Abstract
Free full text

The Interaction and Colocalization of Sam68 with the Splicing-associated Factor YT521-B in Nuclear Dots Is Regulated by the Src Family Kinase p59fyn
Abstract
Alternative pre-mRNA splicing patterns can change an extracellular stimulus, but the signaling pathways leading to these changes are still poorly characterized. Here, we describe a tyrosine-phosphorylated nuclear protein, YT521-B, and show that it interacts with the nuclear transcriptosomal component scaffold attachment factor B, and the 68-kDa Src substrate associated during mitosis, Sam68. Northern blot analysis demonstrated ubiquitous expression, but detailed RNA in situ analysis revealed cell type specificity in the brain. YT521-B protein is localized in the nucleoplasm and concentrated in 5–20 large nuclear dots. Deletion analysis demonstrated that the formation of these dots depends on the presence of the amino-terminal glutamic acid-rich domain and the carboxyl-terminal glutamic acid/arginine-rich region. We show that the latter comprises an important protein–protein interaction domain. The Src family kinase p59fyn-mediated tyrosine phosphorylation of Sam68 negatively regulates its association with YT521-B, and overexpression of p59fyn dissolves nuclear dots containing YT521-B. In vivo splicing assays demonstrated that YT521-B modulates alternative splice site selection in a concentration-dependent manner. Together, our data indicate that YT521-B and Sam68 may be part of a signal transduction pathway that influences splice site selection.
INTRODUCTION
The members of the signal transduction and activation of RNA (STAR) or GRP33/SAM68/GLD1 domain-containing protein family are proposed mediators connecting signal transduction pathways and RNA metabolism (Vernet and Artzt, 1997). The prototype of these proteins is Sam68, the 68-kDa Src substrate associated during mitosis (Taylor and Shalloway, 1994). Sam68 and its related protein family members contain an RNA binding domain that has been referred to as the STAR domain (Vernet and Artzt, 1997), GSG (GRP33/SAM68/GLD1) (Jones et al.), or SGQ (SAM68/GLD1/Quaking homology domain) (Lin et al., 1997). This domain contains an RNA binding KH domain module flanked by two Qua1 and Qua2 domains that are required for high-affinity RNA binding. In addition, Sam68 contains an RGG box that has been implicated in RNA binding (Kiledjian and Dreyfuss, 1992), as well as proline- and tyrosine-rich regions involved in binding to Src homology 3 (SH3) and SH2 domains. Sam68 thus associates with a variety of signaling molecules, including members of the Src family tyrosine kinases, growth factor receptor-bound protein 2 (GRB-2), and phospholipase Cγ-1 (Richard et al., 1995). The RNA binding ability (Wang et al., 1995) and oligomerization of Sam68 are inhibited by p59fyn (Chen et al., 1997), a member of the Src family of kinases. These data suggest that Sam68 functions as a multifunctional SH3 and SH2 adapter protein with the ability to link cytosolic signaling pathways to downstream effects involved in RNA metabolism, such as alternative splicing.
Alternative splicing is an important mechanism for creating different protein isoforms from a single gene. In many cases, stop codons are introduced by alternative splicing, which usually changes the carboxyl terminus of proteins. This can affect the physiological function of a protein, as shown by several examples: 1) creation of soluble instead of membrane-bound receptors (Baumbach et al., 1989; Eipper et al., 1992; Toksoz et al., 1992; Zhang et al., 1994; Hughes and Crispe, 1995; Tabiti et al., 1996); 2) altered ligand affinity (Sugimoto et al., 1993; Xing et al., 1994; Suzuki et al., 1995); 3) protein truncations producing inactive variants (Swaroop et al., 1992; van der Logt et al., 1992; Duncan et al., 1995; Sharma et al., 1995; Eissa et al., 1996); and 4) changes of endocytotic pathways (Wang and Ross, 1995). In addition, inclusion or skipping of alternative exons can add or delete protein modules that change the affinity toward ligands (Danoff et al., 1991; Giros et al., 1991; Guiramand et al., 1995; Strohmaier et al., 1996), modulate enzymatic activity (O’Malley et al., 1995), create different hormones (Amara et al., 1982; Courty et al., 1995), and change properties of ion channels (Sommer et al., 1990; Kuhse et al., 1991). Finally, numerous transcription factors are subject to alternative splicing, which contributes to control of gene expression (reviewed by Lopez, 1995).
Alternative splicing pathways are not static, because the use of alternative exons can change during development (for summary, see Stamm et al., 1994), or in response to outside stimuli. For example, insulin administration influences the incorporation of the alternative exon 11 of the insulin receptor (Sell et al., 1994) and activates exon βII inclusion in the PKC gene (Chalfant et al., 1998); serum deprivation alters usage of the serine/arginine-rich protein 20 (SRp20) exon 4 (Jumaa and Nielsen, 1997); and neuronal activity changes the alternative splicing pattern of clathrin light chain B, NMDA receptor 1, and c-fos (Daoud et al., 1999). Concanavalin A has been shown to change splicing patterns of the splicing factor htra2-beta1 (Beil et al., 1997), as well as the splicing patterns of the class 1b major histocompatibility complex molecule Qa-2 (Tabaczewski et al., 1994). However, the pathways that transduce the signal to the splicing machinery have yet to be established, despite numerous examples that demonstrate a change in alternative splicing in response to external stimuli.
Our aim was to look for novel components of the spliceosomal complex; hence we used known splice factors as baits in a yeast two-hybrid approach (Fields and Song, 1989). Using the human tra2-beta1 protein, we identified a nuclear protein, YT521-B, and demonstrate that it can change alternative splicing patterns in a concentration-dependent manner. Furthermore, we show that YT521-B partially resides in subnuclear compartments, and we identified the protein domains necessary for this localization. Further yeast two-hybrid screens identified Sam68 as a YT521-B interacting protein, and we verified this interaction in vivo by coimmunoprecipitation experiments and the subnuclear colocalization of Sam68 and YT521-B. Because the association and localization of YT521-B and Sam68 were influenced by p59fyn-induced tyrosine phosphorylation, we suggest that the activity of Src family kinases could influence alternative splicing through Sam68 and its interaction with YT521-B.
This is the first report of a nuclear Sam68-associated protein that is involved in RNA splicing, and our findings suggest that Sam68 may influence RNA metabolism not only by means of its RNA binding domain but also through its interaction with other nuclear proteins.
MATERIALS AND METHODS
Two-Hybrid Screen and Cloning
Molecular cloning was performed using standard protocols (Sambrook et al., 1989), and RT-PCR was performed as described (Hartmann and Stamm, 1997). A matchmaker two-hybrid rat brain postnatal day 5 as well as an embryonic day 16 library (Stratagene, La Jolla, CA) were screened using htra2-beta pGBT9 (Beil et al., 1997) as bait. The yeast Gal4 two-hybrid screen was performed according to the method of Fields and Song (1989) using the strain HF7c. For each screen, ~6 × 106 transformants were screened with 100 μg of bait DNA and 50 μg of prey DNA. To test interaction of YT521-B with other proteins, 1 μg of YT521-B fused to the Gal4 activation domain and 1 μg of scaffold attachment factor B (SAF-B), htra2-beta, heterogeneous nuclear ribonuclear protein G (hnRNP-G), Sam68, and YT521-B fused to the Gal4 binding domain were cotransformed and plated onto triple dropout plates lacking leucine, tryptophane, and histidine. Surviving colonies were restreaked on triple dropout plates supplemented with 10 mM 3-amino-triazole.
DNA was isolated from the His-autotrophic and lacZ-positive colonies (Hoffman and Winston, 1987), electroporated into HB101, and selected on ampicillin plates. Resistant colonies were restreaked onto M9 plates without leucine to remove bait plasmids (Hollenberg et al., 1995). DNA from the autotrophic bacteria was isolated and sequenced using an Applied Biosystems (Foster City, CA) sequencer. DNA sequences were analyzed with the Genetics Computer Group (Madison, WI) Wisconsin package (Genetics Computer Group, 1994).
Two cDNA clones, YT521TH-pADgal4 and YT521ΔN10-pADgal4, were obtained. Because neither the clone YT521TH-pADgal4 nor YT521ΔN10-pADgal4 contained the complete cDNA, rapid amplification of cDNA ends (RACE) was used to amplify the missing 5′ end (Marathon cDNA amplification kit; Clontech, Cambridge, United Kingdom). Comparison with the full-length RACE clones showed that YT521TH-pADgal4 lacked the first 253 amino acids, whereas YT521ΔN10-pADgal4 lacked the first 10 amino acids.
YT521TH-B and YT521ΔN10 were cloned as an EcoRI–SacII fragment into pEGFP-C2 (Clontech) and as an EcoRI–PstI fragment into pGBT9 (Clontech). Restriction sites were introduced by PCR amplification with the oligonucleotides yt521eco-f1 (ccgaattctatgaacaggatgagagagatc), yt521-sacII-r (ccccgcggacattatcttcgataacgacctctttcc), and yt521pstI-r (tctgcagggtttttctggttgact). Deletion variants were generated either by PCR (primers available upon request; deletion clones are indicated in Figure Figure1)1) and subcloned into PCR-topo vector (Invitrogen, San Diego, CA) or by using a unique KpnI site (YT521-BΔNLS4, YT521-B THΔNLS4).
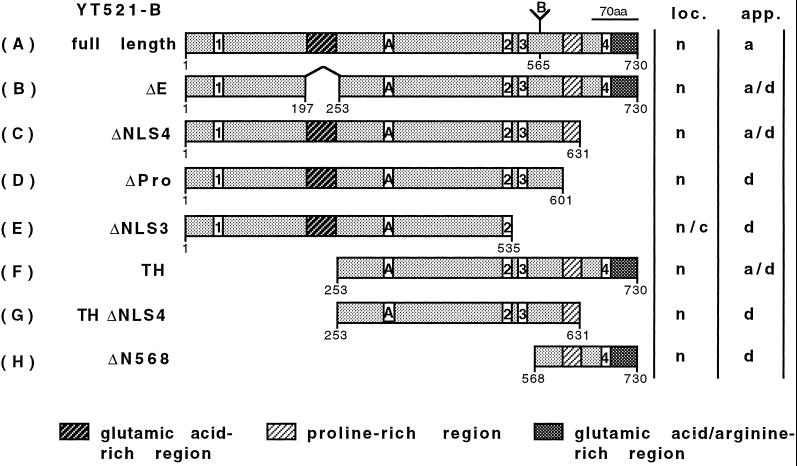
Domain structure of YT521-B and its deletion constructs. The domain structure of the full-length YT521-B clone is shown on top (A). The amino-terminal glutamic acid-rich region, the carboxy-terminal proline-rich region, and the glutamic acid/arginine-rich region are indicated by shaded boxes. Boxes with numbers indicate the four nuclear localization signals. (A and B) Insertions most likely generated by alternative splicing. Numbers underneath the boxes indicate the locations of the amino acids. The deletion clones are shown schematically and named on the left (B–H). The intracellular localization (loc.; n, nuclear; c, cytosol) and staining appearance (app.; a, aggregates; d, diffuse) of the respective EGFP-tagged proteins is indicated on the right.
RNA Isolation and PCR
RNA from whole-tissue homogenates was isolated according to the method of Feramisco et al. (1982). Briefly, frozen tissue was homogenized in guadinium thiocyanate/phenol solution at 60°C. RNA was precipitated from the aqueous phase after several phenol/chloroform extractions. Oligonucleotides for amplification of insertion A were pht6exr1 (cactcattcctgctggaagc) and pht6exf1 (gagctcgaggcatatcaccca) and for insertion B were phtexbr1 (ccgggtaaggaggcattcct) and phtexbf1 (ggcgtcgaccagaagattat). PCR was performed with a touchdown program (Don et al., 1991) (denaturation, 1 min, 94°C; annealing, 1 min, 65–55°C, lowering 0.5°C in each cycle; extension, 2 min, 72°C; 20 cycles, followed by 10 cycles with annealing at 55°C). After analyzing the products on 2% ethidium bromide-stained agarose gels, they were subcloned into pCR2.1-vector (Invitrogen) and sequenced.
Northern Blot
YT521TH (Figure (Figure1)1) cDNA was labeled with [γ32P]dCTP using the random priming method (Amersham, Arlington Heights, IL). A human multiple-tissue Northern blot (Clontech) was probed according to the manufacturer’s instructions.
In Situ Hybridization
One nanogram of the cloned YT521-B probe was PCR amplified using primers specific for the C terminus of YT521-B, anchored by either the T7 or Sp6 RNA-polymerase recognition sequence. A 12.5-μl PCR mix with 0.2 ng of plasmid was performed for 28 cycles with 94°C denaturing for 10 s, 52°C annealing for 10 s, and 74°C elongation for 20 s. PCR product was purified using QiaQuick (Qiagen, Hilden, Germany). Transcription using either T7 or Sp6 RNA-polymerase was performed according to the manufacturer’s protocol (Boehringer Mannheim, Indianapolis, IN), using 250 ng of the respective PCR product as template. Dried cryostate sections were fixed for 20 min in paraformaldehyde in PBS and washed three times for 5 min in PBS. After treatment with protease (10 μg/ml in 50 mM Tris-HCl and 5 mM EDTA, pH 7) for 10 min at 37°C, the fixation (5 min) and subsequent wash steps were repeated. The sections were acetylated (0.25% acetic anhydride and 0.1 M triethanolamine-HCl) for 10 min, rinsed twice in PBS for 5 min, dehydrated through a graded ethanol dilution, and air dried. Sections were incubated with 2 × 106 cpm of the 35S-UTP (Amersham)-labeled probe in 100 μl of hybridization mix (50% formamide, 0.3 M NaCl, 20 mM Tris-HCl, pH 7, 5 mM EDTA, 10 mM phosphate buffer, 10% dextran sulfate, 1× Denhardt’s solution, 0.2% Nonidet-P40, 50 μg/ml tRNA, and 200 μg/ml single-strand salmon sperm DNA) overnight at 57°C. After hybridization, sections were washed in 5× SSC with 10 mM DTT at 55°C for 2 min, followed by a stringent wash in 2× SSC with 10 mM DTT at 65°C for 30 min. For RNase treatment, sections were first rinsed in RNase buffer (0.5 M NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH 7) for 5 min at room temperature, incubated with 1 μg/ml RNase in RNase buffer for 30 min at 37°C, and washed again in RNase buffer for 5 min at 37°C. After two additional washes in 2× and 0.1× SSC for 10 min each at room temperature, sections were dehydrated through graded ethanol solutions containing 0.3 M ammonium acetate, air dried, covered with a thin sheet of film emulsion (NBT-2; Eastman Kodak, Rochester, NY), and exposed at 4°C for 5–7 d. They were developed in Kodak D19 developer and fixed in 24% sodium thiosulfate. Sections were counterstained with Hemalaune, dehydrated, and mounted in DePeX (Gurr; BDH, Poole, United Kingdom).
Immunofluorescence
Immunofluorescence was performed using amino-terminal enhanced green fluorescent protein (EGFP) or Flag-tagged YT521-B cDNA constructs. Two micrograms of the constructs were transfected overnight in 100,000 baby hamster kidney (BHK) cells using Superfect (Qiagen) according to the manufacturer’s protocol. Counterstaining was performed using (4-(4-(dihexadecylamino)styryl)-N-methylquinolinium iodide (Molecular Probes, Eugene, OR) for 2 h in fresh medium. Cells were fixed in 4% paraformaldehyde for 20 min, washed twice with 1× PBS for 10 min, and analyzed with laser confocal fluorescence microscopy. For the Flag construct, a polyclonal rabbit anti-Flag (Santa Cruz Biotechnology, Santa Cruz, CA) antibody was diluted 1:250 in 1× PBS and 3%BSA and incubated overnight at 4°C. The secondary antibody (Cy3-conjugated rabbit anti-goat immunoglobulin G; Sigma, St. Louis, MO) was incubated at room temperature for 4 h.
Cell Culture and Immunoprecipitation
The day before transfection, 3.0 × 105 HEK293 cells per 3.5-cm plate were seeded in 3 ml of DMEM and 10% FCS and incubated at 37°C in 5% CO2 for 17–24 h. Transient transfections of adherent HEK293 cells with 1 μg of total cDNA (EGFP-YT521-B, pRK5-p59fyn, pRK5-p59fynK299A, and pRK5-p59fynY531F) constructs were performed using the calcium phosphate method (Chen and Okayama, 1987). Cells were lysed in 200 μl of radioimmunoprecipitation assay (RIPA) buffer (1% NP40, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 10 mM Na-phosphate, pH 7.2, 2 mM EDTA, 50 mM NaF, 5 mM β-glycerolphosphate, and freshly added 4 mM sodium orthovanadate, 1 mM DTT, 1 mM PMSF, 20 μg/ml aprotinin, and 100 U/ml benzonase) for 30 min on ice. Precipitates were cleared by centrifugation. Fifty microliters of this total cellular lysate were transferred into a 1.5-ml tube, mixed with an equal amount of 1× Laemmli buffer, boiled, and stored at −20°C overnight. One hundred fifty microliters of the lysate were diluted fourfold in RIPA rescue (10 mM Na-phosphate, pH 7.2, 1 mM NaF, 5 mM β-glycerolphosphate, 20 mM NaCl, and freshly added 2 mM sodium orthovanadate, 1 mM DTT, 1 mM PMSF, and 20 μg/ml aprotinin). Immunoprecipitations were performed overnight at 4°C with agitation using anti-GFP antibody (Boehringer Mannheim) and protein A-Sepharose (Pharmacia, Piscataway, NJ; protein A-Sepharose:Sepharose, 1:1, resuspended in RIPA rescue buffer), followed by three washes in 50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Triton X-100, and freshly added 2 mM sodium orthovanadate, 100 mM NaF, 1 mM PMSF, and 20 μg/ml aprotinin (Nayler et al., 1997). The proteins were subsequently analyzed on SDS-PAGE followed by Western blotting and ECL (New England Nuclear, Boston, MA) using anti-tra2 (Daoud et al., 1999), anti-SF2 (Ak96; Oncogene Science, Uniondale, NY), anti-mAb 104 (American Type Culture Collection, Manassas, VA), anti-p62 (Santa Cruz), anti-GFP (mono-clonal, Boehringer Mannheim; polyclonal, Clontech), and antiphosphotyrosine 4G10 (Santa Cruz) antibodies.
In Vivo Splicing Assays
In vivo splicing was performed essentially as described by Cáceres et al. (1994). Briefly, 2 μg of the reporter gene were transfected together with an increasing amount (0, 0.5, 1, 1.5, and 2 μg) of YT521-B-constructs in 300,000 HEK293 cells using the calcium phosphate method (see above). Empty vector (pEGFP-C2; Clontech) was added to ensure that equal amounts of DNA were transfected. Transfection was performed at 37°C in 3% CO2 overnight. RNA was isolated 17–24 h after transfection using an RNeasy mini kit (Qiagen), following the manufacturer’s instructions. RNA was eluted in 40 μl of RNase-free H2O. For reverse transcription, 2 μl of isolated RNA were mixed with 5 pmol of antisense minigene-specific primer in 0.5 μl H2O, 2 μl of 5× RT buffer, 1 μl of 100 mM DTT, 1 μl of 10 mM dNTP, 3 μl of H2O, 0.25 μl of RNase inhibitor, and 0.25 μl of H− reverse transcriptase. The tubes were incubated for 45 min in a 42°C water bath.
Two microliters of the RT reaction were mixed with 2.5 pmol of sense and antisense primer each, 10× PCR reaction buffer, 200 μM dNTP, 2 mM MgCl2, and 0.1 μl Taq-polymerase. For the SRp20 minigene, the PCR conditions were initial denaturation for 2 min at 94°C; 30 cycles: 30 s denaturation at 94°C, annealing at 55°C for 1 min, extension at 72°C for 1 min, after 30 cycles a final extension at 72°C for 20 min, and cooling to 4°C in a Biometra (Göttingen, Germany) trio block thermocycler.
For the tra2-minigene, the conditions were 20 s denaturation at 94°C, 20 s annealing at 65°C, 40 s extension at 72°C for 33 cycles, followed by a final extension at 72°C for 20 min, and cooling to 4°C in a Perkin-Elmer (Norwalk, CT) thermocycler.The PCR reaction products were analyzed on a 0.3- to 0.4-cm-thick 2% agarose Tris borate-EDTA gel.
RESULTS
Cloning of YT521-B
To identify novel components of the spliceosomal complex (Corden and Patturajan, 1997), we have been performing yeast two-hybrid screens using known splicing factors as baits. Recently, we isolated a human homologue of the Drosphila transformer-2 splicing factor (Dauwalder et al., 1996), htra2-beta1 (Beil et al., 1997; Nayler et al., 1998a). We then performed yeast two-hybrid screens using htra2-beta1 as bait and isolated two overlapping cDNAs from a postnatal day 5 rat brain library. A similar cDNA, YT521, was recently isolated by Imai et al. (1998) using rat transformer-2-beta1 (RA301) as a bait. Both our cDNAs still lacked the start codon, and the complete 5′ end of the gene was isolated using the RACE method. Comparison with the full-length sequence revealed that one clone lacked the first 253 amino acids (YT521TH; Figure Figure1),1), and the other (YT521ΔN10) lacked the first 10 amino acids. Our isolates differ from the previously published YT521 clone and several mouse expressed sequence tags (Lennon et al., 1996) (AA646154 and AA183061) by the presence and absence of two sequence elements that we termed insertion A and insertion B, respectively (Figure (Figure1).1). We therefore named our cDNA clone YT521-B. Both insertions are probably generated by alternative splicing. Database searches detected the presence of four putative nuclear localization signals, and sequence inspection revealed an amino-terminal glutamic acid-rich region and a carboxyl-terminal proline-rich stretch. In addition, the carboxyl-terminal domain contains a region that is rich in arginine and glutamic acid residues. The overall domain structure of YT521-B is illustrated in Figure Figure1.1. The charges of the protein display a bipolar distribution. The region encompassing the start codon to the end of the glutamic acid-rich region is acidic (pI = 4.5), whereas the remaining protein is basic (pI = 9). Overall, the protein is acidic, with an isoelectric point of 5.6.
Alternative Variants
Because the sequence of our full-length YT521-B clone differs from the previously published clones by two insertions (Figures (Figures11 and Figure Figure2,2, A and B), we asked whether the expression of these isoforms is tissue specific. Using RNA derived from different rat tissues, we performed RT-PCR with primers flanking either insertion A or insertion B (Figure (Figure2,2, C and D). We found that RNA lacking insertion A is predominantly expressed in brain. Interestingly, the exclusion of insertion A is developmentally regulated in the brain and increases as development proceeds. In contrast, cDNAs containing or lacking insertion B are equally distributed in various tissues and are not developmentally regulated. We conclude that the YT521 gene generates several different isoforms using alternatively spliced exons, one of which seems to be developmentally regulated.
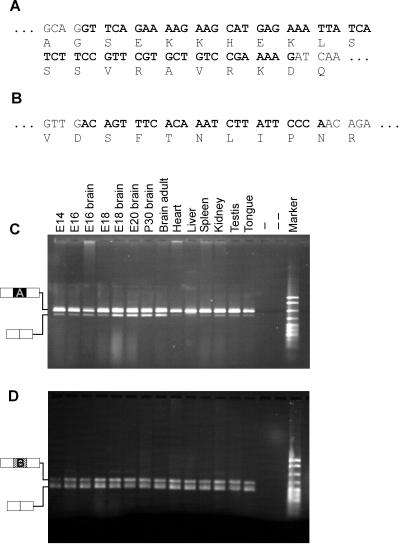
Alternative variants and their tissue distribution. The sequences of insertions A (A) and B (B) are shown in bold, and corresponding amino acids are indicated. RT-PCR was performed using RNA isolated from the tissues indicated to analyze expression of insertion A (C) and B (D). −, absence of reverse transcriptase; − −, absence of template. The structure of the PCR products is indicated on the left of C and D. Marker: pBr322 DNA MspI digest.
−, absence of template. The structure of the PCR products is indicated on the left of C and D. Marker: pBr322 DNA MspI digest.
Expression in Various Tissues
To analyze the expression of YT521-B RNA quantitatively, we performed Northern Blot analysis using poly(A+) mRNA. As shown in Figure Figure3A,3A, we detected a band of ~4.0 kb in all tissues examined. In addition, a smaller form of 3 kb can be seen in testis, and a larger variant of 7.5 kb is seen in spleen, lung, and kidney. This larger transcript is also seen in all other tissues upon longer exposure (our unpublished results). We suspect that the minor transcripts of 3 and 7.5 kb are derived by alternative splicing or by the use of different transcriptional start or termination sites, respectively. These experiments indicate that YT521 RNA is ubiquitously expressed.
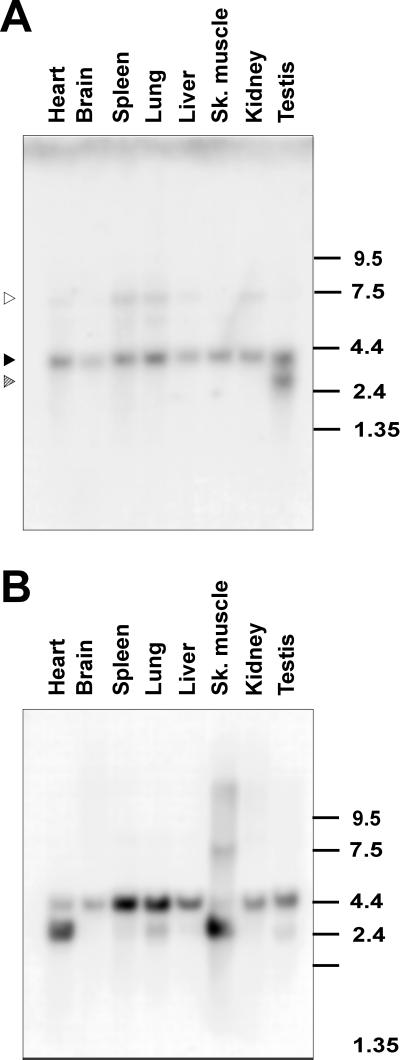
Northern blot analysis of YT521-B expression. (A) Northern blot analysis of YT521-B transcripts in various rat tissues. The location of the 4.0-kb band corresponding to YT521-B is indicated by a solid arrow. The locations of the 3- and 7.5-kb bands corresponding to minor transcript forms are indicated with striped and open arrows, respectively. The numbers on the right indicate the sizes in kilobases. (B) The same filter was rehybridized with an actin probe demonstrating loading in each lane.
However, because Northern blot analysis is limited to whole-tissue homogenates, and YT521 was shown to be regulated by ischemia in the brain (Imai et al., 1998), we sought to obtain a more detailed picture of YT521-B expression within the brain. We therefore performed RNA in situ hybridization on serial rat brain sections. As shown in Figure Figure4,4, YT-521-B could be detected in all brain regions (Figure (Figure4,4, A and B), but at higher magnification, a cell type-specific expression was observed (Figure (Figure4,4, C and D). Silver grains accumulated preferentially over large cells, e.g., in the CA3 region (Figure (Figure4,4, C and D, large arrows) that, given their morphology and location, are most likely neurons. In contrast, no specific hybridization signal was detected over cells with small, dark blue-stained nuclei, which are most likely epithelial cells (Figure (Figure4,4, C and D, small arrows). In summary, we conclude that YT521-B is widely expressed and shows a cell type-specific expression in the brain.
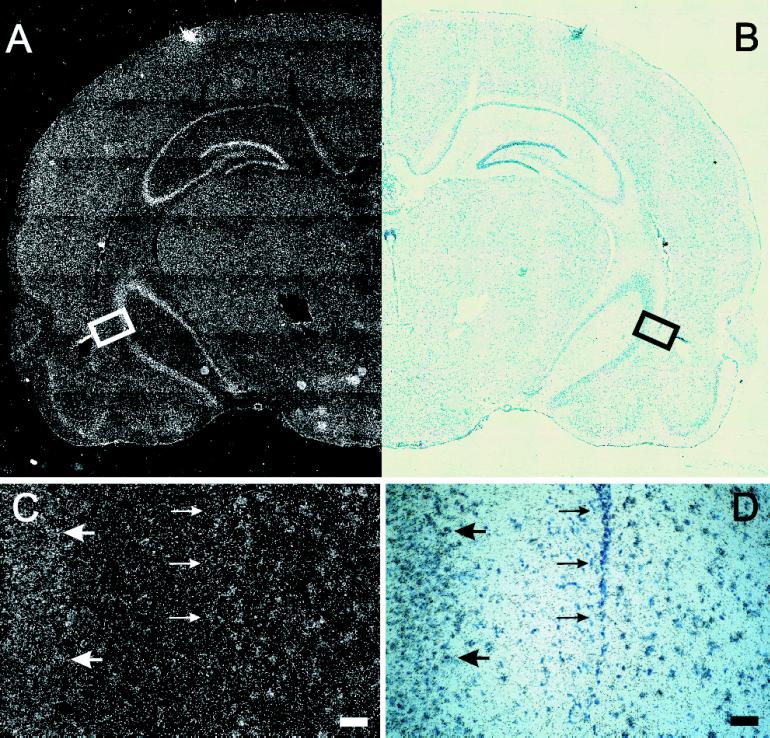
In situ hybridization of YT521-B in the rat brain. (A) Dark-field picture of a coronal section of a brain hemisphere. (B) Same field shown in bright field. In C (darkfield) and D (brightfield), a higher magnification of the box indicated in A and B is shown. Large arrows point to YT521-B-positive cells; small arrows point to YT521-B-negative cells. Bars in C and D, 50 μm.
Intracellular Localization and Domain Structure
To characterize the YT521-B protein further, we investigated its intracellular localization using transiently expressed EGFP-YT521-B fusion proteins in BHK cells. Cells were analyzed by confocal microscopy. The full-length YT521-B protein is exclusively nuclear and is characteristically concentrated in 5–20 evenly distributed dots. In addition, the YT521-B protein is diffusely located throughout the nucleoplasm, excluding the nucleoli (Figure (Figure5A).5A). Sequence inspection revealed four nuclear localization signals located throughout the molecule and four other major motifs within the protein: the N-terminal glutamic acid-rich region, the glutamic acid/arginine-rich region, and a proline-rich region, both located at the C terminus of the protein. To investigate these domains in the context of YT521-B localization, we analyzed several deletion variants that are schematically shown in Figure Figure1.1.
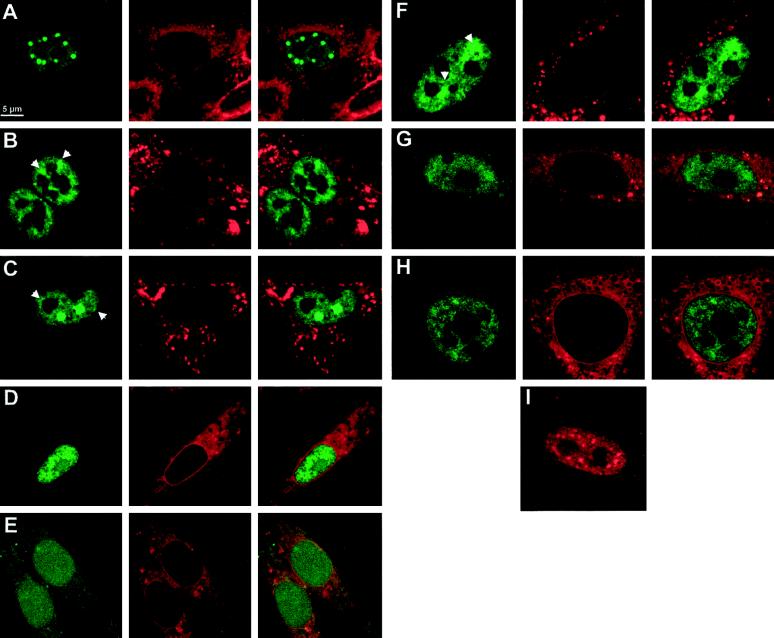
Intracellular localization of YT521-B and its deletion variants. BHK cells were transiently transfected with EGFP-YT521-B as well as various deletion constructs and analyzed by confocal microscopy. The deletion constructs (B–H) are schematically shown in Figure Figure11 in the same order. The staining in green (left column) corresponds to the EGFP-tagged YT521-B variants; the staining in red (middle column) shows membranes counterstained with (4-(4-(dihexadecylamino)styryl)-N-methylquinolinium iodide, and the right column shows the overlay of the green and red stainings (A–H). Arrows point to structures resembling dissolved dots with fuzzy edges. Bar, 5 μm. (I) Staining pattern of FLAG-tagged YT521-B. The following EGFP fusion constructs (A–H) were used: (A) YT521-B; (B) YTD21ΔE; (C) YT521ΔNLS4; (D) YT521ΔPro; (E) YT521ΔNLS3; (F) YT521ΔTH; (G) YT521THΔNLS4; (H) YT521ΔN568; (I) FLAG-tagged YT521-B, detected with anti-FLAG.
First, we deleted the glutamic acid-rich region (amino acids 197–253) and found that the resulting protein YT521ΔE was still nuclear, although the number of intensely stained dots was reduced and the dots became smaller (Figure (Figure5B).5B). Often, the cells contained nuclear structures that looked like dissolving dots (Figure (Figure5B,5B, arrows). Next, we deleted the glutamic acid/arginine-rich region together with the nuclear localization signal 4 (NLS4; amino acids 631–730). The resulting construct, YT521ΔNLS4, is again exclusively expressed in the nucleus. Again, we observed a decrease in the number of dots, and, as seen with the YT521ΔE variant, many cells contained nuclear structures resembling dissolved dots (Figure (Figure5C,5C, arrows). A deletion construct including NLS4 (deletion of amino acids 688–730) was indistinguishable from the YT521ΔNLS4 construct (our unpublished results).
To address the function of the proline-rich region, we removed amino acids 601–631. The resulting protein, YT521ΔPro, was no longer detected in nuclear dots. The staining was now uniform, and the protein was also detected in nucleoli (Figure (Figure55D).
To investigate the role of the nuclear localization signals, four more constructs were used. First, we deleted the two most carboxyl-terminal nuclear localization signals, NLS3 and NLS4 (amino acids 535–601), and found that the resulting protein, YT521ΔNLS3, is uniformly distributed in the nucleus and is also present in the cytosol (Figure (Figure5E).5E). This indicates that NLS1 and 2 alone are not sufficient for exclusive nuclear localization. We then deleted the first 253 amino acids of the protein, removing NLS1 and the glutamic acid-rich stretch. The protein made by this clone, YT521TH, is exclusively in the nucleus (Figure (Figure5F),5F), and the nuclear staining contains structures resembling the dissolving dots observed with the YT521ΔE variant.
We then combined amino- and carboxyl-terminal deletion variants and removed amino acids 631–730 of YT521TH. The resulting protein, YT521THΔNLS4, lacks the amino-terminal part including NLS1, the glutamic acid/arginine-rich region, and the fourth nuclear localization signal and yields a nuclear staining pattern (Figure (Figure5G)5G) resembling that of YT521TH. Finally, we tested the carboxyl-terminal domain alone (amino acids 568–730) containing the proline-rich region, NLS4, and the glutamic acid/arginine-rich domain and found that this protein, YT521ΔN568, is exclusively nuclear, excluding the nucleoli (Figure (Figure55H).
In summary, our deletion analysis demonstrates that the intracellular localization of YT521-B is governed by several domains. The exclusive nuclear localization is due to the NLS3 and NLS4, whereas the subnuclear localization is dependent on several domains, notably the glutamic acid-rich and glutamic acid/arginine-rich regions, which act in concert to localize the protein into 5–20 dots, and the proline-rich region, which appears to be involved in the nucleolar exclusion of the protein.
To exclude the possibility of an EGFP-mediated effect on the cellular localization, we tested the intracellular localization of a Flag-tagged YT521-B construct and analyzed transfected cells by immunofluorescence. Again, nuclear dots were observed, indicating that their formation is independent of either EGFP or a Flag tag (Figure (Figure5I).5I). However, the dots observed with the Flag-tagged construct are slightly smaller than the ones seen with an EGFP-tagged protein, which is most likely a result of the stronger signal to background ratio of GFP detection.
Interaction with Other Proteins in Yeast
To obtain information on the possible function of the YT521-B protein, we determined its binding properties to other nuclear proteins in the yeast two-hybrid system. YT521 and YT521-B were originally isolated owing to their interaction with htra2-beta1, an RS domain-containing spliceosomal protein. To find other YT521-B-interacting proteins, we used YT521-B as a bait in yeast two-hybrid screens and tested a postnatal day 5 and an embryonic day 16 brain library. To our surprise, 70 of 110 interacting clones contained the rat homologue of the Sam68 (Richard et al., 1995) or brain-specific cDNAs highly similar to Sam68 (Stoss, Hartmann, and Stamm, unpublished results). All these interactors are members of the STAR protein family (Vernet and Artzt, 1997). Additional clones included SAF-B (Nayler et al., 1998c), transformer2-α (Dauwalder et al., 1996), and hnRNP-G (Soulard et al., 1993). Based on these initial yeast two-hybrid experiments, we concluded that YT521-B might interact with various components of the spliceosomal complex as well as with members of the STAR protein family.
To define the interaction domains of YT521-B, we used the yeast two-hybrid system and compared the interactions of the full-length YT521-B protein, the deletion mutant YT521TH, which lacks the acidic amino terminus, and YT521THΔNLS3, which lacks the acidic amino terminus, as well as the glutamic acid/arginine-rich and proline-rich domains, with the newly identified interacting proteins (Figure (Figure6).6). We found that deletion of the amino-terminal domain had no effect on the protein–protein interactions with YT521-B, Sam68, and rSAF-B but reduced the interaction with htra2-beta1 and hnRNP-G. In contrast, removing the glutamic acid/arginine-rich domain abolished binding to htra2-beta1, hnRNP-G, Sam68, and rSAF-B, indicating that it is necessary for these observed protein–protein interactions in yeast.
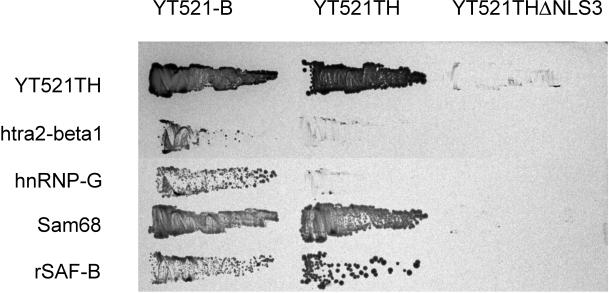
Interaction with proteins in a yeast two-hybrid assay. The interaction between YT521-B, its deletion clones YT521TH and YT521THΔNLS3 (top), and various proteins indicated on the left were tested in the yeast two-hybrid system. Transformants were obtained on plates without 3-amino-triazole and then restreaked on a plate containing 10 mM 3-amino-triazole.
Coimmunoprecipitation of Proteins with YT521-B
We next investigated the observed protein–protein interactions in a mammalian cell system using coimmunoprecipitation experiments. To this end, EGFP-YT521-B constructs were transiently expressed in HEK293 cells, and protein complexes were precipitated with an anti-EGFP antibody. Because the proteins of interest displayed nucleic acid binding activity, we included benzonase in our lysis buffer to avoid nonspecific or indirect interactions. The precipitates were analyzed by Western blotting using antibodies against endogenous proteins. When we used antibodies against Sam68 (Richard et al., 1995) or SAF-B (Nayler et al., 1998c), we were able to demonstrate that these proteins specifically coprecipitate with EGFP-YT521-B under our experimental conditions (Figure (Figure7,7, A and B). Moreover, deletion of the glutamic acid/arginine-rich domain of YT521-B, as seen with the construct YT521ΔNLS4 (Figure (Figure1C),1C), dramatically reduced the binding to Sam68 (Figure (Figure7A)7A) and to SAF-B (Figure (Figure7B),7B), which is in accordance with our initial findings in yeast (Figure (Figure6).6). In contrast, deletion of the glutamic acid-rich region alone (YT521ΔE; Figure Figure1B)1B) had no visible effect (Figure (Figure7,7, A and B).
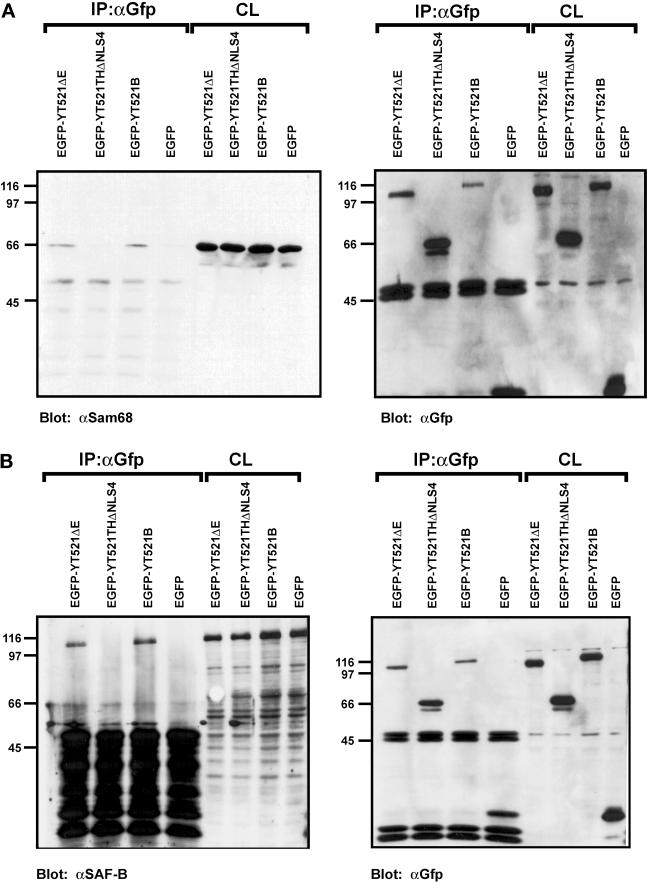
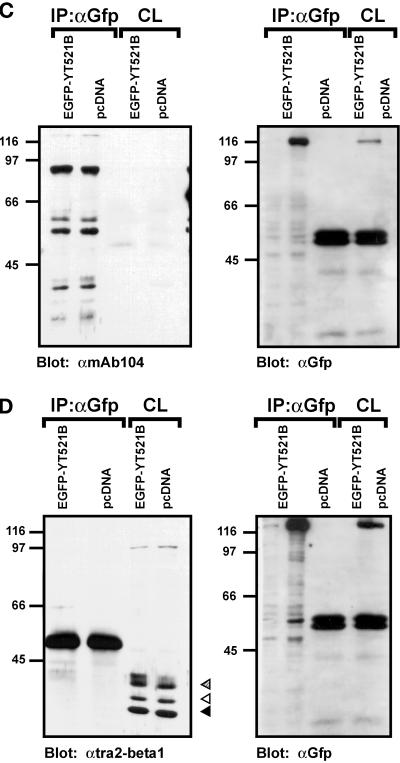
Immunoprecipitation of YT521-B protein complexes. YT521-B or its deletion variants were expressed as EGFP fusion proteins and precipitated with anti-EGFP antibodies. Immunocomplexes were analyzed by Western blot after SDS-PAGE using the antibodies indicated. The endogenous coprecipitating proteins were detected in all experiments. CL, crude lysates; IP, immunoprecipitation. The analysis of the immunoprecipitates is shown on the left. The reblot using anti-GFP antibody to demonstrate proper protein expression is shown on the right. (A) Blot with anti-Sam68. (B) Blot with anti-SAF-B. (C) Blot with mAb104 that recognizes SR proteins. (D) Blot with anti-htra2-beta1. The closed arrow indicates the location of the dephosphorylated protein, whereas the open and striped arrows show the phosphorylated and hyperphosphorylated forms (Daoud et al., 1999).
Using the pan SR protein antibody mAB104 (Figure (Figure7C)7C) (Fu, 1995) and specific antibodies against SF2/ASF (AK96; our unpublished results) and htra2-beta1 (Figure (Figure7D)7D) (Nayler et al., 1998a), we were unable to detect coimmunoprecipitation with YT521-B under these conditions (Figure (Figure7,7, C and D). This contradicts our initial finding in yeast (Figure (Figure6),6), and the previously found association of recombinant SC35, SF2/ASF and rtra2-beta1 with YT521 in far-Western blot analysis (Imai et al., 1998c). However, using the same experimental conditions, we previously demonstrated an in vivo association between SAF-B and RS domain-containing proteins (Nayler et al., 1998). Furthermore, no interaction between RS domain-containing proteins and YT521-B was detected when magnesium was added to the buffer or when a Triton X-100-based lysis buffer was used (Nayler et al., 1997) (our unpublished results). In addition, EGFP-YT521-B did not coimmunoprecipitate with Flag-tagged YT521-B (our unpublished results), indicating that a YT521-B multimerization might not take place under in vivo conditions.
The Association of YT521-B and Sam68 Is Negatively Regulated by Tyrosine Phosphorylation
Because YT521-B interacted with Sam68 in the yeast two-hybrid system and in coimmunoprecipitation experiments, we asked whether the phosphorylation status of Sam68 influenced its binding to YT521-B. To address this question, we immunoprecipitated overexpressed EGFP-YT521-B and analyzed the tyrosine phosphorylation status of the endogenous coimmunoprecipitating Sam68 using anti–phosphotyrosine-specific antibodies. However, we were unable to detect any tyrosine phosphorylation of Sam68 in these experiments (our unpublished results). We then suspected that the endogenous levels of tyrosine-phosphorylated Sam68 were too low, and, accordingly, we increased the phosphorylation levels of Sam68 by cotransfecting the Src family kinase p59fyn or the constitutively active mutant p59fynYF. As a control, the catalytically inactive p59fynKA construct was used. As can be seen in Figure Figure8A,8A, left panel, p59fyn and p59fynYF caused tyrosine phosphorylation of Sam68, which is in agreement with earlier findings (Chen et al., 1997; Di Fruscio et al., 1999). In contrast, the catalytic inactive variant p59fynKA had no effect.
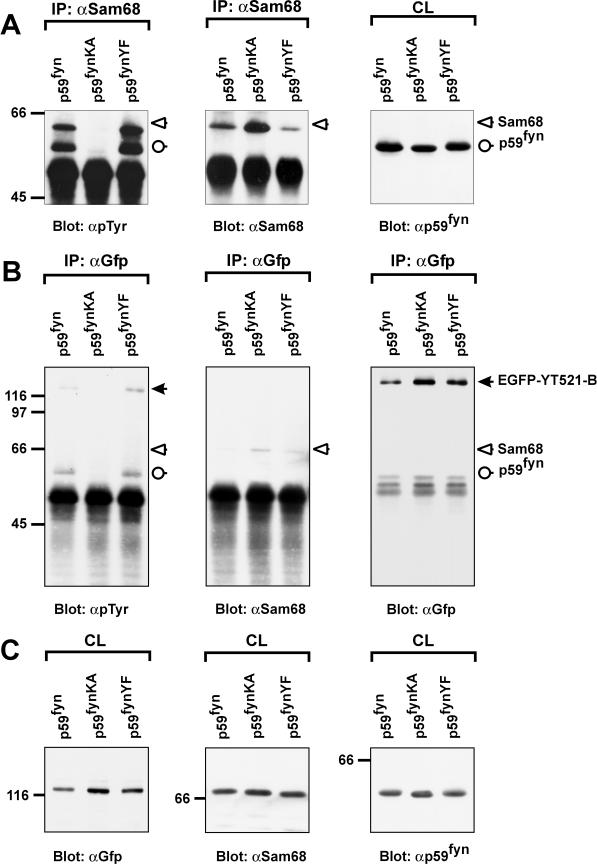
The association of YT521-B and Sam68 is negatively regulated by tyrosine phosphorylation. The catalytically active kinase p59fyn, the catalytically inactive kinase mutant p59fynKA, and the constitutively active kinase mutant p59fynYF were transiently expressed in HEK293 cells together with EGFP-YT521-B. The cells were lysed 24 h after transfection, and immunoprecipitations (IP) using and anti-Sam68 (A) or anti-GFP (B) antibody were performed. Tyrosine phosphorylation was detected after SDS-PAGE and Western blotting using the monoclonal antibody 4G10 (A and B, left panels). Reblots confirmed the successful precipitation of Sam68 (A and B, middle panels) and EGFP-YT521-B (B, right panel) using specific antibodies against those proteins. Aliquots of the crude lysates (CL) were analyzed by SDS-PAGE and Western blotting to confirm expression of EGFP-YT521-B (C, left panel), endogenous Sam68 (C, middel panel), and p59fyn (C, right panel). The detected proteins are labeled, and their positions are indicated with arrows (closed arrow, EGFP-YT521-B; open arrow, Sam68; open circle, p59fyn). The identity of the band corresponding to p59fyn was confirmed by reblotting with anti-p59fyn antibodies (our unpublished results). The molecular mass is indicated in kilodaltons.
Even under those conditions, no detectable levels of tyrosine-phosphorylated Sam68 were observed when overexpressed EGFP-YT521-B was immunoprecipitated (Figure (Figure8B,8B, left panel), although the reblot clearly showed that Sam68 bound to YT521-B (Figure (Figure8B,8B, middle panel). Interestingly, p59fyn and p59fynYF induced tyrosine phosphorylation of EGFP-YT521-B (Figure (Figure8B,8B, left panel), suggesting that YT521-B itself is a tyrosine-phosphorylated protein and a possible substrate of p59fyn or an activated downstream kinase. Together, these data suggest that the heteromerization of YT521-B and Sam68 is inhibited by tyrosine phosphorylation through p59fyn.
Colocalization of YT512-B and Sam68
To test whether the observed binding of YT521-B to Sam68 and SAF-B can take place in vivo, we performed double staining of EGFP-YT521-B-transfected cells with these nuclear proteins (Figure (Figure9).9). As seen in Figure Figure9A,9A, Sam68 colocalized with YT521-B. Both proteins are concentrated in nuclear dots and are also present at lower concentrations in the nucleoplasm. Furthermore, because the association of Sam68 and YT521-B is negatively regulated by p59fyn (see above), we asked whether overexpression of p59fyn also affects the intracellular localization of these proteins. We observed (Figure (Figure9B)9B) that p59fyn completely abolished the nuclear dots formed by YT521-B and Sam68, suggesting that the association between YT521-B and Sam68 is truly influenced by p59fyn activity in vivo. To rule out a disintegration of Sam68 and YT521-B nuclear dots through protein sequestration, we performed the same experiment with the catalytic inactive mutant p59fynKA and found no influence of this protein on the nuclear dots (Figure (Figure9C).9C).
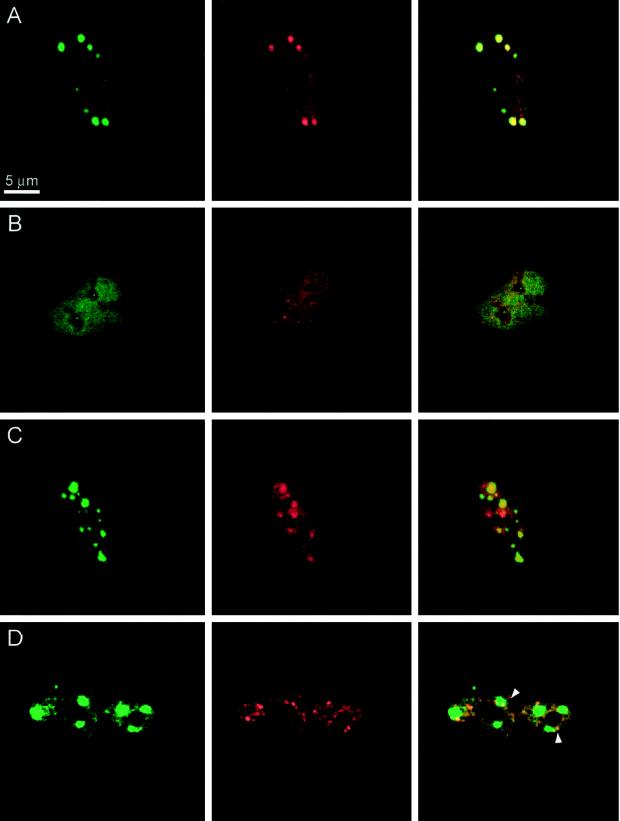
Localization of YT521-B, Sam68, and SAF-B. BHK cells were transfected, fixed, and incubated with the antibodies indicated. The staining in green (left column) is always EGFP-YT521-B. The staining in red (middle column) is either Sam68 or SAF-B, as indicated. The pictures in the right column show the overlay of the red and green signals. Bar, 5 μm. (A) Transfection of EGFP-YT521-B; endogenous Sam68 (red) was detected. (B) Transfection of EGFP-YT521-B and p59fyn; endogenous Sam68 (red) was detected. (C) Transfection of EGFP-YT521-B and p59fynKA; endogenous Sam68 (red) was detected. (D) Transfection of EGFP-YT521-B and FLAG-SAF-B; FLAG-SAF-B was detected with anti-FLAG (red). Arrows indicate colocalization of SAF-B and YT521-B at the periphery of YT521-B nuclear dots.
We then investigated the relationship of YT521-B nuclear dots and SAF-B, which is localized in the nucleoplasm and concentrated in nuclear speckles (Nayler et al., 1998c). As shown in Figure Figure9D,9D, YT521-B dots have frequently contact with SAF-B-containing speckles at their periphery (Figure (Figure9D,9D, arrows), indicating a partial colocalization of YT521-B and SAF-B. However, the majority of the SAF-B-containing speckles do not overlap with YT521-B nuclear dots.
YT512-B Can Change Splice Site Selection in a Concentration-dependent Manner
The nucleoplasmatic localization of YT521-B and its interaction with SAF-B suggest a role for YT521-B in a transcriptosomal complex (Corden and Patturajan, 1997; McCracken et al., 1997; Nayler et al., 1998c). The cellular concentration of YT512 in cells seems to be regulated by several mechanisms. First, we observed that YT521-B is not expressed in every cell (Figure (Figure4).4). Second, overexpression of p59fyn leads to a disintegration of YT512 nuclear dots and a subsequent increase of YT521-B concentration in the nucleoplasm, indicating a possible regulation of local protein concentration through kinases (Figure (Figure99B).
We therefore asked whether YT521-B could modulate splice site selection in vivo in a concentration-dependent manner, analogous to several proteins involved in splicing, such as SR proteins (Cáceres et al., 1994; Wang and Manley, 1995), hnRNPs (Cáceres et al., 1994), and also SAF-B (Nayler et al., 1998c). To investigate this possiblity, we used an SRp20 reporter gene, consisting of the constitutive exons 3 and 5, as well as the alternative exon 4 (Jumaa and Nielsen, 1997). This reporter was transfected with increasing amounts of EGFP-YT521-B in HEK293 cells. Vector DNA (pEGFP) was added to ensure that comparable amounts of DNA were transfected in each experiment. Increasing the amount of transfected EGFP-YT521-B construct led to a decreased incorporation of exon 4, as shown in Figure Figure10A,10A, left, suggesting a protein concentration-dependent modulation of splice site selection by YT521-B. Our binding studies (Figures (Figures66 and and7)7) indicated that the glutamic acid/arginine-rich domain located at the carboxyl-terminal domain of YT521-B is necessary for binding to SAF-B, a factor we had previously shown to be involved in splice site selection (Nayler et al., 1998c). We therefore repeated the experiment using YT521ΔNLS4 (Figure (Figure1C),1C), a deletion mutant lacking the glutamic acid/arginine-rich region. This protein no longer binds to Sam68 or SAF-B (Figure (Figure7).7). As shown in Figure Figure10A,10A, right, this deletion variant had no effect on SRp20 splice site selection. A similar result was obtained with a minigene containing the first four exons of htra2-beta (Nayler et al., 1998c). An increasing amount of EGFP-YT521-B blocked the formation of the beta4 isoform, containing all four exons, in favor of the formation of the beta1 isoform, consisting of exons one, three, and four (Figure (Figure10B,10B, left). Again, no effect was observed when the YT521ΔNLS4 variant was used (Figure (Figure10B,10B, right).
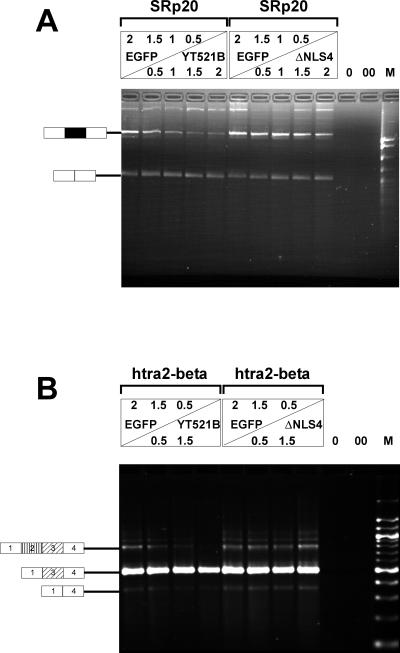
In vivo splicing assays. (A) HEK293 cells were transfected with increasing amounts of EGFP-YT521-B (left), as well as EGFP-YT521ΔNLS4 (right) and the SRp20 minigene. The amount of transfected DNA is indicated and was normalized using pEGFP-C2 (top panel). The RNA was analyzed by RT-PCR. 0, PCR without RT reaction; 00, PCR without template. The structure of the amplified product is indicated on the left. The alternative exon 4 is shown in black. M, pBr322 DNA MspI digest. (B) A similar experiment was performed with a minigene containing the first four exons of the htra2-beta gene. The structures of the amplified products are shown on the left, and alternative exons are indicated by shading. M, 100-bp DNA ladder.
These results indicate that the relative concentration of YT521-B can influence specific alternative splicing decisions, possibly through the binding to transcriptosomal components, such as SAF-B via the glutamic acid/arginine-rich domain.
DISCUSSION
Expression and Sequence Analysis
In this report, we describe the molecular cloning and analysis of the nuclear protein YT521-B. Northern blot analysis indicated that YT521-B is expressed ubiquitously, and, upon closer examination using RNA in situ techniques on serial rat brain slices, we detected that not all cells express YT521-B. From these experiments, we conclude that YT521-B expression in the brain is restricted to certain cell types. Furthermore, at least two regions of the protein are subject to alternative splicing, and one of these regions is subject to developmental control. It remains to be analyzed whether YT521-B isoforms are also controlled in a cell type-specific manner. Sequence inspection shows that YT521-B contains several domains, namely four NLS sequences, a proline- and glutamic acid-rich domain, as well as a glutamic acid/arginine-rich domain.
We tested several deletion clones to analyze the contribution of various domains to the intracellular localization. YT521-B is concentrated in 5–20 nuclear dots and is also present throughout the nucleoplasm, excluding the nucleoli. Removing the proline-rich region (amino acids 601–631) in the deletion clone YT521ΔPro (Figure (Figure5D)5D) resulted in staining of the nucleoli. In addition, a short construct containing amino acids 568–643, which include the proline-rich stretch, was absent from the nucleoli (our unpublished results). Taken together, these data indicate that the proline-rich region is necessary to exclude YT521-B from the nucleoli.
The exclusive nuclear localization of YT521-B is due to the presence of several nuclear localization signals. We tested them independently and found that NLS1 and NLS2 alone are insufficient for nuclear localization (YT521ΔNLS3; Figure Figure5E),5E), whereas the presence of NLS4 or NLS3 results in nuclear staining (YT521ΔN568; Figure Figure5H).5H). This indicates that the nuclear localization is mainly achieved by sequence elements in the carboxyl-terminal part of the protein.
The most striking feature of the observed YT521-B localization is the accumulation of the protein in 5–20 nuclear dots. These dots have sharp borders (Figure (Figure5A)5A) and are dependent on the glutamic acid-rich region (Figure (Figure5B)5B) and the glutamic acid/arginine-rich region (Figure (Figure5C).5C). Both domains appear to act in concert, because deletion of either domain results in fuzzy borders and dissolving dots. We observed these dots in several cell lines (HeLa, HEK293, COS, and BHK; Figure Figure5;5; our unpublished results) using variable amounts of transfected DNA and various incubation times after transfection (our unpublished results). In addition, a Flag-tagged construct (Figure (Figure5I)5I) displayed a staining pattern similar to that of EGFP-tagged constructs. We therefore suggest that the observed nuclear dots might represent a genuine nuclear compartment and not a nonspecific assembly of protein aggregates.
Glutamic Acid/Arginine-rich Domain
The most prominent motif of the YT521-B protein is the glutamic acid/arginine-rich region located at its carboxylterminal end. This domain seems to be important for YT521-B function, because its deletion abolished any detectable binding to the other identified interacting proteins (Figures (Figures66 and and7).7). In addition, we found that it contributes to the subnuclear localization of YT521-B and that it is involved in the YT521-B induced changes of the SRp20 and tra2-beta reporter gene splicing pattern (Figures (Figures5C5C and and10).10). Database searches showed that several other proteins contain a domain with alternating ER or DR repeats (Figure (Figure11).11). Some of them, such as the RD gene (Surowy et al., 1990), U170K (Spritz et al., 1990), SAF-B (Nayler et al., 1998c), the Drosophila shuttle craft protein (Stroumbakis et al., 1996), Caenorhabditis elegans SRp20 (Wilson et al., 1994), tobacco U2 auxiliary factor (Domon et al., 1998), and several RNA helicases (Sukegawa and Blobel, 1995; Ohno and Shimura, 1996), are involved in pre-mRNA splicing or in the formation of the transcriptosomal complex. The alternation of negative and positive charges of the amino acid residues is reminiscent of phosphorylated RS domains. In contrast to SR proteins, in which the RS domain is usually located at the carboxyl-terminal end of the proteins, the position of glutamic acid/arginine-rich regions is variable (Figure (Figure11A).11A). Moreover, RS domains are subject to phosphorylation, thus influencing the subnuclear localization of those proteins (Colwill et al., 1996; Misteli et al., 1997). It is not clear whether the charge distribution of glutamic acid/arginine-rich domains is altered as well. Nevertheless, we suggest that the glutamic acid/arginine-rich region might fulfill an important role in protein-protein interaction.
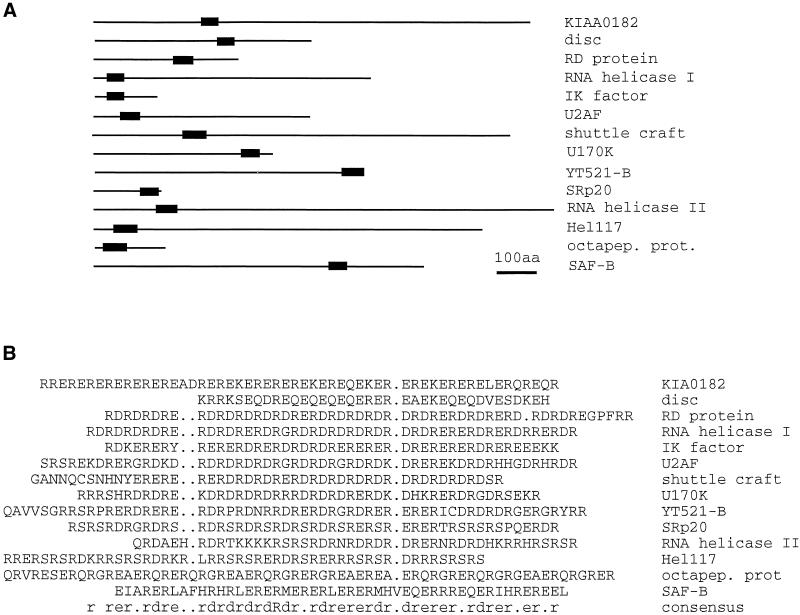
Compilation of proteins containing glutamic acid/arginine-rich sequences. Proteins were retrieved using the last 60 amino acids of YT521-B as well as the sequence (ER)15 and the BLAST algorithm. Similar sequences were assembled using PILEUP. (A) Proteins are shown as lines, whereas the glutamic acid/arginine-rich region is indicated by a black box. Drawing is to scale. Bar, 100 amino acids. (B) Alignment of the glutamic acid/arginine-rich regions of A. KIA0182, hypothetical protein (Nagase et al., 1996); disk, Drosophila disconnected gene (Heilig et al., 1991); RD protein, human RD protein (Surowy et al., 1990); RNA helicase I, RNA helicase isologue from Arabidopsis thaliana (GenBank accession number U78721); IK factor, human IK factor (Krief et al., 1994); U2AF, U2 auxillary factor from tobacco (Domon et al., 1998); shuttle craft, Drosophila shuttle craft protein (Stroumbakis et al., 1996); U170K, human U170K protein (Spritz et al., 1990); YT521-B, this publication; SRp20, C. elegans protein similar to SRp20 (Wilson et al., 1994); RNA helicase II, RNA helicase ATP-dependent RNA helicase HRH1 (Ohno and Shimura, 1996); Hel117, rat RNA helicase (Sukegawa and Blobel, 1995); octapep.prot., mouse octapeptide protein (Di Carlo et al., 1992); SAF-B, rat scaffold attachment factor B (Nayler et al., 1998c); consensus,: amino acid consensus sequence of this alignment, as determined by LINEUP.
Comparison with Other Subnuclear Structures
The striking nuclear staining pattern observed with YT521-B resembled the nuclear dots observed with wild-type ataxin-1 and, more strongly, mutant ataxin-1 (Skinner et al., 1997). However, colocalization studies revealed that the dots formed by YT521-B and ataxin-1 are similar in size, shape, and number but do not overlap (our unpublished results). It is interesting to note that the formation of YT521-B dots is at least in part dependent on the presence of a polyglutamic acid region within the molecule. This region does not appear to be created by a recent trinucleotide (GAA) expansion, because the third codon position is variable. Therefore, the formation of nuclear dot structures is not just confined to proteins such as ataxin-1 (Skinner et al., 1997) or mutant huntingtin (Roizin et al., 1979) but is in fact a rather normal structure in nuclei. It remains to be seen whether other glutamic acid-rich region-containing proteins might give a similar intranuclear distribution.
Sam68 Binds to YT521-B
Based on the amount of positive clones obtained in our yeast two-hybrid screens that are supported by our immunoprecipitation and immunolocalization results (Figures (Figures77 and and9),9), we suggest that Sam68 is an interactor of YT521-B. Sam68 is a member of the STAR family of proteins (Vernet and Artzt, 1997). Sam68 is phosphorylated by p59fyn (Chen et al., 1997) and by Src during mitosis (Taylor et al., 1995). Self-association of Sam68, as well as association with other KH domain-containing proteins such as GRP33, GLD-1, and Qk1, are inhibited by tyrosine phosphorylation (Chen et al., 1997). To study the influence of phosphorylation on the association of Sam68 and YT521-B, we overexpressed p59fyn and its mutant forms, p59fynKA and p59fynYF, together with YT521-B. In these experiments (Figure (Figure8)8) we found that no detectable tyrosine-phosphorylated Sam68 was bound to YT521-B. However, non–tyrosine-phosphorylated Sam68 did bind to YT521-B. Again, it should be pointed out that all immunoprecipitations were performed in the presence of endonuclease (benzonase) to avoid nucleic acid-mediated coimmunprecipitation. Furthermore, and to our surprise, YT521-B itself was tyrosine phosphorylated upon p59fyn overexpression, although it is not clear whether YT521-B is a p59fyn substrate or a substrate of a p59fyn-activated downstream kinase (Figure (Figure8).8). However, phosphorylation of YT521-B itself did not seem visibly to affect the binding to unphosphorylated Sam68. Together, these data show that p59fyn or a p59fyn activated downstream kinase phosphorylate YT521-B, and that tyrosine phosphorylation of Sam68 by p59fyn negatively regulates its association with YT521-B.
The Nuclear Localization of YT521-B Changes upon p59 fyn Overexpression
fyn Overexpression
Immunoflourescence experiments showed a colocalization of YT521-B and Sam68 in nuclear dots (Figure (Figure9A).9A). When investigated in the absence of YT521-B, Sam68 shows a variable nuclear staining pattern that is diffuse nucleoplasmatic in most cells but becomes punctuate upon treatment with transcription inhibitors (McBride et al., 1998; our unpublished results). These Sam68 nuclear structures are smaller than the ones observed upon YT521-B coexpression. Therefore, the localization of Sam68 in the larger YT521-B dots could be a result of the transient expression of YT521-B and a subsequent stabilization of the interaction between the two proteins. This further supports an in vivo interaction between YT521-B and Sam68 and underlines the dynamic behavior of Sam68. Our biochemical analysis (Figure (Figure8)8) indicated that Sam68 and YT521-B interacted in a tyrosine phosphorylation-dependent manner. Therefore, we determined the influence of p59fyn overexpression on the subnuclear structure of Sam68 and YT521-B. As shown in Figure Figure9,9, B and C, p59fyn, but not its catalytically inactive mutant p59fynKA, dissolved the nuclear dots formed by Sam68 and YT521-B. p59fyn-induced phosphorylation seems to specifically affect YT521-B, because no change of ataxin-1 and SAF-B localization was observed (our unpublished results), arguing against a general effect on nuclear architecture. The regulation of the shape and size of Sam68- and YT521-B-containing dots is reminiscent to the control of SR protein-containing speckles by CDC2-like SR protein kinases. Here, overexpression of CDC2-like SR protein kinases dissolves nuclear speckles, the proposed storage compartments of splicing components (Colwill et al., 1996; Nayler et al., 1997, 1998b). It is currently debated whether a release of splicing components from speckles through phosphorylation might be a mechanism to regulate splice site selection (Misteli et al., 1997). Similar to nuclear speckles, the YT521-B dots might serve to concentrate the YT521-B protein to be released upon a phosphorylation signal. As a result, the local nuclear concentration of YT521-B could be regulated by well-characterized signal transduction pathways.
YT521-B Is Involved in Splice Site Selection
The cellular concentration of YT521-B is regulated by its cell type-specific expression and possibly by a phosphorylation-dependent change in local concentation in the nucleoplasm. The cell type specificity is reminiscent of htra2-beta1 (Daoud et al., 1999) and several hnRNPs (Kamma et al., 1995), which were shown to exhibit cell type-specific expression patterns when studied in situ. We therefore performed in vivo splicing experiments to study the effects of increased relative amounts of YT521-B on two reporter gene constructs. In both cases, we observed repression of an alternatively spliced exon when the YT521-B concentration was increased.
We were able to show that YT521-B interacts with the nuclear protein SAF-B (Figures (Figures66 and and7),7), which has been shown to bind to SR proteins and which was implied in the regulation of alternative splicing (Nayler et al., 1998c). In addition, SAF-B-containing speckles are frequently located at the periphery of YT521-B nuclear structures, which could suggest a cross-talk between these compartments. Therefore, YT521-B could change splice site selection through sequestration of nucleoplasmic molecules such as SAF-B that directly bind to SR proteins necessary for pre-mRNA splicing. This model would also explain that a YT521-B variant lacking the glutamic acid–arginine domain, necessary for the protein–protein interaction, had no visible effect on splicing. It is likely that the nuclear concentration of YT521-B, and hence its ability to sequester proteins, is regulated by p59fyn. In summary, our findings suggest that YT521-B can mediate splice site selection in response to cellular kinase activity and could be a molecular basis for the observed dependency of alternative splicing patterns upon outside stimuli.
ACKNOWLEDGMENTS
We thank Claudia Cap for sequencing, James Chalcroft for artwork, Peter Nielsen for providing the SRp20 minigenes, H. Orr for providing ataxin-1 clones, and Gregor Eichele for helping to analyze the in situ hybridizations. This work was supported by the Max-Planck Society and the Human Frontier Science Program (grant RG562/96 to S.S.). We are grateful to Axel Ullrich for financial support and reagents (to O.N.). Sequences from this study were deposited in GenBank (accession number AF144731).
Abbreviations used:
| BHK | baby hamster kidney |
| EGFP | enhanced green fluorescent protein |
| hnRNP | heterogeneous nuclear ribonuclear protein |
| NLS | nuclear localization signal |
| RACE | rapid amplification of cDNA ends |
| RIPA | radioimmunoprecipitation assay |
| SAF-B | scaffold attachment factor B |
| SH | Src homology region |
| SR protein | protein with serine/arginine-rich domain |
| STAR | signal transduction and activation of RNA |
REFERENCES
- Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. [Abstract] [Google Scholar]
- Baumbach WR, Horner DL, Logan JS. The growth hormone-binding protein in rat serum is an alternatively spliced form of the rat growth hormone receptor. Genes & Dev. 1989;3:1199–1205. [Abstract] [Google Scholar]
- Beil B, Screaton G, Stamm S. Molecular cloning of htra2-beta-1 and htra2-beta-2, two human homologues of tra-2 generated by alternative splicing. DNA Cell Biol. 1997;16:679–690. [Abstract] [Google Scholar]
- Cáceres J, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. [Abstract] [Google Scholar]
- Chalfant CE, Watson JE, Bisnauth LD, Kang JB, Patel N, Obeid LM, Eichler DC, Cooper DR. Insulin regulates protein kinase CbetaII expression through enhanced exon inclusion in L6 skeletal muscle cells. A novel mechanism of insulin- and insulin-like growth factor-i-induced 5′ splice site selection. J Biol Chem. 1998;273:910–916. [Abstract] [Google Scholar]
- Chen C, Okayama H. High efficiency transformation of plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. [Europe PMC free article] [Abstract] [Google Scholar]
- Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. [Europe PMC free article] [Abstract] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [Europe PMC free article] [Abstract] [Google Scholar]
- Corden, J.L., and Patturajan, M. (1997). A CTD function linking transcription to splicing. Trends Biochem. Sci. 413–419. [Abstract]
- Courty Y, Rosinski CI, Rougeon F. Various transcripts are generated from the VCSA1 gene by alternative splicing and poly(A) processing in the rat submandibular gland. Gene. 1995;162:291–296. [Abstract] [Google Scholar]
- Danoff SK, Ferris CD, Donath C, Fischer GA, Munemitsu S, Ullrich A, Snyder SH, Ross CA. Inositol 1,4,5,-trisphosphate receptors: distinct neuronal and nonneuronal forms derived by alternative splicing differ in phosphorylation. Proc Natl Acad Sci USA. 1991;88:2951–2955. [Europe PMC free article] [Abstract] [Google Scholar]
- Daoud R, Berzaghi M, Siedler F, Hübener M, Stamm S. Activity dependent regulation of alternative splicing patterns in the rat brain. Eur J Neurosci. 1999;11:788–802. [Abstract] [Google Scholar]
- Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci USA. 1996;93:9004–9009. [Europe PMC free article] [Abstract] [Google Scholar]
- Di Carlo M, Montana G, Romancino DP, Monteleone D. A mouse repeat sequence conserved in eukaryotic genomes. J Submicrosc Cytol Pathol. 1992;24:467–472. [Abstract] [Google Scholar]
- Di Fruscio M, Chen T, Richard S. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc Natl Acad Sci USA. 1999;96:2710–2715. [Europe PMC free article] [Abstract] [Google Scholar]
- Domon C, Lorkovic ZJ, Valcarcel J, Filipowicz W. Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J Biol Chem. 1998;273:34603–34610. [Abstract] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. [Europe PMC free article] [Abstract] [Google Scholar]
- Duncan PI, Howell BW, Marius RM, Drmanic S, Douville EM, Bell JC. Alternative splicing of STY, a nuclear dual specificity kinase. J Biol Chem. 1995;270:21524–21531. [Abstract] [Google Scholar]
- Eipper BA, Green CB, Campbell TA, Stoffers DA, Keutmann HT, Mains RE, Ouafik L. Alternative splicing and endoproteolytic processing generate tissue-specific forms of pituitary peptidylglycine alpha-amidating monooxygenase (PAM) J Biol Chem. 1992;267:4008–4015. [Abstract] [Google Scholar]
- Eissa NT, Strauss AJ, Haggerty CM, Choo EK, Chu SC, Moss J. Alternative splicing of human inducible nitric-oxide synthase messenger-RNA—tissue-specific regulation and induction by cytokines. J Biol Chem. 1996;271:27184–27187. [Abstract] [Google Scholar]
- Feramisco J, Smart JE, Burridge K, Helfman DM, Thomas GP. Coexistence of vinculin and a vinculin-like protein of higher molecular weight in smooth muscle. J Biol Chem. 1982;257:11024–11031. [Abstract] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interaction. Nature. 1989;340:245–247. [Abstract] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [Europe PMC free article] [Abstract] [Google Scholar]
- Genetics Computer Group. Program Manual for the Wisconsin Package, version 8. Madison, WI: Genetics Computed Group; 1994. [Google Scholar]
- Giros B, Martres M-P, Pilon C, Sokoloff P, Schwartz J-C. Shorter variants of the D3 dopamine receptor produced through various patterns of alternative splicing. Biochem Biophys Res Commun. 1991;176:1584–1592. [Abstract] [Google Scholar]
- Guiramand J, Montmayeur JP, Ceraline J, Bhatia M, Borrelli E. Alternative splicing of the dopamine D2 receptor directs specificity of coupling to G-proteins. J Biol Chem. 1995;270:7354–7358. [Abstract] [Google Scholar]
- Hartmann AM, Stamm S. Molecular cloning of a novel alternatively spliced nuclear protein. Biochem Biophys Res Commun. 1997;1353:224–230. [Abstract] [Google Scholar]
- Heilig JS, Freeman M, Laverty T, Lee KJ, Campos AR, Rubin GM, Steller H. Isolation and characterization of the disconnected gene of Drosophila melanogaster. EMBO J. 1991;10:809–815. [Europe PMC free article] [Abstract] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. [Abstract] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. [Europe PMC free article] [Abstract] [Google Scholar]
- Hughes DP, Crispe IN. A naturally occurring soluble isoform of murine fas generated by alternative splicing. J Exp Med. 1995;182:1395–1401. [Europe PMC free article] [Abstract] [Google Scholar]
- Imai Y, Matsuo N, Ogawa S, Tohyama M, Takagi T. Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Mol Brain Res. 1998;53:33–40. [Abstract] [Google Scholar]
- Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16:5077–5085. [Europe PMC free article] [Abstract] [Google Scholar]
- Kamma H, Portman DS, Dreyfuss G. Cell-type specific expression of hnRNP proteins. Exp Cell Res. 1995;221:187–196. [Abstract] [Google Scholar]
- Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. [Europe PMC free article] [Abstract] [Google Scholar]
- Krief P, Augery-Bourget Y, Plaisance S, Merck MF, Assier E, Tanchou V, Billard M, Boucheix C, Jasmin C, Azzarone B. A new cytokine (IK) down-regulating HLA class II: monoclonal antibodies, cloning and chromosome localization. Oncogene. 1994;9:3449–3456. [Abstract] [Google Scholar]
- Kuhse J, Kuryatov A, Maulet Y, Malosio ML, Schmieden V, Betz H. Alternative splicing generates two isoforms of the α2 subunit of the inhibitory glycine receptor. FEBS Lett. 1991;283:73–77. [Abstract] [Google Scholar]
- Lennon GG, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. [Abstract] [Google Scholar]
- Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding. J Biol Chem. 1997;272:27274–27280. [Abstract] [Google Scholar]
- Lopez AJ. Developmental role of transcription factor isoform generated by alternative splicing. Dev Biol. 1995;172:396–411. [Abstract] [Google Scholar]
- McBride AE, Taylor SJ, Shalloway D, Kirkegaard K. KH domain integrity is required for wild-type localization of Sam68. Exp Cell Res. 1998;241:84–95. [Abstract] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. [Abstract] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. The dynamics of a premRNA splicing factor in living cells. Nature. 1997;387:523–527. [Abstract] [Google Scholar]
- Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N. Prediction of the coding sequences of unidentified human genes. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1996;3:17–24. [Abstract] [Google Scholar]
- Nayler O, Cap C, Stamm S. Human transformer-2-beta gene: complete nucleotide sequence, chromosomal localization and generation of a tissue specific isoform. Genomics. 1998a;53:191–202. [Abstract] [Google Scholar]
- Nayler O, Schnorrer F, Stamm S, Ullrich A. The cellular localization of the murine serine/arginine-rich protein kinase CLK2 is regulated by serine 141 autophosphorylation. J Biol Chem. 1998b;273:34341–34348. [Abstract] [Google Scholar]
- Nayler O, Stamm S, Ullrich A. Characterization and comparison of four SR protein kinases. Biochem J. 1997;326:693–700. [Europe PMC free article] [Abstract] [Google Scholar]
- Nayler O, Strätling W, Bourquin J-P, Stagljar I, Lindemann L, Jasper H, Hartmann AM, Fackelmeyer FO, Ullrich A, Stamm S. SAF-B couples transcription and premRNA splicing to SAR/MAR elements. Nucleic Acids Res. 1998c;26:3542–3549. [Europe PMC free article] [Abstract] [Google Scholar]
- O’Malley KL, Harmon S, Moffat M, Uhland-Smith A, Wong S. The human aromatic L-amino acid decarboxylase gene can be alternatively spliced to generate unique protein isoforms. J Neurochem. 1995;65:2409–2416. [Abstract] [Google Scholar]
- Ohno M, Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes & Dev. 1996;10:997–1007. [Abstract] [Google Scholar]
- Richard S, Yu D, Blumer KJ, Hausladen D, Olszowy MW, Connelly PA, Shaw AS. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with src family tyrosine kinases, Grb2, and phospholipase C gamma-1. Mol Cell Biol. 1995;15:186–197. [Europe PMC free article] [Abstract] [Google Scholar]
- Roizin L, Stellar S, Lui JC. Neuronal nuclear-cytoplasmatic changes in Huntington’s chorea: electron microscope investigations. Adv Neurol. 1979;23:95–122. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sell SM, Reese D, Ossowski VM. Insulin-inducible changes in insulin receptor mRNA splice variants. J Biol Chem. 1994;269:30769–30772. [Abstract] [Google Scholar]
- Sharma E, Zhao F, Bult A, Lombroso PJ. Identification of two alternatively spliced transcripts of STEP: a subfamily of brain-enriched protein tyrosine phosphatases. Mol Brain Res. 1995;32:87–93. [Abstract] [Google Scholar]
- Skinner PJ, Koshy BT, Cummings CJ, Klement IA, Helin K, Servadio A, Zoghbi HY, Orr HT. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. (erratum [1998] 391, 307). [Abstract] [Google Scholar]
- Sommer B, Keinänen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Köhler M, Takagi T, Sakman B, Seeburg PH. Flip and Flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. [Abstract] [Google Scholar]
- Soulard M, et al. hnRNP G: sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res. 1993;21:4210–4217. [Europe PMC free article] [Abstract] [Google Scholar]
- Spritz RA, Strunk K, Surowy CS, Mohrenweiser HW. Human U1–70K ribonucleoprotein antigen gene: organization, nucleotide sequence, and mapping to locus 19q13.3. Genomics. 1990;8:371–379. [Abstract] [Google Scholar]
- Stamm S, Zhang MQ, Marr TG, Helfman DM. A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res. 1994;22:1515–1526. [Europe PMC free article] [Abstract] [Google Scholar]
- Strohmaier C, Carter BD, Urfer R, Barde Y-A, Dechant G. A splice variant of the neurotrophin receptor trkB with increased specificity for brain-derived neurotrophic factor. EMBO J. 1996;15:3332–3337. [Europe PMC free article] [Abstract] [Google Scholar]
- Stroumbakis ND, Li Z, Tolias PP. A homolog of human transcription factor NF-X1 encoded by the Drosophila shuttle craft gene is required in the embryonic CNS. Mol Cell Biol. 1996;16:192–201. [Europe PMC free article] [Abstract] [Google Scholar]
- Sugimoto Y, Negishi M, Hayashi Y, Namba T, Honda A, Watabe A, Hirata M, Narumiya S, Ichikawa A. Two isoforms of the EP3 receptor with different carboxyl-terminal domains. Identical ligand binding properties and different coupling properties with Gi proteins. J Biol Chem. 1993;268:2712–2718. [Abstract] [Google Scholar]
- Sukegawa J, Blobel G. A putative mammalian RNA helicase with an arginine-serine-rich domain colocalizes with a splicing factor. J Biol Chem. 1995;270:15702–15706. [Abstract] [Google Scholar]
- Surowy CS, Hoganson G, Gosink J, Strunk K, Spritz RA. The human RD protein is closely related to nuclear RNA-binding proteins and has been highly conserved. Gene. 1990;90:299–302. [Abstract] [Google Scholar]
- Suzuki A, Yoshida M, Ozawa E. Mammalian alpha1- and beta1-syntrophin bind to the alternative splice prone region of dystrophin COOH terminus. J Cell Biol. 1995;128:373–381. [Europe PMC free article] [Abstract] [Google Scholar]
- Swaroop M, Bradley K, Ohura T, Tahara T, Roper MD, Rosenberg LE, Kraus JP. Rat cystathionine beta-synthase. Gene organization and alternative splicing. J Biol Chem. 1992;267:11455–11461. [Abstract] [Google Scholar]
- Tabaczewski P, Shirwan H, Lewis K, Stroynowski I. Alternative splicing of class Ib major histocompatibility complex transcripts in vivo leads to the expression of soluble Qa-2 molecules in murine blood. Proc Natl Acad Sci USA. 1994;91:1883–1887. [Europe PMC free article] [Abstract] [Google Scholar]
- Tabiti K, Cui L, Chhatwal VJS, Moochhala S, Ngoi SS, Pallen CJ. Novel alternative splicing predicts a secreted extracellular isoform of the human receptor-like protein-tyrosine-phosphatase lar. Gene. 1996;175:7–13. [Abstract] [Google Scholar]
- Taylor SJ, Anafi M, Pawson T, Shalloway D. Functional interaction between c-src and its mitotic target, Sam68. J Biol Chem. 1995;270:10120–10124. [Abstract] [Google Scholar]
- Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. [Abstract] [Google Scholar]
- Toksoz D, Zsebo KM, Smith KA, Hu S, Brankow D, Suggs SV, Martin FH, Williams DA. Support of human hematopoiesis in long-term bone marrow cultures by murine stromal cells selectively expressing the membrane-bound and secreted forms of the human homolog of the steel gene product, stem cell factor. Proc Natl Acad Sci USA. 1992;89:7350–7354. [Europe PMC free article] [Abstract] [Google Scholar]
- van der Logt CP, Reitsma PH, Bertina RM. Alternative splicing is responsible for the presence of two tissue factor mRNA species in LPS stimulated human monocytes. Thromb Hemost. 1992;67:272–276. [Abstract] [Google Scholar]
- Vernet C, Artzt K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 1997;13:479–484. [Abstract] [Google Scholar]
- Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [Europe PMC free article] [Abstract] [Google Scholar]
- Wang J, Ross EM. The carboxyl-terminal anchorage domain of the turkey beta 1-adrenergic receptor is encoded by an alternatively spliced exon. J Biol Chem. 1995;270:6488–6495. [Abstract] [Google Scholar]
- Wang LL, Richard S, Shaw AS. p62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–2013. [Abstract] [Google Scholar]
- Wilson R, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. [Abstract] [Google Scholar]
- Xing S, Tong Q, Suzuki T, Jhiang SM. Alternative splicing of the ret proto-oncogene at intron 4. Biochem Biophys Res Commun. 1994;205:1526–1532. [Abstract] [Google Scholar]
- Zhang K, Max EE, Cheah H-K, Saxon A. Complex alternative RNA splicing of epsilon-immunoglobulin transcripts produces mRNAs encoding four potential seccreted protein isoforms. J Biol Chem. 1994;269:456–462. [Abstract] [Google Scholar]
Articles from Molecular Biology of the Cell are provided here courtesy of American Society for Cell Biology
Full text links
Read article at publisher's site: https://doi.org/10.1091/mbc.10.11.3909
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc25688?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
The three YTHDF paralogs and VIRMA are strong cross-histotype tumor driver candidates among m<sup>6</sup>A core genes.
NAR Cancer, 6(4):zcae040, 15 Oct 2024
Cited by: 0 articles | PMID: 39411658 | PMCID: PMC11474903
Reading the m6A-encoded epitranscriptomic information in development and diseases.
Cell Biosci, 14(1):124, 28 Sep 2024
Cited by: 0 articles | PMID: 39342406 | PMCID: PMC11439334
Review Free full text in Europe PMC
The m6A regulators in prostate cancer: molecular basis and clinical perspective.
Front Pharmacol, 15:1448872, 29 Aug 2024
Cited by: 0 articles | PMID: 39268470 | PMCID: PMC11391310
Review Free full text in Europe PMC
The YTH domain-containing protein family: Emerging players in immunomodulation and tumour immunotherapy targets.
Clin Transl Med, 14(8):e1784, 01 Aug 2024
Cited by: 1 article | PMID: 39135292 | PMCID: PMC11319238
Review Free full text in Europe PMC
Regulatory roles of N6-methyladenosine (m<sup>6</sup>A) methylation in RNA processing and non-communicable diseases.
Cancer Gene Ther, 31(10):1439-1453, 05 Jun 2024
Cited by: 1 article | PMID: 38839892
Review
Go to all (126) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (4)
- (1 citation) ENA - AA183061
- (1 citation) ENA - AA646154
- (1 citation) ENA - AF144731
- (1 citation) ENA - U78721
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
p59(fyn)-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1.
Mol Cell Neurosci, 27(1):8-21, 01 Sep 2004
Cited by: 32 articles | PMID: 15345239
The intranuclear localization and function of YT521-B is regulated by tyrosine phosphorylation.
Hum Mol Genet, 13(15):1535-1549, 02 Jun 2004
Cited by: 38 articles | PMID: 15175272
Fyn membrane localization is necessary to induce the constitutive tyrosine phosphorylation of Sam68 in the nucleus of T lymphocytes.
J Immunol, 162(12):7224-7232, 01 Jun 1999
Cited by: 9 articles | PMID: 10358169
SAM68: Signal Transduction and RNA Metabolism in Human Cancer.
Biomed Res Int, 2015:528954, 26 Jul 2015
Cited by: 55 articles | PMID: 26273626 | PMCID: PMC4529925
Review Free full text in Europe PMC





