Abstract
Free full text

Identification of a Chlamydial Protease–Like Activity Factor Responsible for the Degradation of Host Transcription Factors
Abstract
Microbial pathogens have been selected for the capacity to evade or manipulate host responses in order to survive after infection. Chlamydia, an obligate intracellular pathogen and the causative agent for many human diseases, can escape T lymphocyte immune recognition by degrading host transcription factors required for major histocompatibility complex (MHC) antigen expression. We have now identified a chlamydial protease– or proteasome–like activity factor (CPAF) that is secreted into the host cell cytosol and that is both necessary and sufficient for the degradation of host transcription factors RFX5 and upstream stimulation factor 1 (USF-1). The CPAF gene is highly conserved among chlamydial strains, but has no significant overall homology with other known genes. Thus, CPAF represents a unique secreted protein produced by an obligate intracellular bacterial pathogen to interfere with effective host adaptive immunity.
Introduction
Chlamydia has to replicate within a cytoplasmic vacuole of eukaryotic cells and has adapted so well that it can persist in its host for a long period of time. Although chla-mydial infection is known to be responsible for many severe human diseases ranging from trachoma after ocular infections to life-threatening complications after urogenital infections, the mechanism of chlamydial pathogenesis is still not clear. It is thought that the persistent infection is a major cause of chlamydia-induced diseases in humans 1 2. Chlamydia-infected cells are able to continuously release inflammatory cytokines 3. Unfortunately, the tissue-damaging inflammatory responses induced by chlamydial infection often fail to efficiently eliminate the chlamydial organisms that are hidden inside the persistently infected host cells. We have hypothesized that chlamydia may have evolved strategies for evading host defense mechanisms in order to establish and to maintain a persistent infection.
Multicellular organisms have evolved various responses for controlling microbial infections, which, in turn, select for microbial pathogens with the ability to evade these same host defenses. Our previous studies have shown that chlamydia possesses varied strategies for interfering with or preventing both immune recognition and immune effector activity 4 5 6. Chlamydial infection suppressed both MHC class I and class II antigen expression in the infected cells. The inhibition of MHC antigen expression was correlated with the degradation of host transcription factors RFX5 and upstream stimulation factor 1 (USF-1). Furthermore, degradation of both RFX5 and USF-1 was caused by a chlamydial protease– or proteasome–like activity (CPA) in the cytosol of chlamydia-infected cells and the CPA was inhibited only by an irreversible proteasomal inhibitor lactacystin among all the inhibitors tested 4 5. A logical extension of these previous studies is to identify the factor(s) responsible for the chlamydial proteasome-like activity, which is the focus of this study. Using a column chromatography approach, we have purified a chlamydial protein that correlates with the CPA. We designated this chlamydial protein as CPA factor (CPAF).
Materials and Methods
Column Chromatography.
For purification and sequence identification of CPAF, cytosolic protein preparation from ~200 large flasks (150 cm2) of chlamydia-infected HeLa cells (HeLa L2 S100; reference 6) was subjected to the following three consecutive column separations: DEAE, heparin, and Mono Q (all columns were from Amersham Pharmacia Biotech). An AKTA purifier 10 (Amersham Pharmacia Biotech) was used to run the columns. Fractions collected from each column were monitored for both degradation activity in a cell-free assay as we have established previously 4, and total protein profiles on SDS-polyacrylamide gels. Fractions with degradation activity were pooled and both the protein concentration and degradation activity of the pooled fractions were carefully titrated. These measurements were used to calculate the purification efficiency. The pooled fractions were loaded to the next column for further fractionation. However, the pooled fractions collected from DEAE column were diluted 1:2 with H2O to reduce the salt concentration before the heparin column purification. Two dominant protein bands that correlated with the degradation activity from the final Mono Q column were excised for protein sequence identification using both mass spectrometry analysis (Biorealis Biosciences Inc.) and NH2-terminal sequence determination (University of Victoria, British Columbia, Canada). For chromatographic analysis of CPA, a size exclusion column was loaded with HeLa L2 S100 and eluted with PBS as we described previously 4. The eluted fractions were subjected to both the RFX5 degradation activity measurement and Western blot detection of host 20S proteasome and CPAF components.
Western Blot.
The Western blot assay was carried out as we described previously 7. Mouse antibodies were used to detect the NH2-terminal portion of CPAF (CPAFn) fragment (CPAFn54b; mIgG1; data not shown), chlamydial major outer membrane protein (MOMP) (MC22; mIgG3; data not shown), eukaryotic HSP70 (IgG2a; Santa Cruz Biotechnology, Inc.), and 20S proteasome subunits (AFFINITI Research Products Limited). Rabbit antibodies were used to detect RFX5 (Rockland Immunochemicals), USF-1, and USF-2 (Santa Cruz Biotechnology, Inc.).
Cell-free Degradation Assay.
The cell-free degradation assay was performed as described previously 4. To generate fusion proteins for the cell-free assay, chlamydial DNA sequences coding for CPAF or CPAF fragments were cloned into a pGEX vector (Amersham Pharmacia Biotech) and expressed as fusion proteins with the glutathione S-transferase (GST) as fusion partner. The fusion proteins were purified with glutathione-conjugated agarose beads as described in the manufacturer's manual (Amersham Pharmacia Biotech). The following procedure was used to prepare nuclear extracts (NEs) as substrate (containing RFX5 and USF-1 and USF-2) for the cell-free assay. Normal HeLa cells were dounced to break cytoplasmic membranes and the pellets were repeatedly washed with a buffer consisting of 1% NP-40 and 150 mM NaCl in 50 mM Tris, pH 8.0, plus a protease inhibitor cocktail to remove cytosol/membrane proteins as much as possible. The final washed nuclear pellets were extracted with a buffer consisting of 0.5 M NaCl and 1% Triton X-100 in 20 mM Tris, pH 8.0. The NEs thus prepared are essentially free of 20S proteasome components as detected on Western blot (data not shown). The human RFX5 gene from pREP-4/RFX5 plasmid (provided by Dr. P.J. van den Elsen, Leiden University Medical Center, Leiden, The Netherlands) was cloned into the pGEX vector and RFX5 was expressed in the JM109 Escherichia coli strain as a GST fusion protein. The GST-RFX5 fusion protein was purified to homogeneity using glutathione-conjugated agarose beads as described above. The purified GST-RFX5 was used as substrate in the cell-free degradation assays.
Immunofluorescence Staining Assay.
Immunofluorescence detection of CPAF in chlamydia-infected cells was carried out as described previously 6 7. In brief, HeLa cell monolayer was infected with Chlamydia trachomatis L2 for 30 h. The monolayer, after it was fixed with paraformaldehyde (Sigma-Aldrich) and permeabilized with Saponin (Sigma-Aldrich), was costained with Hoechst 32258 (blue), anti-MOMP antibody MC22 (probed with an FITC-conjugated, mouse IgG3-specific secondary antibody), and anti-CPAFn antibody 54b (probed with a Cy3-conjugated, mouse IgG1-specific secondary antibody). Images were acquired individually for each stain in gray using a Cooker digital camera connected to an AX70 Olympus microscope, and the single-color images were merged in frame into the triple-color image using the software Image Pro.
Immunoprecipitation Assay.
The immunoprecipitation assays were carried out as described previously 4 8. For antibody depletion of CPAF and the MOMP, both the anti-CPAFn and anti-MOMP antibodies were used to precipitate proteins in lysates of chlamydia-infected HeLa cells without radiolabeling 4. The intact lysate, the supernatant after antibody precipitation, and the proteins precipitated by the antibodies were all examined for their ability to degrade RFX5 in a cell-free assay. To visualize the proteins precipitated by the above antibodies, a radioimmunoprecipitation assay was performed 8. In brief, HeLa cells infected with chlamydia were metabolically labeled with [S35]methionine/cysteine (ICN Biomedicals) and the proteins in the labeled cell lysate were precipitated with the corresponding antibodies. The supernatant after the first precipitation (I°) was subjected to a second immunoprecipitation (II°) with the same antibodies. Both the anti-CPAFn and anti-MOMP antibodies completely removed the corresponding molecules from the lysates during I° precipitation.
Results
Purification and Sequence Identification of CPAF.
A chlamydial protease–like activity (designated as CPA) has been detected in the cytosol of chlamydia-infected cells and its presence correlated with degradation of host transcription factors 4. To better understand the molecular mechanisms underlying chlamydial evasion of host immune recognition, we used a column chromatography approach to identify the factor(s) responsible for CPA (Fig. 1 A). Although it was not possible to correlate any obvious protein bands with degradation activity in fractions eluted from either DEAE or heparin column, these two purification steps allowed us to remove most of the unwanted proteins and to dramatically enrich the degradation activity (Table ). The final Mono Q column purification resulted in ~1,000-fold increase in specific activity (Table ). Indeed, two predominant protein bands in fractions eluted from a final step Mono Q column correlated with the nuclear protein degradation activity (Fig. 1 A). Both protein bands were excised for sequence identification using mass spectrometry (Fig. 1 B). The sequences of a total of eight tryptic fragments derived from the bottom band matched the sequence of the NH2-terminal portion of a chlamydial protein encoded by open reading frame (ORF) CT858 (sequence data are available from GenBank/EMBL/DDBJ under accession no. AE001359; http://violet.berkeley. edu:4231/ident.html; reference 9), whereas the sequences of 17 tryptic fragments from the top band matched the sequence of the COOH-terminal portion of the same chlamydial protein (Fig. 1 B). These observations indicate that the two purified protein bands are encoded by a single ORF in the chlamydial genome. We have thus designated the bottom band protein as CPAFn and the top as CPAFc (Fig. 1). The sequence identity of CPAF was further confirmed using conventional NH2-terminal sequencing (data not shown). The CPAF gene has no significant overall homology to any other known genes despite its high conservation among chlamydial strains (9 10 11; data not shown). Although a short CPAF sequence (488PICVLINEQDFSCADFFPVVLKDNDRALIVGTRTAGAG525) was found to share a significant homology with the COOH-terminal sequences of tail-specific proteinases from various species (data not shown), the function of the conserved sequence domain is not yet known. Because of the limited homology, the CPAF gene was designated as either hypothetical ORF or “predicted protease containing interphotoreceptor retinoid-binding protein (IRBP) and 13,14-dihydroxy-retinol (DHR) domains” (http://violet.berkeley.edu:4231/ident.html; references 9 10 11).
Table 1
Summary of CPAF Purification
| Purification step | Volume | Total protein | Estimated CPAF activity | Estimated total CPAF activity | Estimated activity recovery | Estimated activity enrichment |
|---|---|---|---|---|---|---|
| ml | mg | hdf/mg | hdf | % | fold | |
| L2 S100 | 100 | 5,310 | 0.094 | 499 | 100 | 1 |
| DEAE | 70 | 214 | 0.97 | 208 | 42 | 10 |
| Heparin | 150 | 13 | 11.49 | 149 | 30 | 122 |
| Mono Q | 5 | 0.5 | 90 | 45 | 9 | 957 |
To monitor the purification efficiency, the pooled fractions from each column purification were titrated for determining both protein concentration and degradation activity. The activity was measured by serial dilution of the pooled fractions and expressed as the highest dilution factors per mg of protein, under which the degradation activity was still detectable. These measurements were further used for calculating the recovery and enrichment of CPAF activity.
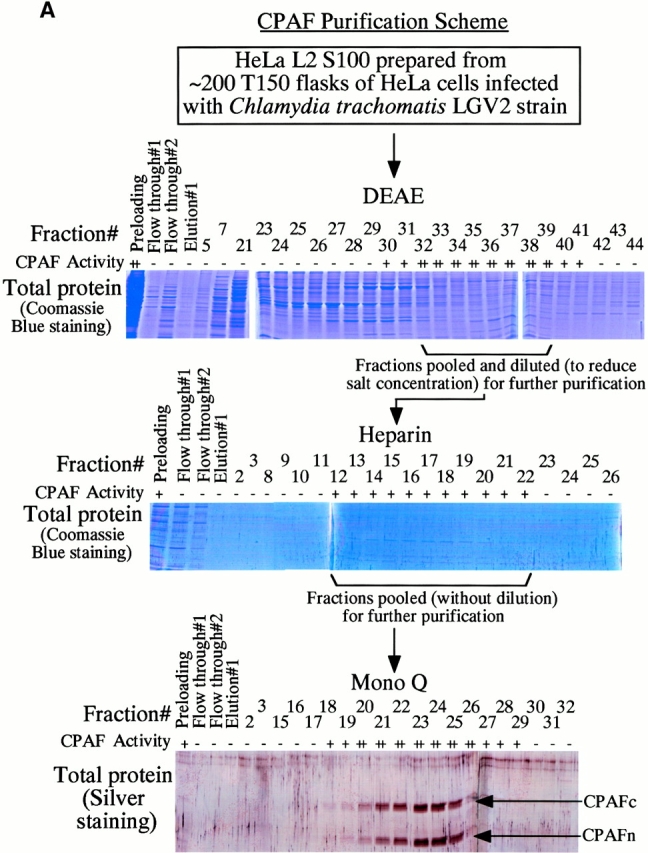
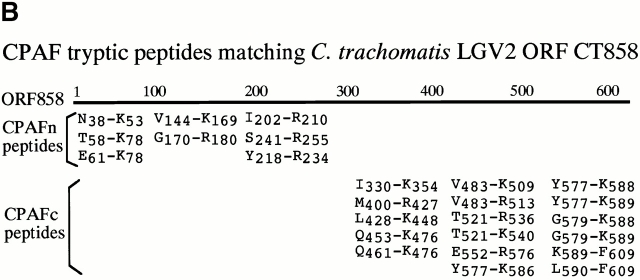
Purification and sequence identification of CPAF. (A) A cytosolic protein preparation from chlamydia-infected HeLa cells (HeLa L2 S100) was subjected to three consecutive column separations as listed in the figure. Fractions collected from each column were monitored for both degradation activity in a cell-free assay and total protein profiles on SDS-polyacrylamide gels. Fractions with degradation activity were pooled and loaded to the next column for further fractionation. Two dominant protein bands that correlated with the degradation activity from the final Mono Q column were excised for protein sequence identification. “+” indicates partial digestion of the substrate and “++” indicates complete loss of substrate. “−” indicates that the substrate band is intact compared with the substrate alone sample. (B) List of tryptic peptides that match the chlamydial ORF CT858-encoded protein (sequence data are available from http://www. ncbi.nlm.nih.gov/Entrez/protein.html under accession no. A71461). Amino acid residues are represented using the single letter codes and numbered according to their positions in CPAF sequence. Tryptic fragments derived from the top band match the COOH-terminal portion whereas those from the bottom band match the NH2-terminal portion of CPAF. The top band was designated as CPAFc and the bottom band as CPAFn.
CPAF Is Both Necessary and Sufficient for Degradation of Host Transcription Factors.
We have previously demonstrated that the chlamydial proteasome–like activity does not reflect the function of host proteasome components 4. When fractions obtained from a size exclusion chromatography column were probed with an antibody recognizing CPAFn, the chlamydial proteasome–like activity measured with RFX5 degradation was found to correlate very well with the presence of the CPAFn fragment but not host proteasome components (Fig. 2 A). To test whether CPAF is necessary for the chlamydial proteasome–like activity, we used the anti-CPAFn antibody to precipitate CPAFn from chlamydia-infected HeLa cell lysate and measured the RFX5 degradation activity of the precipitated pellets as well as the activity of the supernatants after the antibody depletion (Fig. 2 B). The total lysate of chlamydia-infected HeLa cells efficiently degraded RFX5. However, the supernatants remaining after antibody-mediated depletion of CPAFn completely lost the ability to degrade RFX5, whereas RFX5 degradation activity was readily detected in the precipitated material. Depletion of RFX5 degradation activity by anti-CPAFn was specific, because a control antibody recognizing the MOMP failed to precipitate this activity from the lysates. To monitor the efficiency of the antibody precipitation, radiolabeled lysate from chlamydia-infected HeLa cells was similarly precipitated with anti-CPAFn (I° precipitation) and the supernatant after the first precipitation was reprecipitated (II° precipitation) with the same reagent (Fig. 2 C). Autoradiography after SDS-PAGE of the material in both the I° and II° immunoprecipitates revealed that the anti-CPAFn antibody effectively removed both the CPAFn and CPAFc fragments from the lysates during the I° precipitation, because the II° immunoprecipitate showed minimal amounts of CPAF fragments. The control anti-MOMP antibody efficiently precipitated MOMP but not CPAF molecules from the lysates (Fig. 2 C). Together, these experiments demonstrate that CPAF or CPAF-associated material is necessary for RFX5 degradation activity in chlamydia-infected cell lysates.
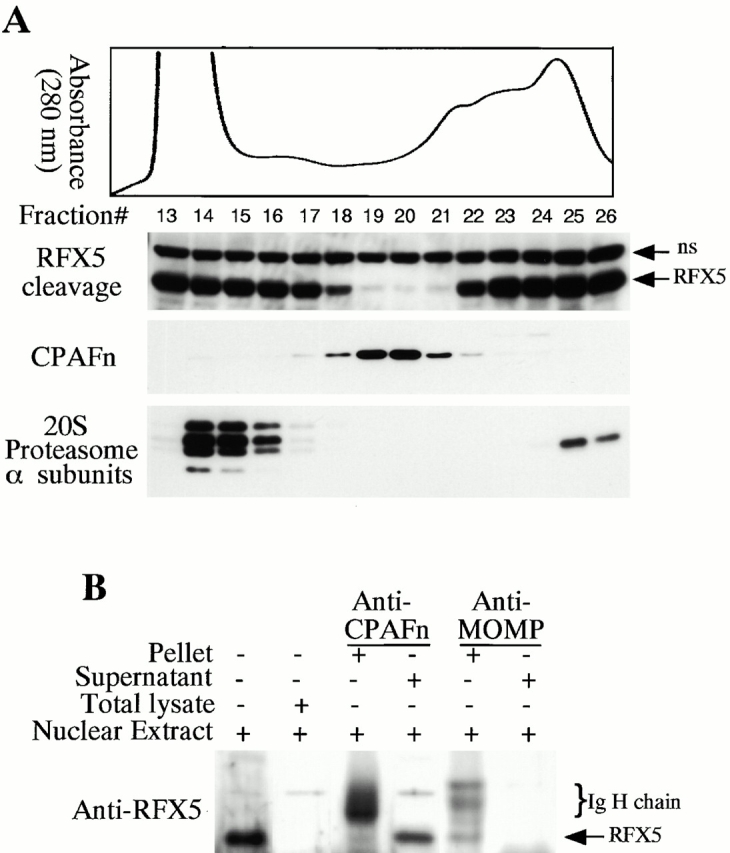
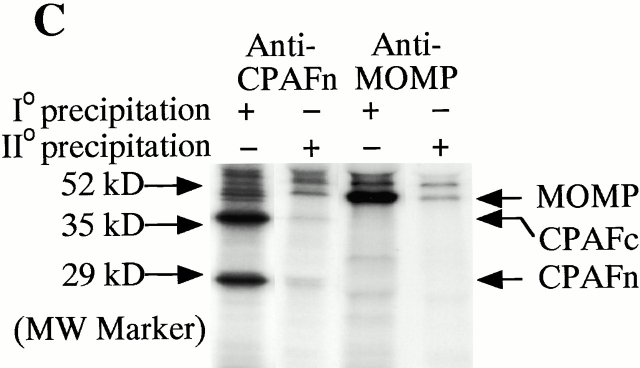
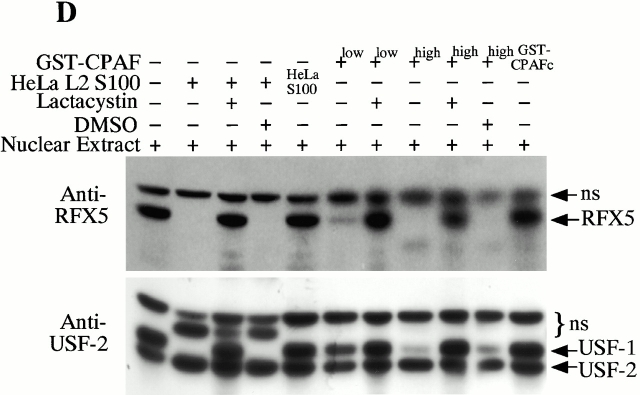
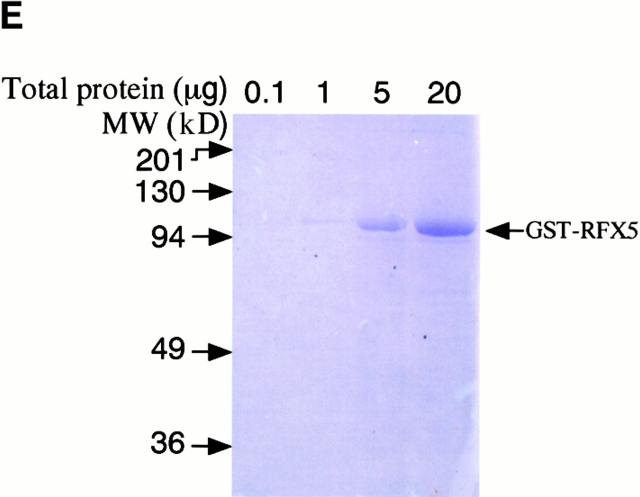
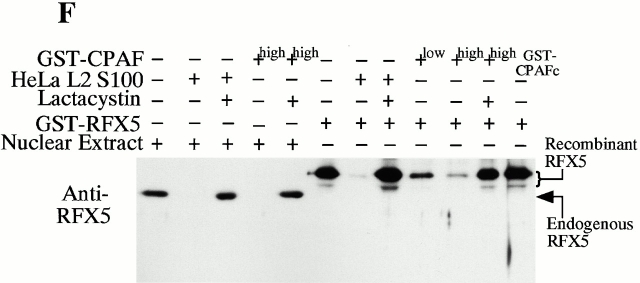
CPAF is both necessary and sufficient for degradation of host transcription factors. (A) HeLa L2 S100 was subjected to Superdex 200 size exclusion column analysis as described previously 4. The fractions were assayed for both RFX5 degradation activity and the presence of either host 20S proteasome subunits or CPAFn fragment. The presence of CPAFn but not host 20S proteasomes correlated with RFX5 degradation activity. (B) The effect of antibody depletion of CPAF on RFX5 degradation activity in chlamydia-infected HeLa cell lysates. Both the anti-CPAFn (54b) and anti-MOMP (MC22) antibodies were used to precipitate proteins from chlamydia-infected HeLa cell lysate. The intact (total) lysate, the supernatant after antibody precipitation, and the proteins precipitated by the antibodies (pellet) were all examined for their ability to degrade RFX5 in a cell-free assay. The anti-CPAFn antibody precipitated RFX5 degradation activity from supernatants to pellets. (C) Radioimmunoprecipitation analysis of proteins precipitated by anti-CPAFn or anti-MOMP antibodies. HeLa cells infected with chlamydia were metabolically labeled and the proteins in the labeled cell lysate were precipitated with antibodies. The supernatant after the first precipitation (I°) was subjected to a second immunoprecipitation (II°) with the same antibodies. Both the anti-CPAFn and anti-MOMP antibodies completely removed the corresponding molecules from the lysates during I° precipitation. The ratio of cell lysate vs. antibody was the same as in Fig. 2 B. (D) Degradation of transcription factors RFX5 and USF-1 by chlamydia-synthesized and bacterium-expressed CPAF in a cell-free assay with or without the inhibitor lactacystin. HeLa L2 S100 (containing chlamydia-synthesized CPAF) was used as positive control, and HeLa S100 was used as a negative control. Bacterium-expressed GST-CPAF was used at a final concentration of either 0.2 (low; sufficient for RFX5 degradation) or 0.6 μM (high; sufficient for both RFX5 and USF-1 degradation). The degradation was inhibitable by the proteasome inhibitor lactacystin (100 μM). A fusion protein containing GST and the COOH-terminal portion of CPAF (GST-CPAFc) failed to degrade any of the nuclear factors even at 4 μM. The anti–USF-2 antibody can detect both USF-1 and USF-2 as described previously (reference 4). Ns, nonspecific binding. (E). Purification of the recombinant human RFX5 from a bacterial expression system. The GST-RFX5 fusion protein was purified with glutathione-agarose beads as described in Materials and Methods. Different amounts of the purified protein were loaded to a 12% polyacrylamide SDS gel. After electrophoresis, the gel was stained with Coomassie blue dye. The GST-RFX5 fusion protein has a MW of ~100 kD. (F). Degradation of the purified human recombinant RFX5 by CPAF in a cell-free assay. The cell-free assay was carried out in the exact same way as described in D, except that 0.5 μg of the purified GST-RFX5 instead of the NEs was used as substrate in some reactions as indicated in the figure. The digestion experiment with the purified GST-RFX5 as substrate was repeated four times and similar results were observed. RFX5 in NEs is defined as endogenous and the bacterium-expressed GST-RFX5 fusion protein defined as recombinant.
To evaluate whether CPAF is sufficient for degrading host transcription factors, the protein encoded by the full-length CPAF gene was expressed as a fusion with GST, using a bacterial system. The purified GST-CPAF was evaluated for its ability to degrade RFX5 in a cell-free assay in comparison to material extracted from infected cells (HeLa L2 S100; Fig. 2 D). The HeLa L2 S100 degraded both the RFX5 and USF-1 transcription factors in NEs, whereas the material from uninfected HeLa cells (HeLa S100) failed to do so. Furthermore, even though the chlamydia proteolytic activity could be fractionated away from host cell 20S proteasomes (Fig. 2 A), the degradation activity of the HeLa L2 S100 was completely inhibited by lactacystin, an irreversible inhibitor of proteasomal proteases. These observations suggest that a chlamydial proteasome–like activity was being measured in this assay, as reported previously 4. More significantly, the purified GST-CPAF was able to degrade both RFX5 and USF-1 and the degradation was inhibitable by lactacystin. A GST fusion protein containing only the COOH-terminal portion of CPAF (GST-CPAFc) failed to degrade either of the host nuclear proteins even at a concentration 20 times higher than that of GST-CPAF (Fig. 2 D). These observations demonstrated that the purified CPAF is sufficient to mediate the degradation of host transcription factors in HeLa cell NEs. However, this set of experiments failed to address whether other components in the NEs also contributed to the degradation activity.
To further confirm that CPAF alone is sufficient for the degradation activity, we expressed the human transcription factor RFX5 in a prokaryotic system as a GST fusion protein. The GST-RFX5 was purified to homogeneity (Fig. 2 E) and the purified protein was used as the substrate for measuring the degradation activity of the cloned CPAF in a cell-free assay (Fig. 2 F). The cloned CPAF completely degraded the endogenous RFX5 in HeLa cell NEs and the degradation activity was inhibited by lactacystin, which is consistent with the observations described above in Fig. 2 D. More importantly, the cloned CPAF also effectively degraded the recombinant human RFX5 purified from a bacterial expression system (Fig. 2 F). Degradation of the recombinant RFX5 was inhibited by lactacystin, suggesting that the same enzymatic activity was responsible for the degradation of both endogenous and recombinant RFX5. A similarly purified control GST-CPAFc fusion protein failed to degrade the recombinant RFX5, suggesting that degradation of RFX5 measured in the assay was not contributed by bacterial contaminants (if there was any) or the GST fusion partner. These observations clearly demonstrate that the chlamydial CPAF alone is sufficient for degrading host RFX5.
Intracellular Distribution of CPAF during Chlamydial Infection.
Although CPAF is encoded by the chlamydial genome, the host transcription factor degradation activity was detected in the cytosol of the infected cells 4, suggesting that CPAF is secreted into host cell cytosol. We next used cell fractionation followed by immunoblotting to further evaluate the subcellular distribution of CPAF during chlamydial infection (Fig. 3 A). The majority of RFX5 degradation activity was associated with the cytosolic fraction of HeLa cells infected with chlamydia (Fig. 3 A, top row; lane HeLa L2 S100), with a small amount of activity in the NE fraction (lane HeLa L2 NE). RFX5 degradation activity correlated very well with the presence of CPAFn fragment. The cytosolic fraction of uninfected HeLa cells had no protease activity (lane HeLa S100). Surprisingly, the purified chlamydial organisms themselves also had no capacity to degrade RFX5. These findings suggest that CPAF is mainly localized in the cytosolic fraction of the infected HeLa cells and does not accumulate to any appreciable extent in chlamydia organisms themselves.
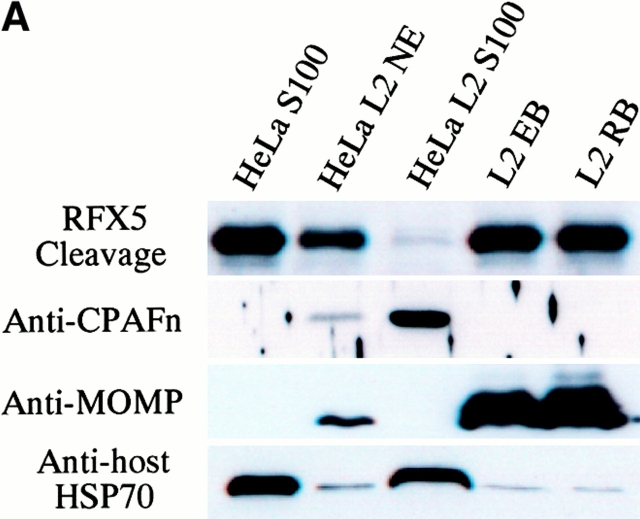
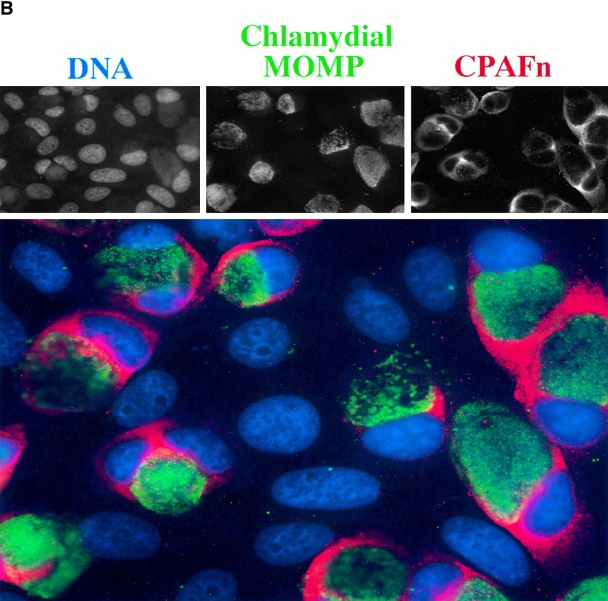
Intracellular distribution of CPAF during chlamydial infection. (A) Infected or uninfected HeLa cells were fractionated into cytosolic (S100) and nuclear (NE) fractions as described in Materials and Methods. The fractionated cellular samples and purified chlamydial organisms were evaluated for RFX5 degradation activity in a cell-free assay (top row) or analyzed for the presence of CPAFn (second row), MOMP (third row), or host heat shock protein 70 (HSP70; last row) using the corresponding antibodies. HeLa L2 S100 contained the highest RFX5 degradation activity, with a small amount of activity detected in the NE prepared from the same infected cells (lane HeLa L2 NE). The purified chlamydial organisms in the form of either the infectious elementary body (lane L2 EB) or the metabolically active but noninfectious reticulate body (lane L2 RB) contained no CPAF or CPAF activity. (B) Immunofluorescence detection of CPAF in chlamydia-infected cells. HeLa cell monolayer was infected with chlamydia organisms for 30 h. The monolayer was costained with Hoechst 32258 for DNA (blue), anti-MOMP antibody MC22 (probed with an FITC-conjugated, mouse IgG3-specific secondary antibody; green), and anti-CPAFn antibody 54b (probed with a Cy3-conjugated, mouse IgG1-specific secondary antibody; red). Images were acquired individually for each stain in gray (top row) using a Cooker digital camera connected to an AX70 Olympus microscope and the single-color images were merged in frame into the triple-color image (bottom) using the software Image Pro. Note that the anti-CPAFn antibody only stained the cytosol (red) of the cells with chlamydial inclusions (green).
An immunofluorescence assay was used to further confirm the intracellular distribution of CPAF (Fig. 3 B). CPAF was only detected in the cells containing chlamydial inclusion bodies but not the adjacent normal HeLa cells growing on the same coverslip, indicating that CPAF is restricted to chlamydia-infected cells. As expected from the cell fractionation studies, CPAF was mainly detected in the cytoplasmic portion of the infected cells, even though CPAF is encoded by the chlamydial genome. These observations suggest that the chlamydia-synthesized CPAF is secreted into the host cell cytoplasm, allowing it to access host proteins.
Discussion
Chlamydial growth occurs strictly within a modified cytoplasmic vacuole of eukaryotic cells 12, and the possibility of communication between chlamydial vacuoles and host cellular compartments has frequently been discussed 13 14 15 16. At least one purpose of such transfer of chlamydia-derived proteins to the invaded cell appears to be protection from host immune recognition 4 5 and attack 6. Our previous studies suggested that chlamydia may secrete a CPAF into the host cell cytosol to degrade transcription factors required for MHC gene activation, thus limiting expression of these key proteins involved in T cell antigen recognition 4 5. We have now identified the gene encoding CPAF that is secreted into host cell cytosol and demonstrated that CPAF is both necessary and sufficient for the degradation of the transcription factors RFX5 and USF-1. Interestingly, CPAF was identified as two separate polypeptides encoded by a single ORF in the chlamydial genome (Fig. 1 A). Both the CPAFn and CPAFc fragments are coprecipitated from chlamydia-infected cell lysates by an antibody that only recognizes the CPAFn fragment (Fig. 2 C). These observations suggest that CPAF may function in the form of intramolecular dimers. Whether CPAF cleavage into CPAFn and CPAFc is necessary for its degradation activity is not yet known. Although the details of the activation and secretion of CPAF remain to be fully determined, it is clear that by secreting a single factor into the cytosol, chlamydia can suppress both MHC class I and class II antigen expression. Many viruses are known to cause suppression or degradation of cellular proteins required for mounting host defense responses 17 18 19. However, these viral strategies are often dependent on the function of host proteasomes 20 21. We have demonstrated that CPAF is both necessary and sufficient for degrading host transcription factors required for MHC antigen expression. Thus, this report identifies a novel molecular mechanism by which a nonviral intracellular pathogen interacts with its host and manipulates immune responses for its benefit. Further characterization of CPAF including its transportation, activation, and substrate accessibility and selectivity is underway.
Acknowledgments
We thank Drs. Ronald N. Germain, Joel Baseman, and Feng Liu for reading the manuscript, and Peter J. van den Elsen for providing the RFX5 plasmid.
This work was supported in part by grants (to G. Zhong) from the Medical Research Council of Canada and the National Institutes of Health (R01 AI47997-01).
Footnotes
Abbreviations used in this paper: CPA, chlamydial protease– or proteasome–like activity; CPAF, CPA factor; GST, glutathione S-transferase; MOMP, chlamydial major outer membrane protein; NE, nuclear extract; ORF, open reading frame; USF, upstream stimulation factor.
References
- Beatty W.L., Morrison R.P., Byrne G.I. Persistent chlamydiaefrom cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 1994;58:686–699. [Europe PMC free article] [Abstract] [Google Scholar]
- Askienazy-Elbhar M., Suchet J.H. Persistent “silent” Chlamydia trachomatis female genital tract infections. Infect. Dis. Obstet. Gynecol. 1999;7:31–34. [Europe PMC free article] [Abstract] [Google Scholar]
- Rasmussen S.J., Eckmann L., Quayle A.J., Shen L., Zhang Y.X., Anderson D.J., Fierer J., Stephens R.S., Kagnoff M.F. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Invest. 1997;99:77–87. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhong G., Liu L., Fan T., Fan P., Ji H. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J. Exp. Med. 2000;191:1525–1534. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhong G., Fan T., Liu L. Chlamydia inhibits interferon gamma–inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 1999;189:1931–1938. [Europe PMC free article] [Abstract] [Google Scholar]
- Fan T., Lu H., Hu H., Shi L., McClarty G.A., Nance D.M., Greenberg A.H., Zhong G. Inhibition of apoptosis in chlamydia-infected cellsblockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 1998;187:487–496. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhong G., Reis e Sousa C., Germain R.N. Production, specificity, and functionality of monoclonal antibodies to specific peptide-major histocompatibility complex class II complexes formed by processing of exogenous protein. Proc. Natl. Acad. Sci. USA. 1997;94:13856–13861. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhong G., Castellino F., Romagnoli P., Germain R.N. Evidence that binding site occupancy is necessary and sufficient for effective major histocompatibility complex (MHC) class II transport through the secretory pathway redefines the primary function of class II–associated invariant chain peptides (CLIP) J. Exp. Med. 1996;184:2061–2066. [Europe PMC free article] [Abstract] [Google Scholar]
- Stephens R.S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R.L., Zhao Q., Koonin E.V., Davis R.W. Genome sequence of an obligate intracellular pathogen of humansChlamydia trachomatis . Science. 1998;282:754–759. [Abstract] [Google Scholar]
- Kalman S., Mitchell W., Marathe R., Lammel C., Fan J., Hyman R.W., Olinger L., Grimwood J., Davis R.W., Stephens R.S. Comparative genomes of Chlamydia pneumoniae and C. trachomatis . Nat. Genet. 1999;21:385–389. [Abstract] [Google Scholar]
- Read T.D., Brunham R.C., Shen C., Gill S.R., Heidelberg J.F., White O., Hickey E.K., Peterson J., Utterback T., Berry K. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–1406. [Europe PMC free article] [Abstract] [Google Scholar]
- Hackstadt T., Fischer E.R., Scidmore M.A., Rockey D.D., Heinzen R.A. Origins and functions of the chlamydial inclusion. Trends Microbiol. 1997;5:288–293. [Abstract] [Google Scholar]
- Hackstadt T., Scidmore M.A., Rockey D.D. Lipid metabolism in Chlamydia trachomatis-infected cellsdirected trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA. 1995;92:4877–4881. [Europe PMC free article] [Abstract] [Google Scholar]
- Stephens R.S. Molecular mimicry and Chlamydia trachomatis infection of eukaryotic cells. Trends Microbiol. 1994;2:99–101. [Abstract] [Google Scholar]
- Hatch T. Chlamydiaold ideas crushed, new mysteries bared. Science. 1998;282:638–639. [Abstract] [Google Scholar]
- Wylie J.L., Hatch G.M., McClarty G. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis . J. Bacteriol. 1997;179:7233–7242. [Europe PMC free article] [Abstract] [Google Scholar]
- Scheffner M., Werness B.A., Huibregtse J.M., Levine A.J., Howley P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. [Abstract] [Google Scholar]
- Shamu C.E., Story C.M., Rapoport T.A., Ploegh H.L. The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J. Cell Biol. 1999;147:45–58. [Europe PMC free article] [Abstract] [Google Scholar]
- Tortorella D., Gewurz B.E., Furman M.H., Schust D.J., Ploegh H.L. Viral subversion of the immune system. Annu. Rev. Immunol. 2000;18:861–926. [Abstract] [Google Scholar]
- Hughes E.A., Hammond C., Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc. Natl. Acad. Sci. USA. 1997;94:1896–1901. [Europe PMC free article] [Abstract] [Google Scholar]
- Tortorella D., Story C.M., Huppa J.B., Wiertz E.J., Jones T.R., Bacik I., Bennink J.R., Yewdell J.W., Ploegh H.L. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential J. Cell Biol. 142 1998. 365 376[published erratum at 145:642] [Europe PMC free article] [Abstract] [Google Scholar]
Articles from The Journal of Experimental Medicine are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1084/jem.193.8.935
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jem/article-pdf/193/8/935/1134479/001922.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101901062
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1084/jem.193.8.935
Article citations
A TriAdj-Adjuvanted Chlamydia trachomatis CPAF Protein Vaccine Is Highly Immunogenic in Pigs.
Vaccines (Basel), 12(4):423, 16 Apr 2024
Cited by: 2 articles | PMID: 38675805 | PMCID: PMC11054031
Requirement of GrgA for Chlamydia infectious progeny production, optimal growth, and efficient plasmid maintenance.
mBio, 15(1):e0203623, 19 Dec 2023
Cited by: 1 article | PMID: 38112466 | PMCID: PMC10790707
Tryptophan residue of plasmid-encoded Pgp3 is important for Chlamydia muridarum to induce hydrosalpinx in mice.
Front Microbiol, 14:1216372, 11 Jul 2023
Cited by: 0 articles | PMID: 37497542 | PMCID: PMC10367112
TargeTron Inactivation of Chlamydia trachomatis gseA Results in a Lipopolysaccharide 3-Deoxy-d-Manno-Oct-2-Ulosonic Acid-Deficient Strain That Is Cytotoxic for Cells.
Infect Immun, 91(7):e0009623, 31 May 2023
Cited by: 2 articles | PMID: 37255490 | PMCID: PMC10353364
GSDMD-mediated pyroptosis restrains intracellular Chlamydia trachomatis growth in macrophages.
Front Cell Infect Microbiol, 13:1116335, 16 Mar 2023
Cited by: 2 articles | PMID: 37009510 | PMCID: PMC10061094
Go to all (264) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - A71461
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Chlamydia pneumoniae secretion of a protease-like activity factor for degrading host cell transcription factors required for [correction of factors is required for] major histocompatibility complex antigen expression.
Infect Immun, 70(1):345-349, 01 Jan 2002
Cited by: 38 articles | PMID: 11748200 | PMCID: PMC127637
Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells.
J Exp Med, 191(9):1525-1534, 01 May 2000
Cited by: 124 articles | PMID: 10790427 | PMCID: PMC2213440
Intramolecular dimerization is required for the chlamydia-secreted protease CPAF to degrade host transcriptional factors.
Infect Immun, 72(7):3869-3875, 01 Jul 2004
Cited by: 29 articles | PMID: 15213129 | PMCID: PMC427400
Chlamydial protease-like activity factor--insights into immunity and vaccine development.
J Reprod Immunol, 83(1-2):179-184, 23 Oct 2009
Cited by: 15 articles | PMID: 19853923 | PMCID: PMC2804894
Review Free full text in Europe PMC
Funding
Funders who supported this work.
NIAID NIH HHS (2)
Grant ID: R01 AI47997-01
Grant ID: R01 AI047997




