Abstract
Free full text

Formation and Characterization of the Trimeric Form of the Fusion Protein of Semliki Forest Virus
Abstract
Enveloped animal viruses infect cells via fusion of the viral membrane with a host cell membrane. Fusion is mediated by a viral envelope glycoprotein, which for a number of enveloped animal viruses rearranges itself during fusion to form a trimeric α-helical coiled-coil structure. This conformational change from the metastable, nonfusogenic form of the spike protein to the highly stable form involved in fusion can be induced by physiological activators of virus fusion and also by a variety of destabilizing conditions. The E1 spike protein subunit of Semliki Forest virus (SFV) triggers membrane fusion upon exposure to mildly acidic pH and forms a homotrimer that appears necessary for fusion. We have here demonstrated that formation of the E1 homotrimer was efficiently triggered under low-pH conditions but not by perturbants such as heat or urea, despite their induction of generalized conformational changes in the E1 and E2 subunits and partial exposure of an acid-specific E1 epitope. We used a sensitive fluorescence assay to show that neither heat nor urea treatment triggered SFV-liposome fusion at neutral pH, although either treatment inactivated subsequent low-pH-triggered fusion activity. Once formed, the low-pH-induced E1 homotrimer was very stable and was only dissociated under harsh conditions such as heating in sodium dodecyl sulfate. Taken together, these data, as well as protein structure predictions, suggest a model in which the less stable native E1 subunit specifically responds to low pH to form the more stable E1 homotrimer via conformational changes different from those of the coiled-coil type of fusion proteins.
Enveloped viruses must carry out a fusion event between the viral membrane and the host cell membrane to deliver the infectious genome to the cytoplasm. For viruses such as human immunodeficiency virus type 1, fusion occurs at the cell surface in a pH-independent fashion whereas viruses such as Semliki Forest virus (SFV), a member of the alphavirus family, or influenza virus, a myxovirus, fuse via a low-pH-dependent reaction in the endosome. Each of these viruses has integral membrane spike proteins incorporated in the virus envelope that drive membrane fusion in response to the appropriate environmental stimulus, such as receptor binding or a pH change. Proper regulation of the fusogenic properties of these viral spike proteins during virus biosynthesis and maturation is critical to successful virus production and infection of cells. The necessary conformational changes of the proteins that carry out the fusion events have been extensively studied for viruses such as influenza virus and SFV.
The influenza virus hemagglutinin (HA) is emblematic of a number of virus fusion proteins with common features. The biosynthetic precursor of HA is a trimer termed HA0 that is activated by proteolytic cleavage to produce a trimer of two disulfide-linked subunits, the receptor-binding HA1 polypeptide and the transmembrane HA2 polypeptide (reviewed in references 26, 61, and 62). Exposure to low pH triggers a conformational change in which a loop region of HA2 refolds to form an extended trimeric α-helical coiled coil, thus translocating the N-terminal HA2 fusion peptide to the top of the molecule, where it can insert itself into the target membrane. A more C-terminal helical region folds back to pack in an antiparallel fashion against the grooves of the coiled-coil trimer, resulting in a six-helix bundle with the HA2 transmembrane domain and fusion peptide at the same end (10). A number of studies suggest that the neutral-pH, nonfusogenic conformation of HA is metastable and that conversion to the fusogenic conformation results in a thermodynamically more stable molecule. (i) The neutral-pH form melts at a significantly lower temperature than the low-pH form, even when assayed at neutral pH (16, 47). (ii) The six-helix bundle is formed spontaneously when a soluble form of HA2 lacking HA1 is expressed in bacteria (15, 16). (iii) The conformational change from the native to the low-pH structure is irreversible (51). (iv) Peptides comprising the loop region of HA2 fold into a low-energy α-helical trimer in solution (12). (v) Both conversion to the low-pH conformation and virus-membrane fusion can be triggered at neutral pH by treatment with heat or the denaturant urea (11, 48). (vi) A series of HA mutants that fuse at less acidic pH also show conformational changes and fusion in response to treatments at lower temperatures, although once formed, the fusogenic conformations of the wild type (wt) and mutants have identical melting curves (48). This last fact demonstrates that destabilization of the metastable native HA in the forward induction pathway is important in regulating the conformational changes, rather than stabilization of the final low-pH conformation of HA. Thus, during biosynthesis or maturation, the influenza virus HA appears to adopt a metastable conformation that is primed to undergo conversion to the more stable fusogenic form. Members of the families Paramyxoviridae, Filoviridae, and Retroviridae, including human immunodeficiency virus type 1, utilize fusion mechanisms which, similar to that of influenza virus, are based on proteolytic activation of a fusion protein precursor and formation of a highly stable structure containing a central coiled-coil trimer and associated antiparallel helices (reviewed in references 6, 27, and 52). Indeed, a very recent paper demonstrated that fusion of the paramyxovirus Sendai virus could similarly be triggered by elevated temperature, again suggesting a transition from a metastable to a more stable form during fusion (59).
SFV fusion is mediated by the E1 spike subunit, a transmembrane glycoprotein of about 50 kDa which binds target membranes at low pH and contains the putative fusion peptide (reviewed in references 21, 32, and 53). During synthesis in the endoplasmic reticulum (ER), E1 associates in a heterodimer with the transmembrane p62 spike subunit and the two proteins traffic together to the plasma membrane. Late in the biosynthetic pathway, p62 is proteolytically processed to the E2 and E3 subunits and the mature virus spike contains trimers of E1/E2 heterodimers. Cleavage of the p62 subunit activates the E1 subunit for fusion at the physiological pH of the endosome, but in contrast to viruses of the influenza type, no processing of the fusogenic E1 subunit occurs. Upon exposure to low pH, the SFV spike protein undergoes a series of very rapid conformational changes, including dissociation of E1/E2 heterodimers (49), exposure of epitopes on E1 (1, 35, 57), and formation of a trypsin-resistant E1 homotrimer (9, 34, 57) that appears to be essential for fusion (36). SFV fusion requires the presence of the specific lipids cholesterol and sphingolipid in the target membrane (33, 63), and these lipids promote rapid and efficient E1 conformational changes and homotrimer formation at low pH (13, 17). Studies of the E1 ectodomain indicate that even in the absence of association with the regulatory E2 subunit, the E1 subunit still requires low-pH treatment and interaction with cholesterol- and sphingolipid-containing target membranes to form stable homotrimers (39). SFV fusion is thus similar to that of influenza virus in its low-pH dependence and involvement of a trimeric fusion protein structure but differs in its requirements for proteolytic cleavage and specific lipids.
In order to determine if SFV fusion, like that of influenza virus, involves a similar conversion from a metastable intermediate, we have here examined the requirements for E1 homotrimer formation and the stability of the homotrimer structure. Our studies showed that heat or urea treatment did not trigger E1 homotrimerization or SFV fusion, although they did significantly alter the structure of the spike protein and inactivate virus fusion and infectivity. The E1 homotrimer formed at low pH was highly resistant to subsequent treatment with protease, elevated temperature, or urea, indicative of its increased stability relative to that of native E1. Taken together, these data suggest that in the virus particle, E1 is in a metastable conformation that can irreversibly and rapidly respond to low pH but that the regulation of the fusogenic response is more complex than that of influenza virus.
(This research was conducted by D.L.G. in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University, New York, N.Y., 2000.)
MATERIALS AND METHODS
Virus, cells, and antibodies.
BHK-21 cells were cultured at 37°C in Dulbecco's modified Eagle medium containing 5% fetal calf serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 10% tryptose phosphate broth (43). The SFV used in these experiments was a well-characterized, plaque-purified isolate (22) and was propagated in BHK-21 cells. Virus was radiolabeled with [35S]methionine and [35S]cysteine and purified as previously described (38) or propagated in the absence of label and purified by banding on tartrate gradients (31). Pyrene-labeled virus was prepared as previously described by growth on BHK-21 cells metabolically labeled with [16C]pyrene (Molecular Probes, Eugene, Oreg.) and purified by centrifugation on a discontinuous sucrose gradient (13). The soluble ectodomain forms of the E1 and E2 spike protein subunits were prepared by protease digestion of [35S]methionine-labeled SFV and purified, unlabeled SFV and purified by concanavalin A chromatography, all as previously described (34, 39).
The E1 acid-conformation-specific monoclonal antibody (MAb) E1a-1 was previously isolated and characterized in our laboratory (1, 35). Monoclonal antibody anti-E1" was obtained as a concentrated ascites preparation from Harm Snippe and has been previously characterized for its reactivity to the low-pH conformation of E1 (1, 39, 57). Immunoprecipitation and gel electrophoresis were performed as previously described (1, 35).
Liposomes.
Liposomes were prepared by extrusion as previously described (13) with phosphatidylcholine (from egg yolk)-phosphatidylethanolamine (derived from egg phosphatidylcholine by transphosphatidylation)-sphingomyelin (bovine brain)-cholesterol at a molar ratio of 1:1:1:1.5 for experiments with virus (“complete” liposomes) (37). For experiments with ectodomains, equimolar amounts of phospholipids and cholesterol were incorporated into the complete liposomes, yielding a molar ratio of 1:1:1:3 (39). Liposomes without cholesterol or without sphingomyelin were prepared similarly, except with omission of either or both lipids. All phospholipids were purchased from Avanti Polar Lipids (Alabaster, Ala.), and cholesterol was from Steraloids (Wilton, N.H.). For assays, liposomes were mixed with ectodomains or SFV at a final concentration of 1 or 0.8 mM, respectively.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) assay of E1 homotrimer formation.
[35S]methionine-cysteine-labeled virus or ectodomains were preincubated with liposomes for 5 min at 20 or 37°C (as indicated), adjusted to pH 5.5 by addition of a precalibrated volume of 0.5 N acetic acid, incubated further as indicated in each figure legend, and then neutralized by addition of 0.5 N NaOH. Analysis on 11% acrylamide gels was performed after samples had been heated in SDS-sample buffer (200 mM Tris [pH 8.8], 4% SDS, 10% glycerol, 0.02% bromophenol blue) at 30°C for 3 min (57), except where indicated in the figure legends. For heat or urea experiments, a mixture of [35S]methionine-cysteine-labeled virus (or ectodomains) and liposomes was heated for 10 min at the indicated temperature or urea treated for the indicated time by addition of an 8 M stock solution in MES-saline buffer (20 mM morpholineethanesulfonic acid [MES; pH 7.0], 130 mM NaCl). Quantitation of spike protein radioactivity was performed by PhosphorImager analysis with ImageQuant v.1.2 software (Molecular Dynamics, Sunnyvale, Calif.).
Trypsin digestion assay of E1 homotrimer formation.
After heat, urea, or acid treatment, samples were incubated with trypsin (type XIII; Sigma Chemical Co., St. Louis, Mo.) at 200 μg/ml in 1% Triton X-100 for 10 min at the indicated temperatures. The digestion was stopped by addition of soybean trypsin inhibitor (type I-S; Sigma) to a final concentration of 400 μg/ml or by addition of phenylmethylsulfonyl fluoride to a final concentration of 5 mM. For the untreated samples, premixed trypsin and inhibitor were added. After digestion, the samples were analyzed by SDS-PAGE either directly or following precipitation with 10% trichloroacetic acid.
Fusion assay.
Lipid mixing during SFV-liposome fusion was assayed by monitoring the decrease in virus pyrene excimer fluorescence (9, 13). Each assay mixture (2 ml) contained purified pyrene-labeled virus (0.6 μM phospholipids as calculated from a phospholipid/protein ratio of 0.45 μmol/mg) and 200 μM liposomes of the indicated lipid composition. The mixtures were stirred continuously in a thermostat-equipped cuvette at the indicated temperature. Fusion was triggered by the addition of a pretitrated volume of 0.3 M MES (pH 4.8) to yield a final pH of 5.5 or by addition of urea (buffered in 5 mM HEPES [pH 7.0]–150 mM NaCl–0.1 mM EDTA) to the indicated final concentration. Pyrene excimer fluorescence was measured in an SLM-8000 fluorometer upgraded to SLM-8100 software (SLM-Aminco, Urbana, Ill.) using excitation and emission wavelengths of 343 and 480 nm, respectively, in the presence of a 470-nm-cutoff filter in the emission beam. For the heat induction experiments, the complete assay mixtures were incubated at the indicated temperatures for 15 min and then cooled to 37°C, and the fluorescence of each sample was measured. Percent fusion was calculated for each sample using the initial excimer fluorescence as 0% and setting 100% fusion as the fluorescence obtained after the addition of the detergent C12E8 to a 10 mM final concentration.
Computer-based protein sequence analysis.
For secondary-structure predictions, the PSIPred, NNPredict, and PHD programs were utilized by submitting the sequence of SFV E1 to servers running the programs over the World Wide Web (3, 29, 40, 45, 46). To search for sequences with the probability of forming α-helical coiled coils, four different programs were used, Coils, Paircoil, Multicoil, and LearnCoil-VMF, each run by submitting the sequence to a World Wide Web server hosting the program (8, 42, 50, 64). For each program, a probability of 0.5 was used as the cutoff value in searching for coiled-coil regions.
RESULTS
Based on recent work, it is clear that viruses from diverse families use a common fusion mechanism involving an α-helical coiled-coil motif. To evaluate if SFV might use similar structural elements to form the E1 homotrimer, a variety of computer programs for predicting secondary structure (PHD, NNPredict, and PSIpred) or for predicting the probability of coiled-coil formation (PairCoil, MultiCoil, Coils, and LearnCoil-VMF) were used to analyze the sequence of SFV E1. Although secondary-structure analysis did indicate two or three probable α helices of 8 to 20 residues in addition to the transmembrane segment, the predominant motif in E1 was predicted to be β sheet. Additionally, none of the programs predicted any coiled-coil regions using any of the accepted window sizes or positional weightings. Using the Coils program with a window of 14, the regions predicted to form α helices show some very low probability of coiled-coil formation but at values significantly below the 0.5 probability normally assigned as the cutoff for an accurate prediction. The inability of these programs to identify any regions of the E1 protein likely to form an α-helical coiled coil is in agreement with previous examinations of SFV and alphavirus E1 (39, 50).
Taken together, sequence analyses thus suggested that the SFV E1 protein did not use an α-helical coiled-coil-based fusion mechanism and therefore that the properties of E1 conversion to the fusion-active state may differ from those of the virus spike proteins exemplified by influenza virus HA. We set out to determine if the fusogenic conformational changes in SFV E1 could be induced in response to general perturbants such as temperature and urea and if such destabilizing treatments could trigger virus-membrane fusion activity. Previous work has demonstrated that E1 homotrimer formation and acid-epitope exposure occur in response to low pH either in vivo or in vitro and that these conformational changes are irreversible (30, 35, 56, 57). Although both epitope exposure and trimerization are triggered regardless of target membrane presence or composition, the most efficient E1 conversion occurs in the presence of fusion-active target membranes containing cholesterol and sphingolipid (13, 17). We therefore included such liposomes in our assays.
Effect of elevated temperature on the SFV E1 subunit.
Mixtures of purified, radiolabeled SFV and fusion-competent liposomes containing cholesterol and sphingolipid were treated at temperatures ranging from 37 to 65°C for 10 min. E1 homotrimer formation was assayed by SDS-PAGE, taking advantage of the resistance of the homotrimer to dissociation when solubilized in SDS-PAGE sample buffer at 30°C (56). Although the control sample treated at pH 5.5 formed a substantial amount of the homotrimer, no homotrimer was present in any of the samples that were maintained at neutral pH at the indicated temperatures (Fig. (Fig.1A)1A) or at temperatures as high as 75°C (data not shown).
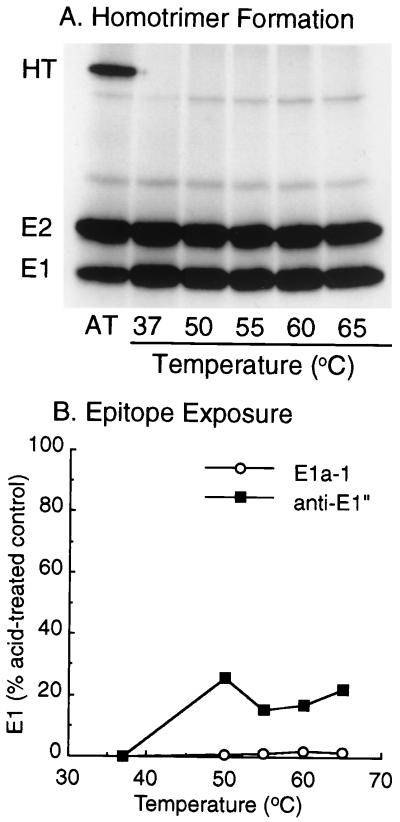
Conformational changes following heat treatment. [35S]methionine- and [35S]cysteine-labeled virus was acid treated at pH 5.5 and 37°C for 5 min (AT) or heat treated at the indicated temperatures for 10 min in the presence of 0.8 mM complete liposomes. (A) Samples were analyzed for homotrimer (HT) formation by incubation at 30°C for 3 min in SDS-sample buffer, followed by SDS-PAGE. (B) Samples were immunoprecipitated with MAb E1a-1 or anti-E1" and analyzed by SDS-PAGE and phosphorimaging, and the data are presented as the ratio of the amount of E1 precipitated after heat treatment divided by the amount of E1 precipitated after acid treatment. A and B are representative examples of two experiments each.
Both MAbs E1a-1 and anti-E1" recognize epitopes that are hidden in the neutral-pH conformation of E1 and rapidly exposed upon acid-pH treatment with kinetics slightly faster than those of virus-membrane fusion (1, 9, 35, 57). The binding site recognized by MAb E1a-1 was recently localized to the region around E1 residue R157, while the epitope for anti-E1" has not been mapped but is spatially related to the E1a-1 site (1). Although the MAbs recognize the same acid-triggered pool of E1 that forms the homotrimer, epitope exposure appears to involve a conformational change(s) distinct from trimerization (36). The induction of epitope exposure by elevated temperature was therefore tested. Radiolabeled virus-liposome mixtures were either treated at acid pH or incubated at various temperatures for 10 min at neutral pH, and the samples were immunoprecipitated with either MAb E1a-1 or anti-E1" (Fig. (Fig.1B).1B). As shown previously (1), both MAbs reacted efficiently with the low-pH-treated virus. By comparison, the reactivity of either MAb with heat-treated E1 was minimal, although in samples incubated at 50°C and higher temperatures, the anti-E1" reactivity approached 15 to 20% of that of acid-treated E1. This difference between the MAbs is in keeping with their recognition of overlapping but nonidentical epitopes.
Effect of urea treatment on the SFV E1 subunit.
Urea was used as a general protein denaturant to destabilize the native structure of E1, and its ability to induce the E1 homotrimer was evaluated. Radiolabeled virus-liposome mixtures were treated with the indicated urea concentrations for 30 min at 37°C and assayed for homotrimer formation by the SDS-PAGE assay (Fig. (Fig.2A).2A). No homotrimer resulted at neutral pH at any urea concentration, while the acid-treated control showed efficient homotrimer production. Similar lack of homotrimer induction was observed with samples treated overnight at room temperature with concentrations of 1.0 to 6.5 M urea (data not shown).

Conformational changes following urea treatment. Radiolabeled virus was mixed with 0.8 mM complete liposomes and either treated at pH 5.5 for 5 min at 37°C (AT) or incubated with the indicated concentrations of urea for 30 min at 37°C. Samples were analyzed for homotrimer (HT) formation (A) and MAb precipitation (B) as described in the legend to Fig. Fig.1.1. A and B are representative examples of two experiments each.
To assay for exposure of acid-specific MAb epitopes, radiolabeled virus-liposome mixtures were treated with the indicated urea concentrations for 30 min at 37°C, the urea concentration was diluted to 1 M, and immunoprecipitation was assessed. Controls showed that the efficiency of immunoprecipitation of acid-treated E1 by either MAb was unaffected by the presence of 1 M urea (data not shown). Only a small amount of exposure of the MAb E1a-1 epitope was induced by urea treatment, about 5 to 10% of that achieved in the acid-treated control (Fig. (Fig.2B).2B). In contrast, anti-E1" immunoprecipitated significant E1 after treatment with urea concentrations as low as 1 M, and after treatment with 5 M urea, the efficiency of E1 reactivity was almost 100% of that induced by control acid treatment. Similar results for both MAbs were obtained if the urea preincubation was carried out overnight (data not shown).
Given that the E1/E2 heterodimer must dissociate in order for the E1 subunit to undergo further conformational changes and form a homotrimer (22, 49), we assayed whether E1 ectodomains, which are already completely dissociated from the complex with E2 (34, 39), could trimerize in response to urea. Similar to the results shown for virus, E1 ectodomains did not form any measurable homotrimer after urea treatment although control samples showed efficient trimerization at acid pH (data not shown).
Spike protein trypsin sensitivity after heat or urea treatment.
The trypsin sensitivity of the SFV spike protein has been extensively used to assay its low-pH-dependent conformational changes (9, 34, 36, 57). The native E1 subunit is fully digested with trypsin at 37°C, while the low-pH-induced E1 homotrimer is resistant to digestion. Native E2 is resistant to mild trypsin digestion at 0°C, while low-pH-treated E2 is acutely sensitive to trypsin digestion. This differential protease sensitivity was used to assay the conformation of the spike protein following acid, heat, or urea treatment and compared with the trypsin profile of the native, untreated spike protein. Samples were digested with trypsin for 10 min at 0 or 37°C, precipitated with trichloroacetic acid, and analyzed by SDS-PAGE (Fig. (Fig.3).3). In agreement with previous results, E2 converted from relative trypsin resistance in the untreated virus sample to relative trypsin sensitivity in the acid-treated sample (compare 0°C lanes). The E1 subunit, in contrast, converted from the relative trypsin sensitivity of the native protein to the relative trypsin resistance of the homotrimeric acid-induced form (compare 37°C lanes). Capsid protein was completely digested under all conditions, demonstrating that active trypsin was present. Similar trypsin digestion experiments were performed after heating the virus for 10 min at 55°C or treating it with 4 M urea for 30 min at 37°C. No trypsin-resistant E1 was observed in response to heat or urea treatment, in keeping with the absence of homotrimer induction observed by mobility on SDS-polyacrylamide gels. Despite the lack of homotrimer formation, both the heat and urea treatments did induce conformational changes in the E1 and E2 subunits, as determined by their dramatically increased trypsin sensitivities. The heat- and urea-treated spike proteins were very sensitive to trypsin even when the protease treatment was performed at 0°C, while the E1 and E2 subunits in both samples were completely digested by trypsin at 37°C.
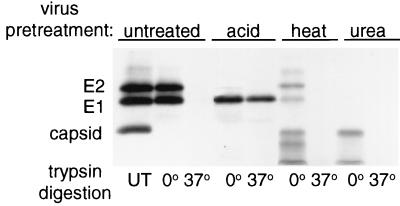
Trypsin digestion of virus after heat or urea treatment. In the presence of 0.8 mM complete liposomes, radiolabeled virus was untreated (UT), acid treated at pH 5 for 5 min at 37°C, heat treated for 10 min at 55°C, or incubated in 4 M urea for 30 min at 37°C. After each of the various treatments, virus was incubated with trypsin (200 μg/ml) and 1% Triton X-100 for 10 min at either 0°C (on ice) or 37°C and the digestion was stopped by addition of soybean trypsin inhibitor (400 μg/ml). Samples were trichloroacetic acid precipitated and analyzed by SDS-PAGE after boiling in sample buffer. The gel shown is representative of two experiments.
As a further test of the effect of urea on the conformation of native E1, virus or purified ectodomains were pretreated with 0 to 5 M urea for 30 min at 37°C in the presence of liposomes. The ability of this pretreated E1 to form homotrimers in response to subsequent treatment at acid pH was assayed (data not shown). Trimerization of viral E1 was unaffected by pretreatment with 1 M urea but showed ~93% inhibition after pretreatment with 2 M urea and 100% inhibition at higher concentrations. Trimerization of the E1 ectodomain was even more sensitive to urea pretreatment, with ~85% inactivation after treatment with 1 M urea.
The overall spike protein trypsin sensitivity after treatment with urea or elevated temperature correlated with the exposure of the epitope recognized by MAb anti-E". Heating exposed ~20% of the anti-E1" epitopes and made both E1 and E2 more susceptible to trypsin proteolysis at 0°C. Urea treatment exposed ~85% of the anti-E1" epitope sites and made E1 and E2 completely susceptible to trypsin digestion at 0°C. Taken together, these data demonstrate that heat or urea treatment produced substantial changes in the native structures of E1 and E2 but failed to induce formation of the E1 homotrimer as assayed by gel mobility or trypsin resistance.
Effect of heating or urea treatment on SFV fusion.
The kinetics of SFV E1 trimerization and its specific promotion by cholesterol and sphingolipid argue strongly that the homotrimer is a critical intermediate in the SFV fusion reaction (9, 13, 39, 49). Importantly, a mutation in the SFV fusion peptide that completely blocks virus fusion and infection also completely inhibits homotrimer formation while the initial response of the mutant spike protein to acid is largely unaffected (36). Thus, the lack of homotrimer induction by heat or urea treatment strongly suggested that unlike that of influenza virus, SFV fusion is not inducible by general destabilizing treatments. However, it was possible that denaturant-treated virus would fuse using a different mechanism. As an unbiased test for the induction of the fusion-active spike protein conformation, we employed a sensitive real-time fluorescence assay that follows the fusion of pyrene-labeled virus with unlabeled target liposomes. This assay has been utilized extensively to study the kinetics of SFV fusion (9, 56) and to characterize the fusion properties of a cholesterol-independent mutant (13). As expected, SFV fused very rapidly with target liposomes containing cholesterol and sphingolipid when treated at pH 5.5, as detected by the decrease of the pyrene excimer peak (Fig. (Fig.4A,4A, curve 4). The final fusion extent was about 75%, compared to complete pyrene dilution by detergent solubilization. This fusion was dependent on the inclusion of cholesterol and sphingolipid in the target membrane (compare to Fig. Fig.4B,4B, curve 4) and on low-pH treatment (compare to Fig. Fig.4A,4A, curve 1). Treatment with 3 or 5 M urea did not trigger significant levels of fusion with either complete or cholesterol- and sphingolipid-deficient liposomes (Fig. (Fig.4A4A and B, curves 2 and 3). Thus, the lack of homotrimer induction by urea treatment correlated fully with the absence of virus-membrane fusion. To assay the effects of urea on the fusion potential of the spike protein, pyrene-labeled virus was pretreated with 3 or 5 M urea and then adjusted to pH 5.5 in the presence of liposomes (data not shown). Such urea-pretreated virus samples were now incapable of acid-triggered fusion activity.
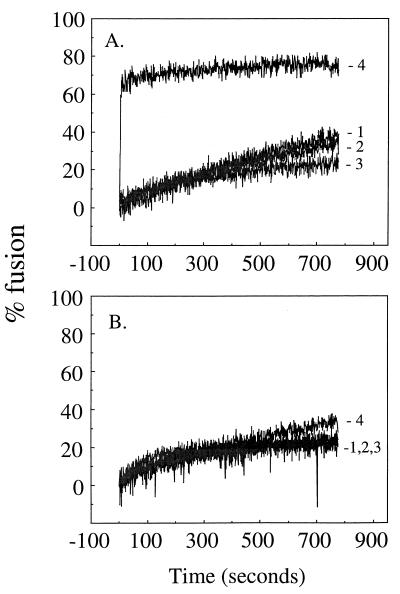
Effect of urea on SFV membrane fusion. Real-time fluorescence recordings of the fusion of pyrene-labeled wt SFV (0.6 μM phospholipid) with unlabeled complete liposomes (200 μM phospholipid) (A) or unlabeled, cholesterol- and sphingolipid-free liposomes (200 μM phospholipid) (B) at 37°C. Acid or urea, as indicated, was added to the reaction mixtures at time zero. Conditions: 1, pH 7.0 without urea; 2, pH 7.0 with 3 M urea; 3, pH 7.0 with 5 M urea; 4, pH 5.5 without urea. After 13 min of incubation, the detergent C12E8 (10 mM final concentration) was added to each reaction mixture to give complete pyrene dilution, defined as 100% fusion. The data shown are representative of three experiments.
To test heat induction of fusion, pyrene-labeled SFV was mixed with cholesterol- and sphingolipid-containing liposomes and incubated at 37, 50, or 65°C for 15 min, and the decrease in the excimer peak was determined. Virus-liposome mixtures that were treated at pH 5.5 at 37°C for 90 s showed ~85% fusion (Fig. (Fig.5,5, first bar), while a negligible fluorescence decrease was induced by heat treatment (Fig. (Fig.5,5, last three bars). Virus-liposome mixtures were also preincubated for 15 min at 37 or 50°C and neutral pH, and the effect of pretreatment on subsequent acid-induced fusion was evaluated. While pretreatment at 37°C was without effect (Fig. (Fig.5,5, bar 2), preincubation at 50°C inactivated virus fusion capacity (Fig. (Fig.5,5, bar 3). Assays of virus infectivity demonstrated that a 15-min preincubation at 52°C inactivated 99% of the viral plaque-forming activity (data not shown). Thus, the lack of urea- or heat-induced fusion and homotrimer formation suggests that only low-pH treatment is capable of inducing the fusion-competent E1 homotrimer. Both heat and urea treatments inactivated virus fusion capacity but appeared to do so by general disruption of the structure of E1 rather than by causing premature triggering of the fusogenic conformation of E1.
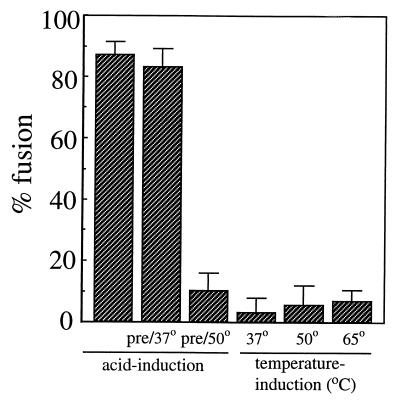
Effect of heat treatment on SFV membrane fusion. Pyrene-labeled wt SFV (0.6 μM) was mixed with unlabeled complete liposomes (200 μM), and the initial excimer fluorescence was determined. For temperature induction of fusion, samples were then incubated at the indicated temperatures for 15 min at pH 7.0 and returned to 37°C and the decrease in the initial pyrene fluorescence was measured. C12E8 detergent (10 mM) was then added to give complete pyrene dilution, and the amount of fusion during temperature induction was calculated using 0% as the initial excimer fluorescence and 100% as the fluorescence after addition of the detergent. For acid induction of fusion, the virus-liposome mixture was pretreated at the indicated temperatures for 15 min at pH 7.0 and then incubated at pH 5.5 for 1.5 min at 37°C. The amount of fusion induced by acid pH was calculated as for the temperature induction samples. Average data and standard deviations of three experiments are shown, except for the 65°C condition (n = 5).
Stability of the E1 homotrimer.
Our data indicated that the native conformation of E1 was quite susceptible to denaturation by heat or urea treatment, resulting in altered E1 trypsin sensitivity (Fig. (Fig.3).3). To better understand the nature of the low-pH conformation of E1, the effects of urea or heat treatment on the E1 homotrimer were evaluated. The homotrimer was preformed by incubating radiolabeled virus with complete liposomes for 3 min at pH 5.5 and 20°C, conditions that give optimal trimer induction and little acid inactivation (9, 13). The virus-liposome mixture was then adjusted to neutral pH, incubated with various concentrations of urea for 30 min at 37°C, digested with trypsin for 3 h at 37°C, and analyzed by SDS-PAGE under conditions that would preserve any remaining homotrimer (Fig. (Fig.6).6). Under these conditions, the stable E1 homotrimer should both display trypsin resistance and migrate in the trimer position in SDS-PAGE. Control samples demonstrated that the homotrimer was efficiently formed during the pretreatment at low pH (Fig. (Fig.6,6, lane C). As observed before, the homotrimer was highly resistant to trypsin digestion while the capsid, E2, and untrimerized E1 proteins were completely degraded (0 M urea lane). The homotrimer structure was stably maintained even after treatment with 5 M urea (1 to 5 M urea lanes), demonstrating the inability of urea to disassemble the trimer or even to destabilize its structure sufficiently to allow partial proteolysis by trypsin. This trypsin resistance was not due to effects on the activity of trypsin, since the other viral proteins were still efficiently digested. Similar trimer stability was observed if the urea treatment was performed overnight or if the samples were assayed directly by SDS-PAGE without trypsin digestion (data not shown). Similar experiments performed on the E1 homotrimer from an SFV mutant with reduced cholesterol dependence (54) showed urea resistance comparable to that of the wt homotrimer (data not shown). This agrees with our earlier finding that although the induction of the mutant homotrimer is less dependent on the presence of cholesterol in the target membrane, once induced, the mutant fusion reaction mechanism appears similar to that of wt SFV (13).
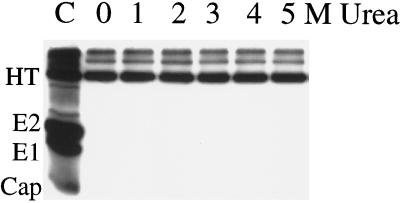
Stability of the E1 homotrimer upon urea treatment. Radiolabeled virus was mixed with complete liposomes (0.8 mM) and acid treated at pH 5.5 for 3 min at 20°C to form the E1 homotrimer. Samples were then incubated as indicated in various concentrations of urea (0 to 5 M) for 30 min at 37°C and digested with trypsin (125 μg/ml) in 0.5% Triton X-100 for 3 h at 37°C. The digestion was stopped by addition of phenylmethylsulfonyl fluoride to a final concentration of 5 mM, and the samples were incubated in SDS-sample buffer at 30°C for 3 min and subjected to SDS-PAGE. C, control undigested reaction mixture showing the E1 homotrimer (HT) and monomeric E1, E2, and capsid proteins. The gel shown is a representative example of three experiments.
Similar experiments were performed to assess the stability of the wt homotrimer when incubated at elevated temperatures. The preformed homotrimer was heated at temperatures of 40 to 65°C for 15 min in a neutral-pH buffer containing 0.5% Triton X-100 to prevent protein aggregation. The samples were then assayed for the presence of the homotrimer by SDS-PAGE. The homotrimer was highly resistant to dissociation by heating to temperatures of up to 65°C (Fig. (Fig.7,7, filled squares). Such heat-treated samples were also digested with trypsin for 1 h at 37°C, and the homotrimer remained completely trypsin resistant (data not shown).
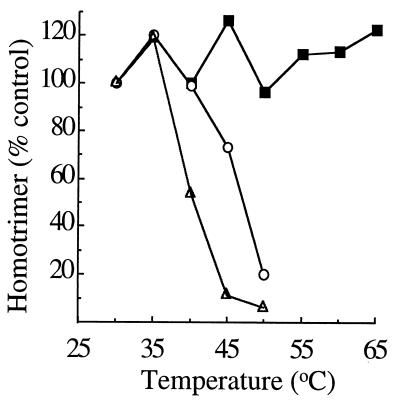
Dissociation of virus or ectodomain homotrimer caused by SDS and heat treatment. Radiolabeled virus was mixed with complete liposomes (1 mM final concentration) and acid treated at pH 5.5 for 3 min at 20°C to form the homotrimer. The samples were then heated at the indicated temperatures for 5 min in either 0.5% Triton X-100 (filled squares) or sample buffer containing 4% SDS (open triangles). Samples were analyzed by SDS-PAGE and phosphorimaging of the homotrimer band and compared to a control homotrimer sample that was not heated. Similarly, radiolabeled ectodomains were mixed with complete liposomes (1 mM final concentration), acid treated at pH 5.5 for 10 min at 37°C, heated at the indicated temperatures for 5 min in sample buffer containing 4% SDS, and analyzed as described above (open circles). Each curve is the average of two experiments.
While our data showed that the preformed E1 homotrimer was highly resistant to treatment with urea or heat, it was known that the trimer could be dissociated by incubation at 70°C in SDS-containing buffer (56). We used this treatment as a way of comparing the stability of homotrimeric virus E1 with that of the homotrimeric E1 ectodomain (Fig. (Fig.7).7). Homotrimers were preformed at low pH as described above, adjusted to a final concentration of 4% SDS, heated at the indicated temperatures for 5 min, and subjected to SDS-PAGE analysis. Trimers of either the viral E1 (open triangles) or the E1 ectodomain (open circles) were stable at 30 and 35°C under these conditions. Disassembly into monomers began after incubation at 40°C or higher temperatures. Viral E1 homotrimers showed ~90 to 95% dissociation after treatment at 45 or 50°C compared to treatment at 30°C. The homotrimers formed from ectodomains appeared slightly more stable, with ~25% dissociation after incubation at 45°C and ~80% dissociation after incubation at 50°C.
To characterize the destabilization of the homotrimer produced by heating in SDS, samples were incubated for 5 min at 30, 35, or 40°C in buffer containing 4% SDS. The SDS was then diluted to 0.5% in a 1% Triton X-100 solution, and the samples were digested with trypsin for 1 h at 37°C. As expected, the E2 and capsid proteins and monomeric E1 were completely digested (data not shown). In contrast, the E1 homotrimer incubated at either 30 and 35°C remained completely trypsin resistant, with recovery comparable to that of undigested controls. The sample treated with SDS at 40°C contained less homotrimer, as expected, but the remaining homotrimer maintained full trypsin resistance. Thus, the E1 homotrimer appeared to disassemble in a cooperative fashion since no partial homotrimer cleavage products were recovered. No trypsin-resistant monomeric E1 was observed, indicating that E1 derived from SDS-dissociated homotrimers was fully trypsin sensitive, similar to native E1.
DISCUSSION
Taken together, the data presented here for SFV and those previously published for influenza virus demonstrate that induction of the fusion-active conformation of the spike protein and of the membrane fusion reaction itself clearly differs between these two viruses. HA fusion is triggered by exposure to temperatures greater than ~58°C or by treatment with 3 to 3.5 M urea for 15 min at 37°C (11, 48, 58). These conditions also induce an HA conformation that appears very similar to that produced by low-pH treatment (11). In contrast, comparable urea or temperature treatments, or treatments at even higher concentrations or temperatures, caused no induction of either SFV fusion or E1 trimer formation. The pyrene fusion assay used here would detect both complete fusion and hemifusion, indicating that not even the initial lipid mixing step of SFV fusion was induced by such destabilizing conditions. Despite these differences in induction of the fusion-active form of the spike proteins, the SFV E1 protein was clearly similar to influenza virus HA in its ability to assume two possible conformations, a native, less stable conformation that can transition irreversibly to a highly stable, low-pH-induced conformation that is involved in fusion. Although there are many differences between these two viruses, three general features seem particularly important in considering the basis for the similarities and differences between their low-pH-dependent fusion proteins: the subunit location of proteolytic cleavage, the use of the coiled-coil fusion motif, and the requirement for specific lipids in fusion. These will be considered in turn.
Regulation of fusion proteins acts to protect them from nonproductive conformational changes that inactivate their fusion capacity while simultaneously ensuring that this fusion capacity is set to rapidly respond to the proper stimuli. Studies of influenza virus fusion have established that the HA0 precursor is inactive in fusion, which aids in protecting the fusion protein from the low-pH environment of the exocytic pathway (61, 62). Proteolytic processing to the mature HA1/HA2 form activates HA's fusion capacity and generates the free amino-terminal fusion peptide (14). Interestingly, other virus spike proteins that appear to use a coiled-coil-based fusion mechanism are also synthesized as precursors and activated by proteolytic processing, suggesting a similar mechanism of regulation of their fusion proteins (27, 52). The cleaved influenza virus HA is metastable and converts to the fusogenic extended coiled-coil conformation when triggered either by low pH, e.g., of the endosome during infection, or by destabilization with urea or heat in vitro. It is not yet clear how the metastable state of HA is set up during its biogenesis. Bacterial expression of HA2 in the absence of HA1 produces the stable, low-pH trimeric form, again indicating that this is HA2's lowest-energy state (15, 16). Under normal infection conditions, cellular chaperones might assist in the folding of nascent HA0 directly into a metastable trimer. Alternatively, during biosynthesis, HA0 might fold into its most stable conformation and the cleavage reaction might act to generate the metastable HA1/HA2 conformation (see reference 11 for a discussion).
One obvious difference between the influenza virus and SFV fusion proteins is that SFV E1 is not itself proteolytically processed during biosynthesis, and thus the putative fusion peptide remains ~80 residues from the E1 N terminus. Instead, SFV fusion is activated by the cleavage of p62, the companion subunit to E1 (41, 49). E1 associates with p62 within the ER, and in its absence, the SFV E1 subunit is misfolded and does not exit the ER (4, 7, 53). Evidence suggests that p62 acts as a chaperone to aid in the folding of E1 and possibly to protect it from the acidic pH of the Golgi complex. Proteolytic processing of p62 late in the exocytic pathway generates mature E1/E2 heterodimers capable of responding to the low pH of endosomes during infection. However, again in contrast to influenza virus, SFV mutants blocked in p62 processing are still able to mediate membrane fusion, although with a much lower pH threshold than wt SFV (28, 41, 49). Taken together, the data suggest that the fusion activity of E1 is regulated at least partially by its association with E2/p62 (22, 55). Cryoelectron microscopic studies suggest that there are clear changes in the E1/E2 heterodimer and spike trimer interactions after p62 cleavage (19), although it is not clear to what extent the secondary or tertiary structure of E1 rearranges following cleavage. The fact that purified E1 ectodomains are monomeric and not associated with E2 ectodomains but still require low pH to undergo conformational changes (39) suggests that dimer associations do not account for all of the regulation of E1 and that the fusogenic subunit has an independent pH-induced trigger.
Computer analysis of the SFV E1 sequence showed no regions predicted to form the extended α-helical coiled coil found in the low-pH structure of HA and in other viral fusion proteins of the influenza virus type. This suggests that a second important difference between the influenza virus and SFV fusion mechanisms is in the structural basis of the conformational changes that occur during the formation of the fusion-active trimer. Interestingly, the membrane fusion mechanism of the flavivirus tick-borne encephalitis virus (TBE) (reviewed in reference 24) appears similar in some respects to that of SFV. TBE fusion is mediated by the E protein, which has an internal fusion peptide and is activated for fusion by cleavage of a companion subunit (23, 25). Fusion is low pH dependent and appears to require the formation of E protein trimers (2). The crystal structure of the native TBE E protein dimer shows few α-helical regions (44), and the E protein trimer thus may form by a non-coiled-coil mechanism. The stability and induction properties of the TBE E protein trimer have not yet been characterized using the types of destabilizing treatments first performed on HA. It remains to be seen if the SFV and TBE proteins are members of a novel group of fusion proteins with similar, non-coiled-coil-based mechanisms and, if so, which other virus spike proteins may also be of this fusion protein type.
A dramatic difference between the influenza virus and SFV fusion mechanisms is in the lipid dependence of fusion. Influenza virus fusion does not require the presence of specific lipids in the target membrane (60), and the binding of the bromelain-generated HA ectodomain (BHA) to membranes is essentially unaffected by their lipid composition (18). In contrast, SFV fusion, E1-membrane binding, and E1 low-pH-dependent conformational changes are all promoted by the presence of the specific lipids cholesterol and sphingolipid in the target membrane (reviewed in reference 33). Although fusion-competent liposomes were included in all of the treatments with denaturants described here, these conditions did not support SFV fusion or E1 trimerization. This result suggests the possibility that the specificity of SFV's low-pH induction lies in the role of low pH in triggering interactions with particular target lipids.
In spite of these and other differences between HA and SFV E1, the native conformations of both proteins convert to a more stable form during fusion. How might these transitions take place? Studies on the fusion mechanism of influenza virus suggest a model for the regulation of HA's conversion from the native to the low-pH-induced conformation (5, 20). In this model, the conversion of HA would be regulated by kinetic partitioning between its two conformations. The native HA would be kinetically trapped during biogenesis in the less stable, “spring-loaded” conformation due to a high energy barrier between the two states. Upon treatment with either low pH, heat, or urea, the energy barrier to conversion from the native HA to the substantially more stable fusogenic trimer would be lowered and irreversible conversion and membrane fusion would occur. The kinetic model contrasts with a model based on thermodynamic partitioning between the native and fusogenic states, a situation in which the stability of the two conformations would depend on pH and the distribution of HA would transition reversibly between the native and low-pH conformations, depending on the pH (5).
Although our data suggest that only treatment at low pH, not treatment with general denaturants, is capable of triggering the formation of the E1 homotrimer and SFV fusion, we nevertheless favor a fusion mechanism for SFV that involves a kinetic rather than a thermodynamic transition. Clearly, the E1 homotrimer is substantially more stable than native E1. The conformational change from the native protein to the E1 trimer appears irreversible, even when triggered in the absence of target membranes and thus in the absence of fusion. The model of kinetic trapping that has been developed to explain the existence of the metastable state of influenza virus HA appears reasonable to explain the data for SFV as well. One difference in the kinetic models between the two viruses could be that for SFV the “activation barrier” preventing metastable E1 from forming an E1 trimer would need to be especially high and perhaps require regulation via specific changes induced by low pH as opposed to the more general effects of heat or denaturants. Although a thermodynamic partitioning between native E1 and the E1 trimer cannot strictly be ruled out, based on the available evidence it seems a less likely model.
A more accurate explanation for the control of the fusion activity of SFV requires a better understanding of the role of E2 in regulating E1, the importance of cholesterol and sphingolipid in the target membrane, and in particular the structural basis of the heterodimeric and homotrimeric associations of E1. Our laboratory is currently conducting genetic and biochemical experiments that address these questions. It is an intriguing possibility that significant differences in both the structure of SFV E1 and its functional requirements could identify a mechanism for virus-membrane fusion fundamentally different from the paradigm established by influenza virus.
ACKNOWLEDGMENTS
We thank Philip Aisen for the use of his fluorometer and Matthew Scharff and the Einstein hybridoma facility for advice and assistance with enzyme-linked immunosorbent assays. We thank Jan Wilschut and Jolande Smit for helpful advice on the pyrene fusion assay. We also thank Christina Eng for assistance with tissue culture and all of the members of our laboratory for helpful discussions, novel suggestions, and critical reading of the manuscript.
This work was supported by a grant to M.K. from the Public Health Service (R01 GM52929), by the Jack K. and Helen B. Lazar Fellowship in Cell Biology, and by Cancer Center core support grant NIH/NCI P30-CA13330. D.L.G. was supported through the Medical Scientist Training Program (NIH T32 GM07288).
ADDENDUM IN PROOF
In agreement with the data on fusion induction of viruses of the influenza type, a recent publication (R. G. Paterson, C. J. Russell, and R. A. Lamb, Virology 270:17–30, 2000) on fusion of the paramyxovirus SV5 suggested that the temperature of fusion induction correlates with the stability of the native fusion protein structure.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.74.17.7772-7780.2000
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc112306?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.74.17.7772-7780.2000
Article citations
Therapeutic alphavirus cross-reactive E1 human antibodies inhibit viral egress.
Cell, 184(17):4430-4446.e22, 01 Aug 2021
Cited by: 22 articles | PMID: 34416147 | PMCID: PMC8418820
Small-Molecule Inhibitors of Chikungunya Virus: Mechanisms of Action and Antiviral Drug Resistance.
Antimicrob Agents Chemother, 64(12):e01788-20, 17 Nov 2020
Cited by: 15 articles | PMID: 32928738 | PMCID: PMC7674028
Review Free full text in Europe PMC
Versatile targeting system for lentiviral vectors involving biotinylated targeting molecules.
Virology, 525:170-181, 02 Oct 2018
Cited by: 7 articles | PMID: 30290312 | PMCID: PMC6269213
Venezuelan Equine Encephalitis Virus Capsid-The Clever Caper.
Viruses, 9(10):E279, 29 Sep 2017
Cited by: 17 articles | PMID: 28961161 | PMCID: PMC5691631
Review Free full text in Europe PMC
The Bridges and Blockades to Evolutionary Convergence on the Road to Predicting Chikungunya Virus Evolution.
Annu Rev Virol, 4(1):181-200, 01 Sep 2017
Cited by: 13 articles | PMID: 28961411
Review
Go to all (45) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein.
J Virol, 68(11):6940-6946, 01 Nov 1994
Cited by: 77 articles | PMID: 7933075 | PMCID: PMC237130
Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein.
J Virol, 76(3):1194-1205, 01 Feb 2002
Cited by: 25 articles | PMID: 11773395 | PMCID: PMC135824
Role of spike protein conformational changes in fusion of Semliki Forest virus.
J Virol, 67(12):7597-7607, 01 Dec 1993
Cited by: 42 articles | PMID: 8230478 | PMCID: PMC238226
Mechanisms of enveloped virus entry into cells.
Mol Biol Med, 7(1):17-31, 01 Feb 1990
Cited by: 70 articles | PMID: 2182968
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P30 CA013330
Grant ID: P30CA13330
NIGMS NIH HHS (4)
Grant ID: R01 GM052929
Grant ID: R01GM52929
Grant ID: T32 GM007288
Grant ID: T32GM07288




