable this nonmotile species to forage for nutrients under adverse conditions.
Two signaling pathways that regulate yeast filamentous growth have been defined. The first involves components of the mitogen-activated protein (MAP) kinase pathway that also functions during mating in haploid cells (8, 28, 35). These components include the kinases Ste20, Ste11, Ste7, and Kss1 and the transcription factor Ste12. In addition, the transcription factor Tec1 forms heterodimers with Ste12 that regulate expression of Tec1 itself and additional targets, such as the cell surface flocculin Flo11 required for invasive and filamentous growth (3, 11, 31, 34). Early elements of the pheromone response pathway, including the pheromones, their receptors, and the coupled heterotrimeric G protein, are not expressed in diploid cells and are not required for filamentous differentiation (28). Instead, the MAP kinase pathway is activated by Cdc42, Ras2, and the 14-3-3 proteins Bmh1 and Bmh2 (38, 39, 49), possibly in response to the Sho1 osmosensing receptor (46).
A second signaling pathway functions in parallel with the MAP kinase pathway to regulate pseudohyphal differentiation. This pathway involves a novel G protein-coupled receptor, Gpr1, which is required for both pseudohyphal differentiation (33) and, in conjunction with Ras2, vegetative growth (63). The Gpr1 ligand has not yet been identified. The Gpr1 receptor is coupled to a heterotrimeric G protein α subunit, Gpa2, which is also required for pseudohyphal differentiation and plays a role in nutrient sensing (26, 32). Early studies suggested Gpa2 might stimulate cyclic AMP (cAMP) production by adenylyl cyclase (41). Consistent with this, cAMP stimulates pseudohyphal differentiation and suppresses the filamentation defects of gpr1 and gpa2 mutant strains (26, 32, 33). A recent study has confirmed that Gpa2 regulates cAMP production by adenylyl cyclase in response to nutritional signals (7). Dominant activated Gpa2 mutants or cAMP suppresses the pseudohyphal defect of mutant strains lacking MAP kinase cascade components (32). In summary, a second signaling pathway comprised of the Gpr1 receptor, the Gpa2 Gα protein, and cAMP regulates pseudohyphal growth in parallel to and independently from the MAP kinase pathway.
The target of cAMP in yeast is the cAMP-dependent protein kinase, protein kinase A (PKA). The yeast PKA kinase is similar to mammalian PKA and consists of a regulatory subunit encoded by a single gene, BCY1, and three catalytic subunits encoded by the TPK1, TPK2, and TPK3 genes (6, 57, 58). In both yeast and mammals, PKA in resting cells is an inactive tetramer composed of two regulatory subunits bound to two active subunits. In response to external signals that increase intracellular cAMP levels, cAMP binds to the regulatory subunit and triggers conformational changes that release the active catalytic subunits. Hydrolysis of cAMP by cAMP phosphodiesterases, the products of the PDE1 and PDE2 genes in yeast, restores PKA to the resting, inactive state (43, 54).
The yeast cAMP-dependent protein kinase is required for vegetative growth (58). Triple mutants lacking the Tpk1, Tpk2, and Tpk3 catalytic subunits are inviable, whereas mutant strains expressing any one of the three Tpk subunits are all viable. These findings led to the model that the three PKA catalytic subunits are largely redundant for function. The PKA catalytic subunits share a conserved C-terminal kinase domain attached to unique N-terminal regions. Tpk1 and Tpk3 share 88% identity in the kinase domain, whereas Tpk2 is more divergent (77 and 75% identity with Tpk1 and -3, respectively). Several candidate PKA targets for vegetative growth have recently been identified. For example, the Msn2 and Msn4 transcription factors are regulated by PKA and repress expression of genes that regulate vegetative growth (4, 18, 56). The Rim15 protein kinase is also phosphorylated and inhibited by PKA and regulates entry into meiosis and stationary phase (48, 60).
In parallel, studies of PKA constitutively activated by cAMP, bcy1 mutation, or activated Ras2 revealed roles in regulating stationary phase, meiosis, and sporulation (5, 59). Activation of PKA prevents glycogen accumulation, heat shock resistance, and survival during nutrient limitation, all hallmarks of entry into stationary phase. Similarly, activation of PKA inhibits sporulation. Thus, activation of PKA promotes vegetative growth in response to nutrients, whereas inactivation of PKA in response to nutrient limitation regulates sporulation and entry into stationary phase.
Several observations suggest PKA might also regulate yeast pseudohyphal differentiation. The dominant active Ras2val19 mutant protein enhances filamentous growth (14), whereas overexpression of the cAMP phosphodiesterase Pde2 inhibits filamentation (62). In addition, exogenous cAMP enhances filamentous growth (32).
Here we report that the cAMP-dependent protein kinase regulates yeast pseudohyphal differentiation. First, we show that mutation of the PKA regulatory subunit Bcy1 enhances filamentous growth. Second, we demonstrate that the PKA catalytic subunits play distinct roles in regulating filamentous growth: the Tpk2 subunit activates filamentous growth, whereas the Tpk1 and Tpk3 subunits primarily inhibit filamentous growth. The unique activating function of the Tpk2 subunit is linked to structural differences in the catalytic region of the kinase and not to differences in gene regulation or the unique amino-terminal region of the protein. Genetic epistasis experiments support a model in which Tpk2 functions downstream of the Gpr1 receptor and the Gα protein Gpa2. Importantly, activation of PKA by mutation of the Bcy1 regulatory subunit restores pseudohyphal growth in mutants lacking elements of the MAP kinase pathway, including ste12, tec1, and ste12 tec1 mutant strains. Thus, the MAP kinase and PKA pathways independently regulate filamentous growth. Further analysis reveals that the PKA pathway regulates the switch to unipolar budding and invasion, whereas the MAP kinase pathway is required for cell elongation and invasion. Finally, our studies define a role for the PKA pathway in activating pseudohyphal growth via transcriptional regulation of the cell surface flocculin Flo11 by the Flo8 transcription factor, and both Flo11 and Flo8 were previously shown to be required for pseudohyphal growth (27, 29, 31). Taken together, our studies reveal an intimate role for the cAMP-dependent kinase in the regulation of yeast dimorphism and suggest this role has been evolutionarily conserved in diverse yeast species and fungi, including pathogens of both plants and animals.
MATERIALS AND METHODS
Media and growth conditions.
Standard yeast media and genetic manipulations were used as described previously (55). Limiting nitrogen media contains 0.17% yeast nitrogen base without amino acids or ammonium sulfate (32), 2% dextrose, 2% Bacto agar, and 50 μM ammonium (SLAD [14]) or 500 μM ammonium (SMAD [1]). SLARG medium, used to induce the dominant active GPA2-2 (Gly132Val) allele, contains 0.5% galactose and 2% raffinose (32).
Yeast strains and plasmids.
The yeast strains used in this study are listed in Table Table11 and are all derived from the Σ1278b strain background. The Δbcy1::G418, Δtpk1::G418, Δtpk2::G418, and Δtpk3::G418 mutations were created by the PCR-mediated gene disruption technique with the G418 resistance cassette from plasmid pFA6-KanMX2 (61). The Δsok2::HygB, Δrim15::HygB, Δflo1::HygB, Δflo5::HygB, Δflo8::HygB, and Δflo11::HygB mutations were generated by PCR-mediated gene disruption with a hygromycin B resistance cassette (17). Independently derived haploid strains (created in strains MLY40α and MLY41a [Table 1]) were mated to produce the homozygous diploid strains (Table (Table1).1). To construct multiply mutant strains, haploid strains with single or double gene deletions were crossed, sporulated, and dissected. Tetrads in which G418 resistance segregated 2 resistant:2 sensitive were chosen and confirmed to contain the expected double or triple G418-resistant gene deletions by PCR analysis of genomic DNA. Sterile ste12 haploid strains were complemented with plasmid pSC4 (STE12 URA3 CEN) before mating, while all of the haploid bcy1 mutant strains were complemented with plasmid pXP1 (2μm BCY1) to increase mating efficiency. The pSC4 and pXP1 plasmids were ejected from homozygous diploid strains by selection on 5-fluoroorotic acid medium. When necessary, a control URA3 plasmid was introduced to complement the ura3-52 mutation and allow growth on SLAD medium.
TABLE 1
Characteristics of the yeast strains used in this study
study
| Strain | Genotype | Source or reference |
|---|---|---|
| MLY40α | ura3-52 MATα | Lorenz and Heitman (32) |
| MLY41a | ura3-52 MATa | Lorenz and Heitman (32) |
| MLY61a/α | ura3-52/ura3-52 MATa/α | Lorenz and Heitman (32) |
| XPY1a/α | Δbcy1::G418/Δbcy1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY1α | Δbcy1::G418 ura3-52 MATα | This study |
| XPY4a/α | Δtpk1::G418/Δtpk1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY5a/α | Δtpk2::G418/Δtpk2::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY5α | Δtpk2::G418 ura3-52 MATα | This study |
| XPY6a/α | Δtpk3::G418/Δtpk3::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY12a/α | Δtpk1::G418/Δtpk1::G418 Δtpk2::G418/Δtpk2::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY13a/α | Δtpk2::G418/Δtpk2::G418 Δtpk3::G418/Δtpk3::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY14a/α | Δtpk1::G418/Δtpk1::G418 Δtpk3::G418/Δtpk3::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY59a/α | Δbcy1::G418/Δbcy1::G418 Δtpk2::G418/Δtpk2::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY69a/α | Δbcy1::G418/Δbcy1::G418 Δste12::G418/Δste12::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY70a/α | Δbcy1::G418/Δbcy1::G418 Δtpk1::G418/Δtpk1::G418 Δtpk2::G418/Δtpk2::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY71a/α | Δbcy1::G418/Δbcy1::G418 Δtpk2::G418/Δtpk2::G418 Δtpk3::G418/Δtpk3::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY73a/α | Δbcy1::G418/Δbcy1::G418 Δgpa2::G418/Δgpa2::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY74a/α | Δbcy1::G418/Δbcy1::G418 Δgpr1::G418/Δgpr1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY75a/α | Δbcy1::G418/Δbcy1::G418 Δtec1::G418/Δtec1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY75α | Δbcy1::G418 Δtec1::G418 ura3-52 MATα | This study |
| XPY76a/α | Δbcy1::G418/Δbcy1::G418 Δste12::G418/Δste12::G418 Δtec1::G418/Δtec1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY77a/α | Δste12::G418/Δste12::G418 Δtec1::G418/Δtec1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY78a/α | Δbcy1::G418/Δbcy1::G418 Δphd1::G418/Δphd1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY80a/α | Δsok2::HygB/Δsok2::HygB ura3-52/ura3-52 MATa/α | This study |
| XPY81a/α | Δsok2::HygB/Δsok2::HygB Δtpk2::G418/Δtpk2::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY88a/α | Δbcy1::G418/Δbcy1::G418 Δphd1::G418/Δphd1::G418 Δtec1::G418/Δtec1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY89a/α | Δphd1::G418/Δphd1::G418 Δtec1::G418/Δtec1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY90a/α | Δrim15::HygB/Δrim15::HygB ura3-52/ura3-52 MATa/α | This study |
| XPY95a/α | Δflo8::HygB/Δflo8::HygB ura3-52/ura3-52 MATa/α | This study |
| XPY95α | Δflo8::HygB ura3-52 MATα | This study |
| XPY99a/α | Δflo8::HygB/Δflo8::HygB Δbcy1::G418/Δbcy1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY99α | Δflo8::HygB Δbcy1::G418 ura3-52/ura3-52 MATα | This study |
| XPY103a/α | Δflo1::HygB/Δflo1::HygB ura3-52/ura3-52 MATa/α | This study |
| XPY105a/α | Δflo5::HygB/Δflo5::HygB ura3-52/ura3-52 MATa/α | This study |
| XPY107a/α | Δflo11::HygB/Δflo11::HygB ura3-52/ura3-52 MATa/α | This study |
| XPY107α | Δflo11::HygB ura3-52 MATα | |
| XPY119a/α | Δflo11::HygB/Δflo11::HygB Δbcy1::G418/Δbcy1::G418 ura3-52/ura3-52 MATa/α | This study |
| XPY119α | Δflo11::HygB Δbcy1::G418 ura3-52 MATα | This study |
| MLY132a/α | Δgpa2::G418/Δgpa2::G418 ura3-52/ura3-52 MATa/α | Lorenz and Heitman (32) |
| MLY182a/α | Δphd1::G418/Δphd1::G418 ura3-52/ura3-52 MATa/α | Lorenz and Heitman (32a) |
| MLY183a/α | Δtec1::G418/Δtec1::G418 ura3-52/ura3-52 MATa/α | Lorenz and Heitman (32a) |
| MLY216a/α | Δste12::G418/Δste12::G418 ura3-52/ura3-52 Δleu2::hisG/Δleu2::hisG MATa/α | Lorenz and Heitman (32) |
| MLY226a/α | Δmsn2::G418/Δmsn2::G418 ura3-52/ura3-52 MATa/α | Lorenz and Heitman (32a) |
| MLY227a/α | Δmsn4::G418/Δmsn4::G418 ura3-52/ura3-52 MATa/α | Lorenz and Heitman (32a) |
| MLY228a/α | Δmsn2::G418/Δmsn2::G418 Δmsn4::G418/Δmsn4::G418 ura3-52/ura3-52 MATa/α | Lorenz and Heitman (32a) |
| MLY232a/α | Δgpr1::G418/Δgpr1::G418 ura3-52/ura3-52 MATa/α | Lorenz et al. (33) |
| DSY1a/α | Δyak1::G418/Δyak1::G418 ura3-52/ura3-52 MATa/α | Surratt and Heitman (56a) |
The plasmids used in this study are listed in Table Table2.2. The plasmid-borne BCY1 (pXP1), TPK1 (pXP2), TPK2 (pXP3), and TPK3 (pXP4) genes were obtained by PCR amplification from genomic DNA of strain MLY61a/α and cloned in the polylinker of the multicopy plasmid YEplac195 (12).
TABLE 2
Characteristics of the plasmids used in this study
study
| Plasmid | Construct | Source or reference |
|---|---|---|
| YEplac195 | 2μm URA3 | Gietz and Sugino (12) |
| pXP1 | BCY1 in YEplac195 | This study |
| pXP2 | TPK1 in YEplac195 | This study |
| pXP3 | TPK2 in YEplac195 | This study |
| pXP4 | TPK3 in YEplac195 | This study |
| pXP5 | Hybrid gene pTPK1-TPK2 in YEplac195 | This study |
| pXP6 | Hybrid gene pTPK2-TPK1 in YEplac195 | This study |
| pXP7 | Hybrid gene TPK1-TPK2 in YEplac195 | This study |
| pXP8 | Hybrid gene TPK2-TPK1 in YEplac195 | This study |
| pSEYC68 | CEN URA3 pGal1, 10 | S. Elledge |
| pML160 | GPA2-2 (Gly132Val) in pSEYC68 | Lorenz and Heitman (32) |
| pML218 | TEC1 driven by the promoter of TDH1 | Lorenz and Heitman (32a) |
| pMW2 | CEN URA3 RAS2 (Gly19Val) | Ward et al. (62) |
| pCG38 | 2μm URA3 PHD1 | Gimeno and Fink (13) |
Photomicroscopy.
All single-colony photographs were taken directly from petri plates by using a Nikon ECLIPSE E400 microscope with a ×10 primary objective and a ×2.5 trinocular camera adaptor for a final magnification of ×25.
Construction of TPK chimeric genes.
Hybrids between the TPK1 and TPK2 genes were constructed by PCR overlap (20) and cloned into the multicopy plasmid YEplac195 (12). To make the pTPK1-TPK2 hybrid gene, primers 5′-CGGGATCCCGAAGCTGTGCTGCTATTC and 5′-GCCCTTTCTGCAACGAATTCCATACCCAAAAAAAAGATTCTTTCAC were used to generate the promoter portion of the TPK1, and primers 5′-GTGAAAGAATCTTTTTTTTGGGTATGGAATTCGTTGCAGAAAGGGC and 5′-CTAGTCTAGACTAGGAGGACTTAAAGCATGTCG were used to amplify the structural and 3′ untranslated region (UTR) region of the TPK2 gene. The products of the first-round PCRs were gel purified and mixed as template to amplify the hybrid pTPK1-TPK2 gene with primers 5′-CGGGATCCCGAAGCTGTGCTGCTATTC and 5′-CTAGTCTAGACTAGGAGGACTTAAAGCATGTCG. The resulting ~2-kb PCR product was gel purified, digested with BamHI and XbaI, and cloned into the multicopy plasmid YEplac195. For the construction of the pTPK2-TPK1 hybrid gene, primers 5′-CGGGATCCCAAGCATCTGTACCTCCAC and 5′-CTCCATTTTGTTCTTCAGTCGACATACCGACAATTTTCAACAGTATG were used to amplify the promoter portion of the TPK2 gene, and primers 5′-CATACTGTTGAAAATTGTCGGTATGTCGACTGAAGAACAAAATGGAG and 5′-GCTGCAGCCGGTGAAAGCTTCTCATC were used to generate the structural and 3′-UTR region of the TPK1 gene. The PCR products were gel purified and combined as the template for a second round of PCR with primers 5′-CGGGATCCCAAGCATCTGTACCTCCAC and 5′-GCTGCAGCCGGTGAAAGCTTCTCATC. The PCR product was gel purified, digested with BamHI and PstI, and cloned into plasmid YEplac195. For the construction of the TPK1-TPK2 hybrid gene, primers 5′-CGGGATCCCGAAGCTGTGCTGCTATTC and 5′-GTCATGTAGTGTATATTTGCCCACTGTAACTCTCGCTTG were used to generate the promoter portion and the coding sequence for the amino-terminal region of the TPK1 gene, and primers 5′-CAAGCGAGAGTTACAGTGGGCAAATATACACTACATGAC and 5′-CTAGTCTAGACTAGGAGGACTTAAAGCATGTCG were used to amplify the coding sequence for the carboxyl-terminal portion and the 3′-UTR of the TPK2 gene. The PCR products were gel purified and combined as the template for a second-round PCR to amplify the TPK1-TPK2 gene with primers 5′-CGGGATCCCGAAGCTGTGCTGCTATTC and 5′-CTAGTCTAGACTAGGAGGACTTAAAGCATGTCG. The PCR product of this reaction was gel purified and cloned into the TA-cloning PCR2.1 vector. The construct was digested with EcoRI, and the desired TPK1-TPK2 hybrid gene was then subcloned into YEplac195. For the construction of the TPK2-TPK1 hybrid, primers 5′-CGGGATCCCAAGCATCTGTACCTCCAC and 5′-GTTCTTGTAAACTATACTTCCTTTGGATACGAGAGATTTC were used to amplify the promoter and the coding sequence for the amino-terminal portion of the TPK2 gene, and primers 5′-GAAATCTCTCGTATCCAAAGGAAGTATAGTTTACAAGAAC and 5′-GCTGCAGCCGGTGAAAGCTTCTCATC were used to amplify the coding sequence for the carboxyl-terminal portion and 3′-UTR of the TPK1 gene. The PCR products were gel purified and combined as the template for a second-round PCR to amplify the TPK2-TPK1 gene with primers 5′-CGGGATCCCAAGCATCTGTACCTCCAC and 5′-GCTGCAGCCGGTGAAAGCTTCTCATC. The product of this PCR was gel purified, digested with BamHI and PstI, and cloned into the multicopy plasmid YEplac195. The junctions of the resulting chimeric genes were confirmed by DNA sequencing.
Cell morphology and budding pattern assays.
Cell shape determination was performed based on a method by Mösch and Fink (38) with minor modifications. Diploid strains were grown on SLAD medium for 16 h at 30°C. Cells were washed off the plates and collected for analysis of the proportion of elongated pseudohyphal cells (PH), ovate yeast form cells (YF), and round yeast form cells (round YF) by microscopic examination. Cells were photographed at a magnification of ×200, and at least 300 cells were counted for each strain.
For budding pattern assays, cells were collected from the diploid strains as described above. Daughter cells were micromanipulated on SLAD medium and incubated for 5 to 6 h at 30°C. The positions of the second and third buds with respect to the first bud were studied by microscopy for three- and four-celled microcolonies. The proportion of the first two buds emerging from opposite poles (bipolar budding) versus those from the same pole (unipolar budding) was analyzed. At least 200 microcolonies were counted for each strain. Calcofluor white staining of the wild-type (MLY61a/α) strain and Δtpk2/Δtpk2 (XPY5a/α) and Δste12/Δste12 (MLY216a/α) mutant strains grown in liquid SLAD medium revealed cells with a unipolar (two or more bud scars at the same pole and opposite to the birth scar) or bipolar budding pattern (one or more bud scars at opposite poles) and no cells with an axial budding pattern, as expected for diploid yeast strains. Cells were photographed at a ×100 magnification, and the number of cells exhibiting a bipolar or unipolar budding pattern were counted.
Northern analysis.
Total RNA was isolated with the QIAGEN RNeasy Mini kit or by acid phenol extraction, separated by electrophoresis, and transferred overnight by capillary action to nylon membranes (VWR Scientific Products). DNA fragments to be used as probes (200-bp PCR products lying 5′ to the TPK1, TPK2, and TPK3 open reading frames (ORFs), a 300-bp PCR product 5′ to the FLO11 open reading frame, and a 500-bp fragment 5′ to the ACT1 open reading frame) were gel purified and radiolabeled by random priming (Boehringer Mannheim). Hybridization, washes, stripping, and reprobing were performed as described previously (53). The radioactive bands were visualized by autoradiography and quantitated with a Molecular Dynamics PhosphorImager. The TPK2 gene was induced to similar extents (~4-fold) by nitrogen starvation in two independent experiments.
RESULTS
PKA regulates pseudohyphal growth.
We recently found that exogenous cAMP stimulates pseudohyphal differentiation in S. cerevisiae (32). This finding, and other previous studies (14, 62), suggested that cAMP-dependent protein kinase (PKA) might regulate pseudohyphal growth.
To test whether PKA regulates yeast filamentous growth, we disrupted the gene encoding the PKA regulatory subunit Bcy1 in the Σ1278b strain background commonly used for studies of filamentous growth. Isogenic BCY1/BCY1 wild-type and bcy1/bcy1 mutant strains were assayed for filamentous growth on medium containing limiting concentrations of ammonium ions. As shown in Fig. Fig.1,1, the bcy1 mutation significantly enhanced filamentous growth on medium containing 50 μM ammonium sulfate (SLAD medium). Moreover, when the ammonium sulfate concentration was increased 10-fold to 500 μM (SMAD medium), filamentation of the BCY1/BCY1 wild-type strain was inhibited, whereas the isogenic bcy1/bcy1 mutant strain continued to exhibit pseudohyphal differentiation (Fig. (Fig.1).1). Introduction of the wild-type BCY1 gene complemented the enhancing effects of the bcy1 mutation and restored pseudohyphal growth to the wild-type level (data not shown). Because the PKA catalytic subunits are released in a constitutively active form in mutants lacking the Bcy1 regulatory subunit, these findings indicate that PKA regulates filamentous growth.
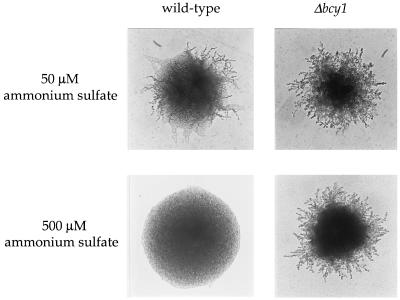
Deletion of the PKA regulatory subunit BCY1 enhances filamentous growth. Homozygous wild-type (MLY61a/α) and Δbcy1/Δbcy1 (XPY1a/α) mutant diploid strains were incubated on low-ammonium sulfate (SLAD; 50 μM) and medium-ammonium sulfate (SMAD; 500 μM) media for 3 days at 30°C. Colonies were photographed originally at a ×25 magnification in this and the following figures.
PKA catalytic subunits play distinct roles in regulating filamentous growth.
To establish the functions of the PKA catalytic subunits in filamentous growth, the genes encoding Tpk1, Tpk2, and Tpk3 were individually deleted in the Σ1278b strain background. The resulting isogenic wild-type and tpk1/tpk1, tpk2/tpk2, and tpk3/tpk3 mutant strains were grown on medium containing 50 or 200 μM ammonium sulfate to assay filamentous growth. As shown in Fig. Fig.2,2, a clear and striking finding was that the tpk2/tpk2 mutant strain was severely reduced in pseudohyphal growth. This finding suggests that the Tpk2 catalytic subunit is required for pseudohyphal growth and plays a positive signaling role. The tpk2/tpk2 mutant strain does form some rudimentary filaments, which is also the case with most other mutants with defects in filamentous growth (28, 32).
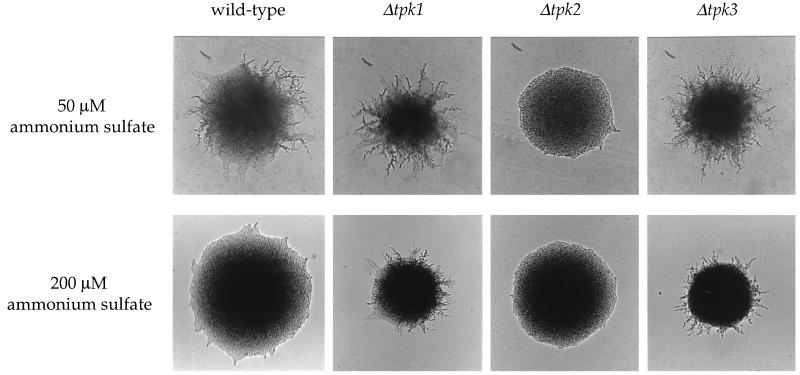
The tpk2 mutation reduces pseudohyphal growth, whereas tpk1 and tpk3 mutations enhance filamentous growth. Homozygous wild-type (MLY61a/α), Δtpk1/Δtpk1 (XYP4a/α), Δtpk2/Δtpk2 (XPY5a/α), and Δtpk3/Δtpk3 (XPY6a/α) mutant strains were incubated on SLAD medium with 50 or 200 μM ammonium sulfate, as indicated, and incubated for 3 days at 30°C.
In contrast, the tpk1/tpk1 and tpk3/tpk3 mutant strains exhibited the opposite phenotype, namely enhanced filamentous growth. This was apparent on standard pseudohyphal medium containing 50 μM ammonium sulfate (Fig. (Fig.2).2). Moreover, when the ammonium sulfate level was increased fourfold to 200 μM, filamentation of the wild-type strain was suppressed, whereas tpk1 and tpk3 mutant strains continued to filament to a significant degree (Fig. (Fig.2).2). These observations suggest that the Tpk1 and Tpk3 catalytic subunits play a negative role to inhibit filamentous growth.
Analysis of strains overexpressing Tpk1, Tpk2, or Tpk3 from multicopy plasmids provides additional support for distinct roles of the PKA catalytic subunits. When the TPK2 gene was introduced on a 2μm plasmid into the wild-type Σ1278b diploid strain, pseudohyphal differentiation was enhanced, further supporting the hypothesis that Tpk2 plays a stimulatory role (data not shown). In contrast, 2μm plasmids expressing the Tpk1 or Tpk3 catalytic subunits had the opposite effect and inhibited filamentous growth (data not shown), supporting the conclusion that Tpk1 and Tpk3 play an inhibitory role. In control experiments, we confirmed that the TPK2, TPK1, and TPK3 plasmids were functional, and each complemented to restore, or inhibit, pseudohyphal growth in tpk2, tpk1, or tpk3 mutant strains, respectively (data not shown).
Taken together, these findings support a model in which the Tpk2 catalytic subunit plays a positive signaling role and the Tpk1 and Tpk3 subunits play an inhibitory signaling role, to regulate filamentous differentiation. Thus, the three PKA catalytic subunits do not play a redundant role in filamentous growth. These findings are in accord with a recent report by others (51). The distinct roles of the three catalytic subunits are well correlated with the higher level of identity shared by Tpk1 and Tpk3 compared to the more divergent Tpk2 subunit.
Point of Tpk1 and Tpk3 inhibitory action.
Analysis of tpk double mutant strains revealed that the tpk2 mutation is epistatic to the tpk1 and tpk3 mutations. Thus, the tpk2/tpk2 tpk3/tpk3 and tpk1/tpk1 tpk2/tpk2 double mutant strains exhibited the filamentation defect conferred by the tpk2 mutation, whereas the tpk1/tpk1 tpk3/tpk3 double mutant strain exhibited the enhanced filamentation phenotype of the tpk1 and tpk3 single mutant strains (Fig. (Fig.3A).3A). By Northern blot analysis, the TPK2 gene was expressed to the same extent in the wild-type strain and in tpk1/tpk1, tpk3/tpk3, and tpk1/tpk1 tpk3/tpk3 mutant strains, and thus Tpk1 and Tpk3 are not involved in the regulation of TPK2 expression (data not shown). These observations suggest that the negative function of Tpk1 and Tpk3 is exerted on components upstream or downstream of PKA.
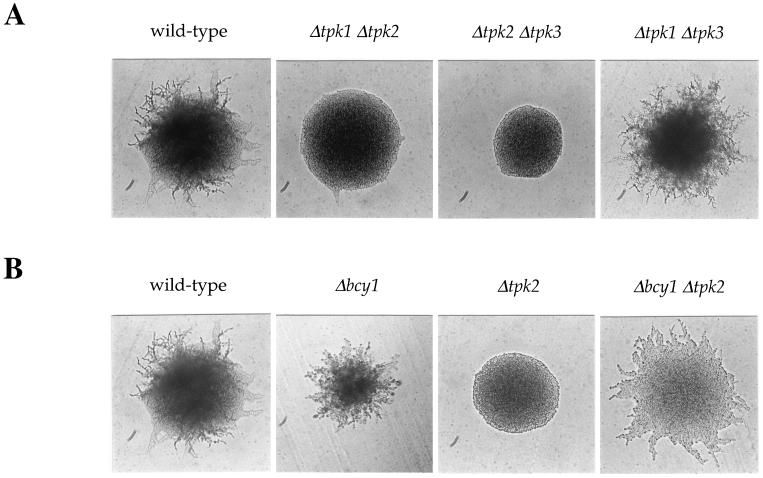
Epistasis analysis of tpk and bcy1 mutations. (A) The Δtpk2 mutation is epistatic to the Δtpk1 and Δtpk3 mutations. Isogenic wild-type (MLY61a/α) and Δtpk1/Δtpk1 Δtpk2/Δtpk2 (XPY12a/α), Δtpk2/Δtpk2 Δtpk3/Δtpk3 (XPY13a/α), and Δtpk1/Δtpk1 Δtpk3/Δtpk3 (XPY14a/α) mutant diploid strains were incubated on SLAD medium for 3 days at 30°C. (B) The Δbcy1 mutation suppresses the filamentation defect conferred by the tpk2 mutation. Isogenic wild-type (MLY61a/α) and Δbcy1/Δbcy1 (XPY12a/α), Δtpk2/Δtpk2 (XPY5a/α), and Δbcy1/Δbcy1 Δtpk2/Δtpk2 (XPY59a/α) mutant diploid strains were grown on SLAD medium for 3 days at 30°C.
Further support for the point of inhibition by Tpk1 and Tpk3 is provided by epistasis analysis with the bcy1 regulatory subunit mutation. Mutation of bcy1 uncouples the PKA pathway from upstream regulatory elements and releases the PKA catalytic subunits in a constitutively active, cAMP-independent form. To address whether the point of inhibitory action of Tpk1 and Tpk3 is upstream or downstream of PKA itself, the bcy1 mutation was used to sever the pathway from upstream regulation.
A bcy1/bcy1 tpk2/tpk2 double mutant strain was constructed and analyzed in which the only active PKA subunits are Tpk1 and Tpk3. If Tpk1 and Tpk3 inhibit by competing with Tpk2 for a downstream target, then activation of Tpk1 and Tpk3 by the bcy1 mutation should not alter the filamentation defect conferred by the tpk2 mutation. In contrast, the bcy1 mutation partially suppressed the tpk2 mutation to restore filamentous growth in the bcy1/bcy1 tpk2/tpk2 mutant strain, although the filaments differed from a wild-type morphology and agar invasion was not restored (Fig. (Fig.3B).3B). This finding suggests that when the Tpk1 and Tpk3 catalytic subunits are released from the Bcy1 regulatory subunit, they no longer exert an inhibitory effect. We propose that the Tpk1 and Tpk3 subunits normally regulate the PKA pathway via a negative feedback loop that inhibits cAMP production. In this model, tpk1 or tpk3 mutations enhance filamentation by increasing cAMP levels and Tpk2 activity, whereas Tpk1 or Tpk3 overexpression inhibits by further reducing cAMP levels and impairing Tpk2 activation. This interpretation is in accord with previous studies that established that the PKA pathway functions as part of a robust feedback loop that, following the initial activation of the pathway, reduces cAMP levels to the basal state (7, 37, 44). In addition, these observations also suggest that when the Tpk1 and Tpk3 catalytic subunits are expressed in a constitutive form, they can play a weak stimulatory role to regulate filamentation. Either Tpk1 or Tpk3 can in part support filamentous growth in the bcy1 tpk2 background, because bcy1/bcy1 tpk1/tpk1 tpk2/tpk2 and bcy1/bcy1 tpk2/tpk2 tpk3/tpk3 triple mutant diploid strains expressing only Tpk1 or Tpk3 still exhibited a modest level of filamentous growth (data not shown). Finally, overexpression of either Tpk1 or Tpk3 in a bcy1/bcy1 tpk2/tpk2 mutant strain enhanced pseudohyphal differentiation (data not shown), indicating that both Tpk1 and Tpk3 can play a positive signaling role when the pathway is uncoupled from regulation by cAMP.
TPK2 expression is induced by nitrogen starvation.
We next focused on the role of the Tpk2 catalytic subunit in stimulating filamentous growth to address why Tpk2 is unique compared to Tpk1 and Tpk3. We first examined the expression pattern of the TPK2 gene. Wild-type diploid cells of the Σ1278b strain were grown in liquid media containing a range of ammonium sulfate concentrations from 5 to 5,000 μM. RNA was isolated and analyzed by Northern blotting for the expression of TPK2 and with actin as a control. Expression of the TPK2 gene was induced ~4-fold in response to nitrogen starvation in two independent experiments (data not shown). By comparison, expression of the TPK1 gene was induced by only ~1.5-fold, and the TPK3 gene was not induced, under similar conditions (data not shown). These findings are consistent with Tpk2 playing a unique role in nitrogen-starved diploid cells.
Tpk2 catalytic domain stimulates filamentous growth.
That expression of the TPK2 gene is induced by nitrogen starvation suggested this might be the unique feature of Tpk2. To test this, we constructed chimeric genes in which the Tpk2 protein was expressed from the TPK1 promoter and the Tpk1 protein was expressed from the TPK2 promoter (Fig. (Fig.4A).4A). These chimeric genes were introduced into the wild-type Σ1278b diploid strain and analyzed for the phenotype conferred by overexpression of Tpk2 (enhanced filamentation) or Tpk1 (reduced filamentation). Expression of Tpk2 by either its own promoter or the promoter of the TPK1 gene stimulated filamentous growth to similar extents (Fig. (Fig.4B).4B). Expression of Tpk1 from either promoter inhibited filamentous growth (Fig. (Fig.4B).4B). Thus, differences in the TPK1 and TPK2 gene promoters and transcriptional regulation do not underlie the unique regulatory functions of Tpk1 and Tpk2.
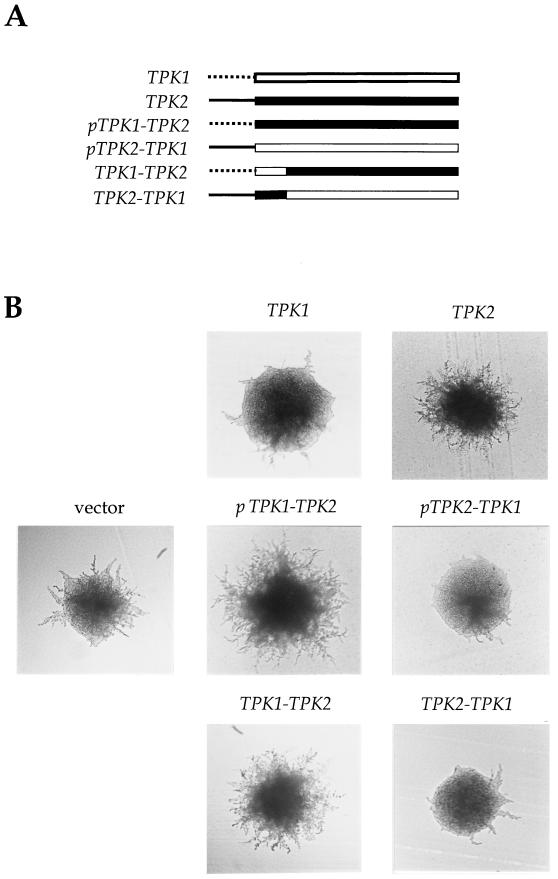
The unique activating function of Tpk2 maps to the C-terminal kinase domain. (A) The structures of the wild-type TPK1 (open bars) and TPK2 (solid bars) genes and four chimeric TPK genes are illustrated. The pTPK1-TPK2 hybrid gene consists of the promoter region of TPK1 (dashed line) and the ORF and 3′-UTR of the TPK2 gene. The pTPK2-TPK1 hybrid gene consists of the promoter region of TPK2 (solid line) and the ORF and 3′-UTR of the TPK1 gene. The TPK1-TPK2 hybrid gene consists of the promoter plus the coding sequence for the unique amino-terminal portion of the TPK1 ORF (aa 1 to 79) and the carboxyl-terminal portion of the TPK2 ORF (aa 63 to 281) and 3′-UTR. The TPK2-TPK1 hybrid gene consists of the promoter and coding sequence of the amino-terminal portion of the TPK2 ORF (aa 1 to 62) and the carboxyl-terminal portion of the TPK1 ORF (aa 80 to 378) and 3′-UTR. (B) The wild-type TPK1 and TPK2 genes and the hybrid TPK genes were expressed from the high-copy plasmid YEplac195 in wild-type yeast strain MLY61a/α and assayed for filamentous growth following incubation at 30°C for 3 days on SLAD medium.
We next tested the sequences of the Tpk1 and Tpk2 proteins. The three PKA catalytic subunits share 75 to 88% sequence identity in the carboxy-terminal kinase domain, whereas unique amino-terminal regions of ~60 to 80 amino acids (aa) are appended to the catalytic regions of each subunit. One hypothesis was that these unique amino-terminal regions might determine the activity of Tpk2 compared to that of Tpk1 and Tpk3. To address this, we constructed chimeras exchanging the unique amino-terminal regions of the Tpk1 and Tpk2 proteins (Fig. (Fig.4A).4A). Surprisingly, expression of the Tpk1-Tpk2 chimeric protein enhanced filamentation to the same extent as wild-type Tpk2, whereas expression of the Tpk2-Tpk1 chimeric subunit inhibited filamentation similar to Tpk1 expression (Fig. (Fig.4B).4B). We conclude that the unique activating function of the Tpk2 subunit maps to the catalytic region and not to the unique amino-terminal region. By additional chimera studies, the unique region of Tpk2 was mapped to the amino-terminal one-half of the catalytic region, between residues 62 and 226 of Tpk2. In summary, subtle sequence differences in the Tpk1 and Tpk2 catalytic regions underlie the unique activity of Tpk2, possibly by altering affinity for substrates that regulate filamentous growth.
Tpk2 regulates the switch to unipolar budding and invasive growth.
Yeast pseudohyphal growth consists of several physiological events, including cell elongation, a switch from bipolar to unipolar budding, mother-daughter cell adhesion, and invasive growth. We addressed which of these facets are effected by the Tpk2 subunit of PKA by analyzing the isogenic wild-type strain and the tpk2/tpk2 and ste12/ste12 mutant strains (see Materials and Methods). This analysis revealed that the tpk2 mutation impairs the switch to unipolar budding and also agar invasion, whereas cell elongation occurred normally in response to nitrogen starvation (Fig. (Fig.55 and Table Table3).3). On the other hand, the ste12 mutation inhibited cell elongation and agar invasion, but had little or no effect on the switch to a unipolar budding pattern (Fig. (Fig.5,5, Table Table3,3, and data not shown), in accord with previous reports (28, 39, 50). We note that robust activation of PKA, either by exogenous cAMP or the bcy1 mutation, results in filaments largely composed of round cells (Fig. (Fig.1)1) (32), further indicating that the PKA pathway does not promote cell elongation. In the microcolony budding assays, bipolar budding in wild-type cells produced quite linear chains of four cells, whereas the ste12/ste12 and tpk2/tpk2 mutant strains produced chains that were often perturbed at the central mother-daughter cell junction (Fig. (Fig.5),5), suggesting that the MAP kinase and PKA pathways may also regulate cell adhesion required for the integrity of pseudohyphal filaments.
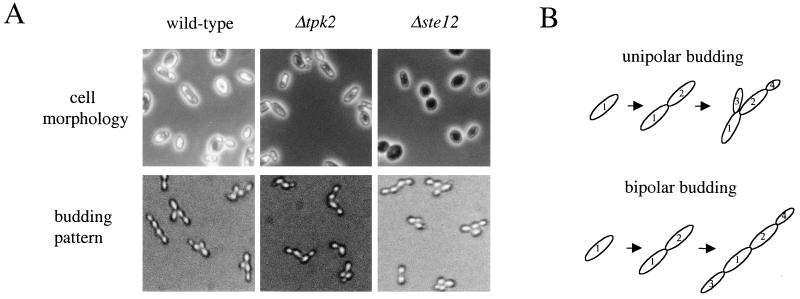
PKA pathway regulates unipolar budding, while the MAP kinase pathway is required for cell elongation. (A) Isogenic wild-type (MLY61a/α), Δtpk2/Δtpk2 (XPY5a/α), and Δste12/Δste12 (MLY216a/α) strains were incubated on SLAD medium for 16 h at 30°C. Cells were collected and studied for cell morphology (upper panels) and budding pattern (lower panels). Cell elongation in response to nitrogen starvation occurs in the wild-type and tpk2 mutant strain, whereas the number of elongated cells is severely reduced in the ste12 mutant strain. For the microcolony budding pattern assay, daughter cells were micromanipulated on SLAD medium and incubated at 30°C for 5 to 6 h, at which time, three- and four-cell microcolonies were photographed and scored for budding pattern. Cells were also stained with Calcofluor white and photographed to score the pattern of chitin bud scars (not shown [see Materials and Methods]). (B) The patterns of cell division that give rise to four-celled microcolonies by either the bipolar or the unipolar budding patterns are depicted.
TABLE 3
Quantitative analysis of cell morphology and budding pattern
pattern
| Straina | Relevant genotype | % with cell shapeb:
| % with budding patternc:
| |||
|---|---|---|---|---|---|---|
| Long PH | Oval YF | Round YF | Bipolar | Unipolar | ||
| MLY61a/α | Wild type | 45.2 | 53.1 | 1.7 | 39.9 | 60.1 |
| XPY5a/α | Δtpk2::G418/Δtpk2::G418 | 53.7 | 45.5 | 0.8 | 76.6 | 23.4 |
| MLY216a/α | Δste12::G418/Δste12::G418 | 12.0 | 69.1 | 18.9 | 56.1 | 43.9 |
In summary, this analysis revealed an interesting specialization of function whereby the PKA pathway functions to switch budding pattern, the MAP kinase cascade regulates cell elongation, and both pathways are required for agar invasion and cell adhesion, providing evidence that the MAP kinase and PKA pathways have both independent and shared functions in the regulation of pseudohyphal growth.
Tpk2 functions downstream of Gpr1, Gpa2, and Ras2.
The point of action of Tpk2 with respect to other known regulators of pseudohyphal growth was addressed by epistasis analysis. First, we used the bcy1 mutation to activate PKA and test if this suppresses mutations in the Gpr1 receptor or the linked Gα protein Gpa2. In isogenic bcy1/bcy1 gpr1/gpr1 and bcy1/bcy1 gpa2/gpa2 double mutant strains, the bcy1 mutation suppressed the filamentation defect conferred by either the gpr1 or the gpa2 mutation (data not shown), suggesting that Tpk2 functions downstream of Gpr1 and Gpa2. We also tested whether the tpk2 mutation would block activation of filamentation in response to dominant activated alleles of GPA2 or RAS2. As shown in Fig. Fig.6,6, the tpk2 mutation completely suppressed filamentation in response to the Gpa2-gly132val dominant active mutant. On the other hand, the tpk2 mutation only partially suppressed filamentation in response to the dominant active Ras2-gly19val mutant protein (Fig. (Fig.6).6). These findings suggest that Gpr1 and Gpa2 signal upstream of Tpk2, in accord with a role in stimulating cAMP production by adenylyl cyclase. These findings also support a model in which Ras2 signals to activate both the MAP kinase and the PKA pathways (32, 40, 49).
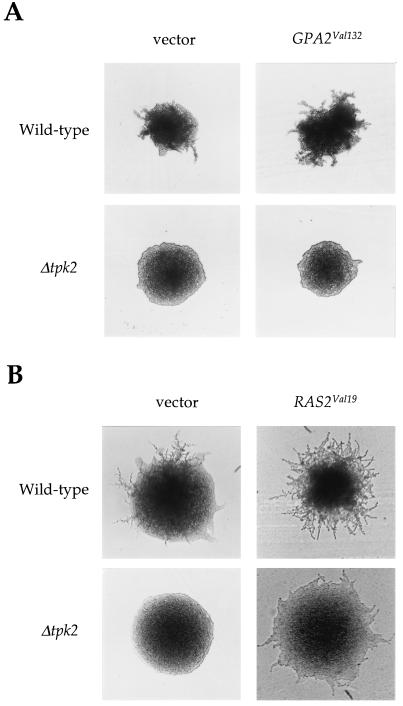
Tpk2 acts downstream of Gpa2 and in part downstream of Ras2. (A) The Δtpk2 mutation is epistatic to the activated GPA2Val132 allele. Wild-type (MLY61a/α) and Δtpk2/Δtpk2 (XPY5a/α) mutant diploid strains containing a control vector (pSEYC68) or expressing the dominant active GPA2-2 allele (pML160), which is under the control of a galactose-inducible promoter, were incubated on SLARG medium for 5 days at 30°C. (B) Wild-type (MLY61a/α) and Δtpk2/Δtpk2 (XPY5a/α) mutant diploid strains containing a control plasmid or expressing the dominant active RAS2Val19 allele (pMW2) were grown on SLAD medium at 30°C for 3 days.
PKA regulates filamentous growth independently of Ste12 and Tec1.
We next addressed the point of action of PKA with respect to the MAP kinase pathway regulating pseudohyphal growth. To address this, we performed epistasis analysis with the bcy1 mutation (to activate PKA) in mutants lacking Ste12, Tec1, or both Ste12 and Tec1.
An isogenic series of strains lacking Ste12, Tec1, or both Ste12 and Tec1 and containing either wild-type BCY1 or the bcy1 mutant allele were constructed. Remarkably, the bcy1 mutation dramatically suppressed the filamentation defect of the ste12/ste12, tec1/tec1, and ste12/ste12 tec1/tec1 mutant strains on SLAD medium with 50 μM ammonium sulfate (Fig. (Fig.7).7). Thus, PKA can drive filamentous growth in the absence of Ste12, Tec1, or both Ste12 and Tec1. Because Ste12 and Tec1 are the transcription factor targets of the MAP kinase pathway, the MAP kinase pathway is, in part, dispensable for filamentous growth when PKA is activated. ste12 or tec1 mutant colonies that also lacked bcy1 formed filaments that were largely composed of round rather than filamented cells, again consistent with a role for the MAP kinase pathway in cell elongation. In summary, the bcy1 mutation suppresses the filamentous growth defect conferred by ste12 and tec1 single and double mutations, demonstrating that the PKA pathway can regulate pseudohyphal growth independently of the MAP kinase cascade.
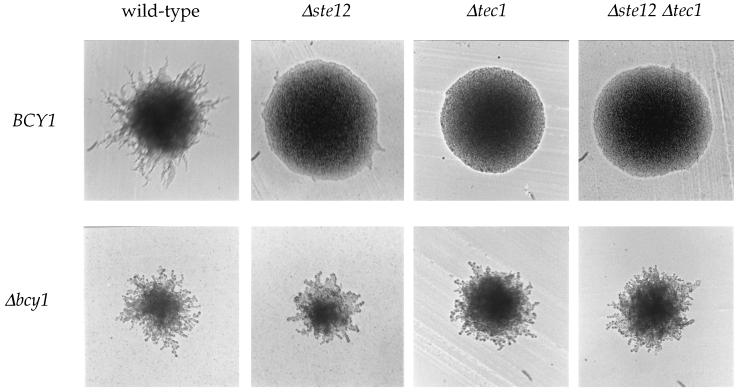
Activated PKA restores filamentous growth in MAP kinase cascade mutants. Isogenic wild-type (MLY61a/α) and Δste12/Δste12 (MLY216a/α), Δtec1/Δtec1 (MLY183a/α), Δste12/Δste12 Δtec1/Δtec1 (XPY77a/α), Δbcy1/Δbcy1 (XPY1a/α), Δbcy1/Δbcy1 Δste12/Δste12 (XPY69a/α), Δbcy1/Δbcy1 Δtec1/Δtec1 (XPY75a/α), and Δbcy1/Δbcy1 Δste12/Δste12 Δtec1/Δtec1 (XPY76a/α) mutant diploid strains were incubated on SLAD medium at 30°C for 3 days.
Target of PKA for filamentous growth is not Phd1, Msn2, or Msn4.
Several targets of yeast PKA have been identified, and we tested which regulate pseudohyphal growth. Previous studies identified two related transcription factors, Sok2 and Phd1, which regulate filamentation and may function downstream of PKA. Phd1 enhances filamentous growth when overexpressed (13), whereas overexpression of Sok2 suppresses the growth defect of strains with reduced PKA activity and sok2 mutations enhance filamentous growth (62). By comparison, phd1 mutations confer only a modest defect in filamentous growth unless combined with an ste12 mutation (30). These observations suggest Phd1 and Sok2 may antagonistically regulate filamentous growth.
To test if Phd1 and Sok2 were PKA targets for filamentation, we performed epistasis analysis with the BCY1, TPK2, PHD1, and SOK2 genes. First, the bcy1 mutation was shown to enhance filamentous growth to similar extents in either a wild-type or a phd1/phd1 mutant background (Fig. (Fig.8A).8A). Second, introduction of the PHD1 gene on a multicopy plasmid enhanced filamentous growth to similar extents in either a wild-type or a tpk2/tpk2 mutant strain (Fig. (Fig.8B).8B). Third, introduction of the TPK2 gene on a multicopy plasmid enhanced filamentation to the same extent in wild-type and phd1/phd1 mutant strains (data not shown). Thus, PKA and Phd1 independently regulate filamentous growth. The observation that the bcy1 mutation suppressed the filamentation defect in both phd1 tec1 (Fig. (Fig.8A)8A) and phd1 ste12 (data not shown) double mutant strains also excludes models in which PKA regulates both Phd1 and Tec1 or both Phd1 and Ste12. Finally, the sok2 mutation enhanced filamentous growth in either a wild-type or a tpk2/tpk2 mutant strain (data not shown). Thus, PKA functions independently of several transcription factors known to regulate pseudohyphal growth.
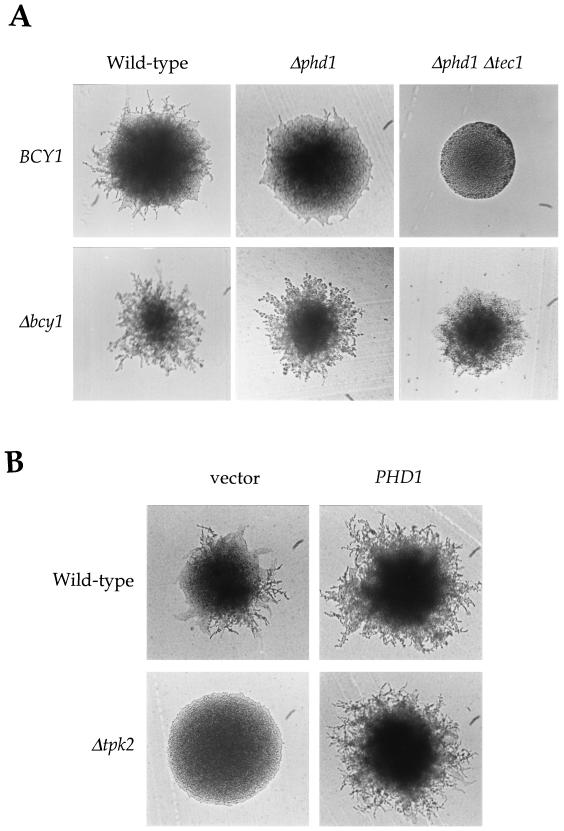
Tpk2 regulates pseudohyphal growth independently of Phd1. (A) Activated PKA pathway induces filamentous growth in the absence of Phd1. Isogenic wild-type (MLY61a/α) and Δphd1/Δphd1 (MLY182a/α), Δphd1/Δphd1 Δtec1/Δtec1 (XPY89a/α), Δbcy1/Δbcy1 (XPY1a/α), Δbcy1/Δbcy1 Δphd1/Δphd1 (XPY78a/α), and Δbcy1/Δbcy1 Δphd1/Δphd1 Δtec1/Δtec1 (XPY88a/α) mutant diploid strains were incubated on SLAD medium at 30°C for 3 days. (B) Overexpression of PHD1 is epistatic to the tpk2 mutation. Wild-type (MLY61a/α) and Δtpk2/Δtpk2 mutant (XPY5a/α) diploid strains containing a control plasmid (vector) or the PHD1 overexpression plasmid (pCG68) were incubated on SLAD medium at 30°C for 3 days.
Recent studies have identified two transcription factors, Msn2 and Msn4, which are regulated by the PKA signaling pathway. PKA regulates cellular localization of the Msn2 protein (18), and msn2 and msn4 mutations bypass the essential function of PKA for vegetative growth (56). To establish whether Msn2 and Msn4 were PKA targets for pseudohyphal growth, we constructed isogenic wild-type and msn2/msn2, msn4/msn4, and msn2/msn2 msn4/msn4 mutant strains in the Σ1278b strain background. The msn2 and msn4 single and double mutations neither activated nor inhibited pseudohyphal differentiation on either SLAD or SMAD medium containing 50 or 500 μM ammonium sulfate (data not shown). Moreover, introduction of the TPK2 gene multicopy plasmid enhanced filamentous growth to a similar extent in the wild-type strain compared to that in the msn2/msn2, msn4/msn4, and msn2/msn2 msn4/msn4 mutant strains (data not shown). Thus, Msn2 and Msn4 are not the targets of PKA for filamentous growth. Finally, mutations in either of two protein kinases implicated in PKA functions, Yak1 and Rim15 (10, 48), had no affect on pseudohyphal growth (data not shown).
PKA regulates filamentation via Flo8 and the cell surface flocculin Flo11.
Several observations suggested the function of the PKA signaling pathway might be linked to expression and function of cell surface proteins involved in cell-cell adhesion (flocculation). Namely, linear microcolonies produced by tpk2/tpk2 mutant strains were often bent, in contrast to linear four-celled microcolonies produced by wild-type cells (Fig. (Fig.5).5). Second, the Flo8 transcription factor and the Flo11 cell surface flocculin were previously found to be required for pseudohyphal growth (27, 29, 31), and the Flo8 protein has five consensus PKA phosphorylation sites. We therefore tested if these are targets of PKA that regulate filamentation.
Mutations in the FLO1 and FLO5 flocculation genes had no effect on filamentous growth (data not shown). In contrast, pseudohyphal differentiation was severely impaired in both flo8/flo8 and flo11/flo11 mutant strains, consistent with previous reports (Fig. (Fig.9A).9A). Most importantly, both mutations were epistatic to the enhancing effects of the bcy1 mutation. Thus, in bcy1/bcy1 flo8/flo8 and bcy1/bcy1 flo11/flo11 double mutant strains, only a few rudimentary filaments were formed on SLAD medium, in marked contrast to the dramatic enhanced pseudohyphal growth of the bcy1/bcy1 mutant strain (Fig. (Fig.9A).9A). These observations suggest that Flo8 and Flo11 function downstream of PKA in a signaling pathway regulating filamentous differentiation. In addition, we found that overexpression of the MAP kinase-regulated transcription factor Tec1 completely suppressed the pseudohyphal defect of flo8/flo8 mutant strains (Fig. (Fig.9A),9A), providing evidence that the FLO11 gene is a common target of the PKA and MAP kinase signaling cascades.
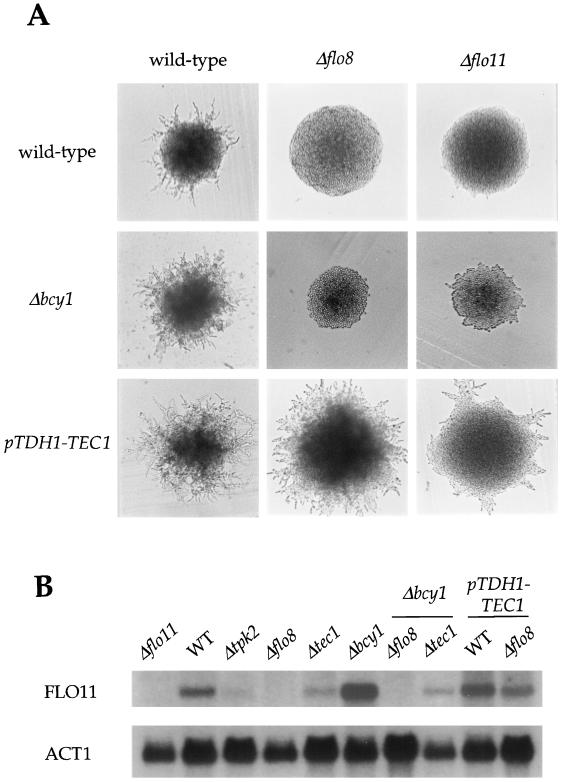
PKA pathway regulates expression of the cell surface flocculin Flo11 via the transcription factor Flo8. (A) flo8 and flo11 mutations block pseudohyphal growth and are epistatic to activated PKA and MAP kinase cascade signaling. Wild-type (MLY61a/α) and Δflo8/Δflo8 (XPY95a/α), Δflo11/Δflo11 (XPY107a/α), Δbcy1/Δbcy1 (XPY1a/α), Δbcy1/Δbcy1 Δflo8/Δflo8 (XPY99a/α), and Δbcy1/Δbcy1 Δflo11/Δflo11 (XPY119a/α) mutant strains containing a control plasmid and wild-type and Δflo8/Δflo8 and Δflo11/Δflo11 mutant strains containing the pTDH1-TEC1 overexpression plasmid were grown on SLAD medium for 3 days at 30°C. (B) The PKA pathway regulates the expression of FLO11 by the transcription factor Flo8. Total RNA was prepared from wild-type (WT) (MLY61α) and Δtpk2 (XPY5α), Δflo8 (XPY95α), Δflo11 (XPY107α), Δtec1 (MLY183α), Δbcy1 (XPY1α), Δbcy1 Δflo8 (XPY99α), and Δbcy1 Δtec1 (XPY75α) mutant strains and wild-type and Δflo8 strains containing the pTDH1-TEC1 plasmid grown in synthetic medium lacking uracil. RNA was fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nylon membrane, and probed with portions of the FLO11 and ACT1 genes.
Studies on the regulation of FLO11 gene expression by Northern analysis confirm that the FLO11 gene is regulated by Flo8 and is a common target of the PKA and MAP kinase signaling cascades (Fig. (Fig.9B).9B). First, FLO11 expression was readily detected in wild-type cells, but no or greatly reduced FLO11 expression was observed in mutant strains lacking Tpk2, Flo8, or Tec1 (Fig. (Fig.9B).9B). Second, FLO11 expression was increased in a bcy1 mutant, indicating activated PKA can promote FLO11 transcription (Fig. (Fig.9B).9B). Third, introduction of a flo8 mutation abolished FLO11 expression in the bcy1 mutant strain (Fig. (Fig.9B).9B). Finally, overexpression of Tec1 in a flo8 mutant strain partially restored FLO11 expression (Fig. (Fig.9B),9B), in accord with the epistasis result showing that Tec1 restores filamentous growth in a flo8 mutant strain (Fig. (Fig.9A).9A). By Northern blotting, FLO8 is expressed at equivalent levels in wild-type and tpk2 mutant strains, suggesting PKA may directly regulate Flo8 (data not shown). Taken together, these genetic epistasis and gene expression studies indicate that PKA regulates expression of the cell surface flocculin Flo11 via the Flo8 transcription factor and that FLO11 expression is regulated in parallel by the MAP kinase cascade via the Tec1 transcription factor (Fig. (Fig.10).10). Finally, we note that Flo8 and Flo11 are required for agar invasion and cell-cell adhesion, but do not mediate the effects of PKA on the bipolar-unipolar budding pattern switch or of the MAP kinase cascade on cell elongation (data not shown). Moreover, overexpression of Tec1 completely suppressed the filamentation defect of a flo8/flo8 mutant and also partially suppressed the defect of a flo11/flo11 mutant strain (Fig. (Fig.9A).9A). These findings suggest that additional targets of both PKA and Tec1 that regulate pseudohyphal growth remain to be identified.
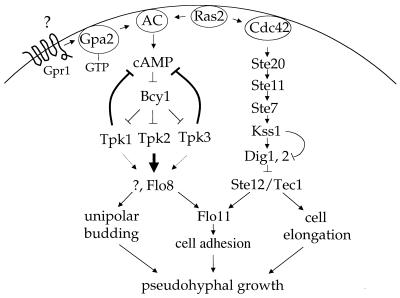
A model for the regulation of pseudohyphal growth by the PKA and MAP kinase pathways. The three catalytic subunits of PKA play distinct roles in regulating yeast pseudohyphal growth. The Tpk2 catalytic subunit plays a positive role to activate filamentous growth, whereas the Tpk1 and Tpk3 catalytic subunits play negative roles to inhibit filamentous growth. Epistasis analysis indicates that PKA signals downstream of the Gpr1 receptor and Gα protein Gpa2. PKA and the MAP kinase cascades function independently to regulate budding pattern and cell elongation, respectively, during filamentous growth. In contrast, PKA (via Flo8) and the MAP kinase cascade (via Ste12 and Tec1) coordinately regulate the cell surface flocculin Flo11, agar invasion, and cell adhesion.
DISCUSSION
Our studies have analyzed the signal transduction pathways that regulate yeast S. cerevisiae pseudohyphal differentiation in response to nitrogen limitation. Our findings support a model in which the cAMP-dependent protein kinase (PKA) plays a central role in regulating filamentation (Fig. (Fig.10).10). First, mutation of the PKA regulatory subunit Bcy1 dramatically enhances filamentation and, in part, bypasses the requirement for nitrogen starvation. Second, we show that the three PKA catalytic subunits play distinct roles in which the Tpk2 catalytic subunit activates filamentous growth, whereas the Tpk1 and Tpk3 subunits inhibit filamentation. This specialization of function is correlated with the sequence of the subunits, in that Tpk1 and Tpk3 are the most closely related (88% identity), whereas Tpk2 is more divergent (75 and 77% identity to Tpk1 and Tpk3). The unique activating function of the Tpk2 subunit was mapped to the catalytic region and not to the unique amino-terminal region or the TPK2 gene promoter. Third, epistasis analysis is in accord with a model in which PKA functions downstream of the novel G protein-coupled receptor Gpr1 and the G proteins Gpa2 and Ras2. Fourth, activated PKA can drive filamentous growth in the absence of the Ste12 and Tec1 transcription factors that are components of the MAP kinase cascade that also regulates filamentous growth. We show that PKA regulates filamentous growth independently of Phd1 and several PKA targets, including the transcription factors Msn2 and Msn4, and the protein kinases Yak1 and Rim15. On the other hand, we define a role for PKA in regulating expression of the cell surface flocculin Flo11 by the transcription factor Flo8, both of which are known to be required for pseudohyphal growth (27, 29, 31).
In conclusion, as outlined in Fig. Fig.10,10, our studies define a signal transduction cascade regulating the cAMP-dependent protein kinase that positively, (and negatively) regulates yeast pseudohyphal differentiation. While this report was in preparation, similar findings were reported with respect to TPK2 and TPK3 function in filamentous growth, suggesting that another target of PKA may be the transcription factor homolog SFL1, and defining Flo11 as a common target of the PKA and MAP kinase cascades (51, 52).
Relationship between the PKA and MAP kinase signaling cascades.
Our studies also provide a perspective on the relationship between the MAP kinase and PKA signal transduction pathways that regulate yeast pseudohyphal growth. First, our studies demonstrate that activated PKA (as a result of a bcy1 mutation) can support filamentous growth in the complete absence of Ste12, Tec1, or both Ste12 and Tec1. These observations are in accord with previous findings that exogenous cAMP restores filamentation in ste20 and ste12 mutant strains and also represses expression of the Fg(TyA)::lacZ reporter gene (32). Taken together, these findings indicate that the function of the MAP kinase pathway can be largely bypassed by activated A kinase, suggesting the two signaling pathways can function independently.
On the other hand, the filaments produced in response to either cAMP or the bcy1 mutation are composed of round, rather than elongated, yeast cells, suggesting the PKA pathway is not responsible for cell elongation during pseudohyphal growth. In accord with this hypothesis, the Tpk2 catalytic subunit is required for invasive growth and the switch to unipolar budding, but not cell elongation. In contrast, the MAP kinase cascade component Ste12 is required for cell elongation and agar invastion, but not for the switch to unipolar budding (see also reference 39). This analysis suggests a division of labor in which both pathways regulate invasive growth and the adhesion of mother and daughter cells, whereas the PKA pathway affects the switch in budding pattern and the MAP kinase pathway affects cell elongation. Finally, our studies define a shared role between the PKA and MAP kinase signaling pathways in regulating expression of the cell surface flocculin Flo11, which is required for invasive and filamentous growth (31) and may mediate the adhesion of mother and daughter cells required for the integrity of pseudohyphal filaments.
PKA catalytic subunits have unique functions.
Our studies reveal that the cAMP-dependent kinase has a novel role in regulating pseudohyphal differentiation in addition to the well-established roles of PKA in vegetative growth, meiosis, and stationary phase. One of the most interesting findings to emerge from our studies is that the three PKA catalytic subunits have unique functions. The more divergent Tpk2 subunit enhances filamentous growth, whereas the two more closely related subunits, Tpk1 and Tpk3, inhibit filamentous growth. In contrast, several previous studies found similar phenotypes in strains expressing any one of the three catalytic subunits (either Tpk1, Tpk2, or Tpk3), leading to the common view that the three PKA catalytic subunits are redundant for function (58). On the other hand, there were some clues that the three Tpk subunits might have distinguishing features. For example, mutant strains expressing only Tpk1 or Tpk3 can utilize acetate as a carbon source, whereas strains expressing only Tpk2 cannot (58). Our findings demonstrate that the three PKA catalytic subunits are specialized with respect to pseudohyphal differentiation to play both positive and negative signaling roles. This dual function may be analogous to the negative and positive signaling roles of the Kss1 MAP kinase in filamentous differentiation (8, 35).
Our studies further reveal that the Tpk1 and Tpk3 catalytic subunits are specialized to inhibit filamentous differentiation. The negative function of PKA may serve to constrain filamentous growth under rich medium conditions in which yeast budding form growth is preferred. In addition, the negative function of Tpk1 and Tpk3 may also serve to return filamentous cells to normal vegetative growth once nutrients are encountered, similar to adaptive mechanisms that inhibit signaling in other cascades. Epistasis analysis suggests that Tpk1 and Tpk3 exert their negative effects upstream of PKA itself. This finding is consistent with previous findings that PKA functions in a negative feedback loop that, following activation of the pathway, represses cAMP synthesis (7, 37, 44). The target of this negative feedback loop has not been defined but may involve adenylyl cyclase itself (7).
cAMP and PKA signaling in yeasts and pathogenic fungi.
cAMP and PKA also regulate mating, meiosis, and sporulation in the fission yeast Schizosaccharomyces pombe (42, 64). Like S. cerevisiae, S. pombe has two mating types that communicate with peptide pheromones. In S. pombe, a second signal, nitrogen starvation, is also required for mating. Two pathways regulate mating in S. pombe. The first is a pheromone-activated MAP kinase cascade analogous to the MAP kinase cascade for mating in budding yeast. The second is a nutrient-sensing pathway involving the Gα protein gpa2, which is a homolog of the S. cerevisiae Gα protein Gpa2 that regulates filamentous growth (22). In S. pombe, gpa2 activates adenylyl cyclase in response to nutrients, raising cAMP levels and thereby inhibiting mating.
The target of cAMP in both fission and budding yeast is PKA, and there is a single PKA catalytic subunit in S. pombe (36). The gpa2-adenylyl cyclase-cAMP-PKA pathway also regulates transcriptional repression in response to glucose (21, 45). The targets of PKA in mating and gene regulation in S. pombe remain to be identified.
cAMP and PKA also play a central role in regulating filamentous growth and virulence of plant fungal pathogens (reviewed in reference 24). For example, in the corn smut Ustilago maydis, mating and environmental signals trigger a dimorphic transition from budding to filamentous growth. Mutations in the adenylyl cyclase gene uac1 result in a constitutively filamentous phenotype that is suppressed by cAMP or mutations in the PKA regulatory subunit ubc1 (15) that also attenuate virulence (16). Similarly, mutation of the Gα protein Gpa3 results in attenuated virulence and constitutive filamentous growth that is suppressed by cAMP (25, 47). Most interestingly, Gpa3 is closely related to the Gpa2 and gpa2 nutrient-sensing Gα proteins from S. cerevisiae and S. pombe. Two genes encoding PKA catalytic subunits have been identified in U. maydis, adr1 and uka1, and disruption of the adr1 gene causes a constitutive filamentous phenotype (9). Taken together, these findings outline a conserved signaling pathway involving Gpa3, adenylyl cyclase, cAMP, and PKA that inhibits the dimorphic transition between budding yeast and filamentous growth and that also regulates virulence of U. maydis.
An analogous signaling cascade regulates mating, differentiation, and virulence of the human fungal pathogen Cryptococcus neoformans. The Gα protein GPA1 regulates C. neoformans mating, induction of virulence factors, and virulence in response to nutritional deprivation (1, 2), and gpa1 mutant phenotypes are suppressed by cAMP. Interestingly, the C. neoformans GPA1 Gα protein is a homolog of the nutrient-sensing Gα proteins Gpa2 in S. cerevisiae, gpa2 in S. pombe, and Gpa3 in U. maydis. The regulatory and catalytic subunits of PKA have been recently identified in C. neoformans (8a), and studies to define roles in differentiation and virulence of this human fungal pathogen are in progress.
Conclusions.
In summary, studies of the yeasts S. cerevisiae and S. pombe, the plant fungal pathogen U. maydis, and the human fungal pathogen C. neoformans have converged to define a novel signaling cascade involving the G protein-coupled receptor Gpr1, a highly conserved Gα protein, adenylyl cyclase, cAMP, and the cAMP-dependent protein kinase which regulates mating, differentiation, and virulence. The targets of these PKA-regulated pathways remain to be defined, but promise to further our understanding of the conserved signaling pathways regulating differentiation and virulence of diverse yeasts and pathogenic fungi.
ACKNOWLEDGMENTS
We thank Maria Cardenas, Steve Garrett, Mike Lorenz, and John Pringle for advice and discussions; John McCusker and Alan Goldstein for the hygromycin resistance cassette; Namjin Chung for experimental advice; and Mike Lorenz for strains and plasmids.
Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.19.7.4874
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/mcb/19/7/4874.full.pdf
Free to read at mcb.asm.org
http://mcb.asm.org/cgi/content/abstract/19/7/4874
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/reprint/19/7/4874
Free after 4 months at mcb.asm.org
http://mcb.asm.org/cgi/content/full/19/7/4874
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.19.7.4874
Article citations
Gain- and loss-of-function alleles within signaling pathways lead to phenotypic diversity among individuals.
iScience, 27(10):110860, 31 Aug 2024
Cited by: 0 articles | PMID: 39381740 | PMCID: PMC11460476
Conserved signaling modules regulate filamentous growth in fungi: a model for eukaryotic cell differentiation.
Genetics, 228(2):iyae122, 01 Oct 2024
Cited by: 0 articles | PMID: 39239926
Review
Protein Kinase A Negatively Regulates the Acetic Acid Stress Response in S. cerevisiae.
Microorganisms, 12(7):1452, 17 Jul 2024
Cited by: 0 articles | PMID: 39065219 | PMCID: PMC11278818
A single gene mutation underpins metabolic adaptation and acquisition of filamentous competence in the emerging fungal pathogen Candida auris.
PLoS Pathog, 20(7):e1012362, 08 Jul 2024
Cited by: 1 article | PMID: 38976759 | PMCID: PMC11257696
To each its own: Mechanisms of cross-talk between GPI biosynthesis and cAMP-PKA signaling in Candida albicans versus Saccharomyces cerevisiae.
J Biol Chem, 300(7):107444, 04 Jun 2024
Cited by: 0 articles | PMID: 38838772 | PMCID: PMC11294708
Review Free full text in Europe PMC
Go to all (259) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae.
Genetics, 154(2):609-622, 01 Feb 2000
Cited by: 157 articles | PMID: 10655215 | PMCID: PMC1460933
Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog.
EMBO J, 16(23):7008-7018, 01 Dec 1997
Cited by: 206 articles | PMID: 9384580 | PMCID: PMC1170304
Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae.
Mol Microbiol, 33(5):904-918, 01 Sep 1999
Cited by: 388 articles | PMID: 10476026
Review
The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae.
EMBO J, 17(5):1236-1247, 01 Aug 1998
Cited by: 265 articles | PMID: 9482721 | PMCID: PMC1170472





