Abstract
Free full text

Complete In Vitro Assembly of the Reovirus Outer Capsid Produces Highly Infectious Particles Suitable for Genetic Studies of the Receptor-Binding Protein
Abstract
Mammalian reoviruses, prototype members of the Reoviridae family of nonenveloped double-stranded RNA viruses, use at least three proteins—ς1, μ1, and ς3—to enter host cells. ς1, a major determinant of cell tropism, mediates viral attachment to cellular receptors. Studies of ς1 functions in reovirus entry have been restricted by the lack of methodologies to produce infectious virions containing engineered mutations in viral proteins. To mitigate this problem, we produced virion-like particles by “recoating” genome-containing core particles that lacked ς1, μ1, and ς3 with recombinant forms of these proteins in vitro. Image reconstructions from cryoelectron micrographs of the recoated particles revealed that they closely resembled native virions in three-dimensional structure, including features attributable to ς1. The recoated particles bound to and infected cultured cells in a ς1-dependent manner and were approximately 1 million times as infectious as cores and 0.5 times as infectious as native virions. Experiments with recoated particles containing recombinant ς1 from either of two different reovirus strains confirmed that differences in cell attachment and infectivity previously observed between those strains are determined by the ς1 protein. Additional experiments showed that recoated particles containing ς1 proteins with engineered mutations can be used to analyze the effects of such mutations on the roles of particle-bound ς1 in infection. The results demonstrate a powerful new system for molecular genetic dissections of ς1 with respect to its structure, assembly into particles, and roles in entry.
Mammalian orthoreoviruses (reoviruses) provide useful models to study how viruses from the Reoviridae family in particular, and viruses lacking lipid envelopes in general, enter their host cells and initiate infection. Reovirus virions are 85-nm particles comprising the segmented double-stranded RNA genome enclosed by two concentric icosahedral protein capsids. The outer capsid mediates viral entry into the cytoplasm of host cells, where viral replication occurs. Outer capsid protein ς1 (50 kDa, 36 copies) forms trimers that extend from the fivefold axes of virions and mediates viral attachment to cellular receptors. μ1 (76 kDa, 600 copies), found in virions mostly as fragments μ1N (4 kDa) and μ1C (72 kDa), participates in viral penetration of the cellular membrane barrier during entry. ς3 (41 kDa, 600 copies), the major surface protein of virions, interacts closely with μ1, thereby controlling its conformational status. λ2 (144 kDa, 60 copies) forms pentameric turrets that surround the fivefold axes and bridge the inner and outer capsids. λ2 is involved in viral mRNA synthesis and assembly of the outer capsid onto virus particles but is not known to participate in entry. Several recent articles discuss the structure of reovirus virions and functions ascribed to the capsid proteins (30, 34, 48).
Reovirus entry into cells is a multistep process characterized by programmed disassembly of virions into at least two types of subvirion particles, each with specialized roles in infection (28, 34, 48). After binding to a receptor(s) at the cell surface, virions are taken up into the endocytic pathway. There, lysosomal proteinases act upon them to produce intermediates that resemble infectious subvirion particles (ISVPs) generated by in vitro proteinase treatment of virions (2, 14, 42). ISVPs lack ς3, contain a cleaved form of μ1C, and may possess a conformer of ς1 different from that in virions (9, 26, 29, 35, 40). These ISVP-like particles initiate penetration of cellular membranes, culminating in delivery of particles into the cytoplasm (10, 32, 42). Concomitantly with membrane penetration, virus particles are activated to synthesize the viral mRNAs. These transcriptase particles may resemble cores produced by in vitro proteinase treatment of virions or ISVPs (11, 28, 40). Cores lack μ1 and ς1, contain a conformer of λ2 different from that present in virions and ISVPs, and are transcriptionally active (11, 21, 29, 40).
The mechanisms by which outer capsid proteins ς1, μ1, and ς3 mediate the steps in viral entry remain to be fully elucidated. A major obstacle is the lack of a reverse genetics system to produce virions with mutations in these proteins. To mitigate this problem, we recently described a strategy termed “recoating genetics” that permits analysis of infectious particles containing engineered forms of μ1 and ς3 (12, 27). Recoating genetics is enabled by the capacity of the recombinant proteins to bind and “recoat” purified subvirion particles in vitro, generating infectious particles that resemble virions: ς3 binds ISVPs to produce recoated ISVPs, and μ1 and ς3 bind cores to produce recoated cores (r-cores). However, recoated ISVPs and r-cores do not permit molecular genetic studies of the receptor-binding protein ς1 because the former particles contain native ς1 and the latter particles lack ς1 altogether. In this report, we extend recoating genetics to the ς1 protein by generating a new type of recoated cores that contains recombinant ς1, μ1, and ς3 (r-cores+ς1). These particles closely resemble native virions in structure, behavior in entry assays, and infectivity in cultured cells. Experiments with r-cores+ς1 demonstrated their utility for molecular genetic studies of ς1 and provided new insights into the structure and assembly of reovirus particles.
MATERIALS AND METHODS
Cells and viruses.
Spinner-adapted murine L929 fibroblast cells (L cells) (26), murine erythroleukemia cells (MEL cells) (39), Spodoptera frugiperda Sf21 insect cells (12), and Trichoplusia ni Tn High Five insect cells (Invitrogen) (12) were grown as described previously. Purified reovirus virions (26), ISVPs (26), and cores (12) were obtained as described previously. Particle concentrations were measured by A260 (19, 41). Plaque assays to determine infectivities of reovirus preparations were performed as described previously (26). cDNA clones and recombinant baculoviruses for expressing μ1 and ς3 together (12) or ς1 alone (15, 22) have been described previously.
Expression of ς1, μ1, and ς3.
Tn High Five cells were infected with fourth-passage virus stocks at a multiplicity of infection of 5 to 10 PFU/cell, and cells were harvested at 65 h postinfection. Cytoplasmic lysates of baculovirus-infected cells expressing μ1 and ς3 only were prepared by lysis with Triton X-100 as described previously (12). Lysates containing ς1, μ1, and ς3 were generated as follows: cells were separately infected with ς1- and μ1/ς3-expressing baculoviruses (1.5 × 107 cells each), mixed together, resuspended in 1 ml of lysis buffer (100 mM NaCl, 1.5 mM MgCl2, 250 mM sucrose, 10 mM Tris [pH 7.5]), lysed by probe sonication in an ice-ethanol bath (six 25-s pulses at 15 W), and centrifuged at 14,000 × g for 10 min at 4°C. The supernatant was used to prepare r-cores+ς1. Cells were separately infected with the ς1- and μ1/ς3-expressing baculoviruses because coinfection substantially reduced the yield of each protein at the multiplicity of infection used.
Preparation of r-cores, r-cores+ς1, and proteinase-treated r-cores+ς1.
r-cores were prepared from insect cell lysates containing μ1 and ς3 essentially as described previously (12). Briefly, lysates were incubated with purified T1L cores at a ratio of 6 × 106 cell equivalents (400 μl of lysate) per 1012 cores at 37°C for 2 h. The amounts of μ1 (200 μg) and ς3 (100 μg) present in the lysate represented a twofold excess of protein relative to the amounts needed to fully recoat the cores. Reaction mixtures were chilled and then loaded atop 14-ml step gradients, each containing a preformed CsCl gradient (ρ = 1.25 to 1.55 g/cm3, 12 ml) and a 2-ml sucrose cushion (20% [wt/vol]). Gradients were centrifuged for 2 to 16 h in a Beckman SW-28 rotor at 25,000 rpm and 5°C. r-cores were recovered as an optically homogeneous band and were further purified by being loaded onto a preformed CsCl gradient (ρ = 1.25 to 1.45 g/cm3, 4 ml) and centrifuged overnight in a Beckman SW-50.1 rotor at 40,000 rpm and 5°C. The harvested particles were dialyzed extensively against virion buffer. r-cores+ς1 were prepared in the same way as were r-cores, except that lysates containing ς1, μ1, and ς3 were used at a ratio of 1.2 × 107 cell equivalents (400 μl of lysate) per 1012 cores. The amount of ς1 (5 mg) present in the lysate represented a 500-fold excess of protein relative to the amount needed to fully recoat the cores. Concentrations of r-cores and r-cores+ς1 were measured by densitometry of Coomassie blue-stained gels as described previously (12). Proteinase-treated r-cores+ς1 were prepared from r-cores+ς1 by in vitro treatment with chymotrypsin as described previously (12).
Cryo-TEM and image reconstructions.
Purified virions and r-cores+ς1 were embedded in vitreous ice, and micrographs were captured at a nominal magnification of 38,000× using low-dose transmission cryoelectron microscopy (cryo-TEM) procedures (4) on a Philips CM200FEG microscope. Micrographs were digitized at a 7-μm step size on a Zeiss PHODIS microdensitometer. Image pixels were bin averaged to obtain pixel sizes of 14 μm (3.68 Å) for virions and r-cores+ς1 and 21 μm (5.53 Å) for r-cores. The r-core data were obtained from previously captured micrographs (12), except that 194 instead of 228 particles were used and these were reprocessed to reduce the effects of the microscope contrast transfer function in the final density maps. The r-core+ς1 data consisted of 1,078 particles (eight micrographs; defocus values, 1.4- to 2.5-μm underfocus). The virion data consisted of 860 particles (five micrographs; defocus values, 1.4- to 2.8-μm underfocus). Reconstructions were computed using established icosahedral particle procedures (6, 25). Corrections to compensate in part for the effects of the microscope contrast transfer function were applied to image data in the reconstruction program with Wiener filtering as described previously (49). For all three particle types, orientation angles were evenly distributed throughout the asymmetric unit as shown by all inverse eigenvalues being <0.1 (25). Final density maps were calculated to a 24-Å resolution limit, deemed to be near or below the resolution of each of the data sets as measured by various R-factor, correlation coefficient, and phase difference calculations (5). For the r-core+ς1 minus r-core difference map (21, 47), the magnifications and density values of the maps were scaled for maximum correlation of densities between radii 220 and 450 Å. The scaled r-core map was then subtracted from the r-core+ς1 map and displayed at a density threshold approximately three times higher than that used for the other maps, thereby allowing visualization of the most significant portions of the difference map.
RESULTS
In vitro recoating of cores with recombinant ς1, μ1, and ς3 proteins.
To test whether recombinant ς1 can be added to cores in vitro along with μ1 and ς3, a cell lysate containing all three proteins was incubated with purified cores, and virus particles were repurified by CsCl gradient centrifugation. Gel electrophoresis revealed that proteins comigrating with ς1, μ1 and μ1C (henceforth termed μ1/μ1C), and ς3 had bound to cores (Fig. (Fig.1).1). The identities of these proteins were confirmed by immunoblotting with polypeptide-specific antibodies (data not shown). No other proteins from the cell lysate were detected in the recoated particles. The cores recoated with ς1, μ1/μ1C, and ς3 (r-cores+ς1) in this study contained approximately stoichiometric amounts of μ1/μ1C and ς3 relative to virions (data not shown), as previously shown for cores recoated with only μ1/μ1C and ς3 (r-cores) (12). The amount of ς1 in r-cores+ς1 varied between preparations but was always sufficient to be detected by Coomassie blue staining. In several preparations of r-cores+ς1, the amount of ς1 approached that in virions (Fig. (Fig.1),1), suggesting that ς1 can bind to cores in vitro with approximately native stoichiometry.
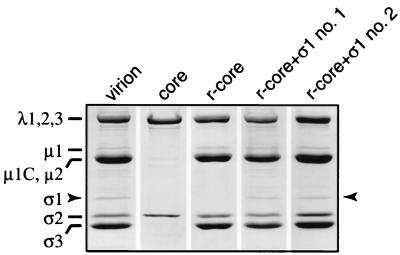
Protein composition of r-cores+ς1. Purified virions, cores, r-cores, and two preparations of r-cores+ς1 (no. 1 and no. 2) (8 × 1010 particles each) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% acrylamide) and Coomassie blue staining. Positions of viral proteins are indicated at left, and the position of ς1 is highlighted with arrows.
r-cores+ς1 resemble virions in structure.
Comparison of the image reconstruction of r-cores+ς1 with those of virions and r-cores revealed that all three types of particles are similar in overall appearance. Each possesses 600 finger-like projections at the particle surface attributed to the 600 subunits of ς3, features beneath the ς3 layer that represent the 600 subunits of μ1, and similar conformational states of the pentameric λ2 turret (Fig. (Fig.2a2a to c). Image reconstructions of virions (Fig. (Fig.2a)2a) and r-cores+ς1 (Fig. (Fig.2c)2c) also contain an extended feature emerging from the center of the λ2 turret that, in virions and ISVPs, has been attributed to ς1 (21). The presence of this feature in r-cores+ς1 and its absence in r-cores (Fig. (Fig.2b)2b) support the conclusion that r-cores+ς1, like virions, contain ς1 bound at the fivefold axes.
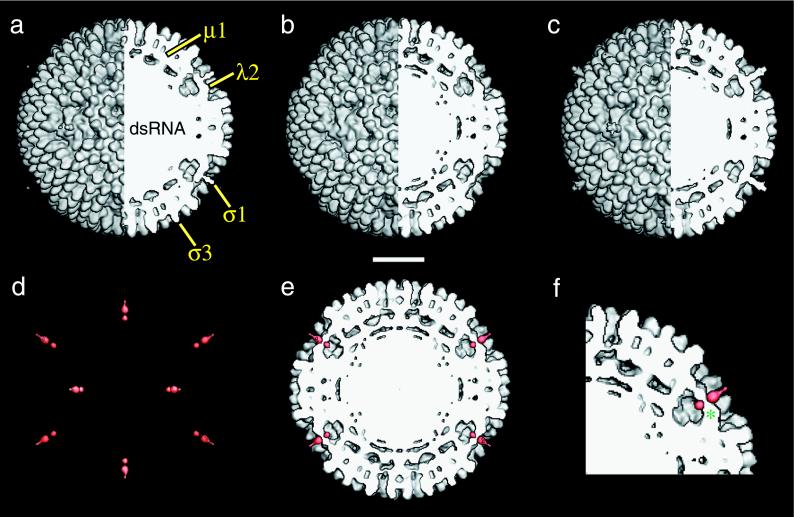
Image reconstruction of r-cores+ς1. (a to c) Composite images comprising external (left half) and cutaway (right half) surface-shaded representations of virions (a), r-cores (b), and r-cores+ς1 (c) are shown. Features attributed to outer capsid proteins and the genome are labeled in panel a. (d) Difference map between r-cores+ς1 and r-cores. Only density features in the front half of the map are shown and are coded in red. (e) The difference map (red) superimposed upon a cutaway section of the r-core map. Features in the difference map at positions above and below the sectioning plane are not shown. (f) Magnified view of a portion of the image in panel e. One segment of the pentameric shutter that closes the λ2 turret is labeled (*). Scale bar, 200 Å.
A feature attributable to ς1 is present within the λ2 turret.
Difference maps between the r-core+ς1 and r-core reconstructions were computed to assess more carefully the effects of ς1 assembly on particle structure (Fig. (Fig.2d).2d). Only two significant density features, both located at the fivefold axes of r-cores+ς1, were seen in the difference map. The absence of significant differences in other locations confirmed that assembly of ς1 into r-cores+ς1 causes no major rearrangements in the core proteins, μ1, ς3, or regions of λ2 distal from ς1. Superposition of the two features upon a cutaway section of the r-core reconstruction (Fig. (Fig.2e2e and f) indicated that the upper feature represents the ς1 fiber extending above λ2 (see above). The lower feature is located in line with the upper feature, just beneath the pentameric “shutter” that closes the top of the λ2 turret. This feature might represent either a portion of ς1 or fivefold-proximal λ2 sequences that have undergone rearrangement upon ς1 binding. However, we consider the latter unlikely because all of the density associated with λ2 in r-cores is also present at similar positions in r-cores+ς1 (i.e., no significant loss of density from the λ2 region was seen in the inverse difference map of the r-core and r-core+ς1 reconstructions [data not shown]). Therefore, we conclude that this lower feature represents the base of the ς1 fiber that protrudes through the λ2 shutter and into the turret cavity.
r-cores+ς1 bind efficiently to RBCs and L cells.
To assess the competence of recombinant ς1 in r-cores+ς1 for binding to cell receptors, we measured the capacity of virions, cores, r-cores, and r-cores+ς1 to induce hemagglutination (HA) of human erythrocytes (RBCs) (Fig. (Fig.3a).3a). ς1 mediates HA via its interactions with one or more glycoprotein on the cell surface (1, 38). Virions induced HA efficiently as expected, while cores and r-cores, which lack ς1, failed to do so even at the highest particle concentration tested (fivefold higher than the maximum shown in Fig. Fig.3a3a [data not shown]). r-cores+ς1 agglutinated RBCs nearly as efficiently as did virions, indicating that these particles containing recombinant ς1 are functional for attachment to the reovirus receptor(s) on human RBCs.
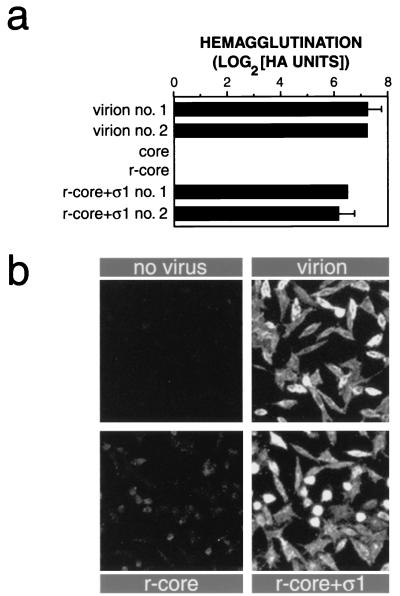
Capacity of r-cores+ς1 to bind to cells. (a) HA induced by purified virions, cores, r-cores, and r-cores+ς1 was determined by endpoint titration using human type A+ RBCs (American Red Cross), as described previously (20). Serial twofold virus dilutions were incubated with RBCs (0.4% [vol/vol]) in a 96-well round-bottomed plate at 4°C for 2 h, and HA was visually detected. The number of HA units induced by a virus preparation is given by the highest number of particles used (2 × 1010 particles) divided by the minimum number of particles required to induce HA. HA activity is expressed as the log2(HA units). Averages ± standard deviations of three replicates are shown. (b) Attachment of purified virions, r-cores, and r-cores+ς1 to L cells at 4°C was determined by indirect immunofluorescence as described previously (18). Cells on coverslips were incubated with purified virus (5 × 105 particles/cell) at 4°C for 1 h. Unbound virus was removed by washing with ice-cold phosphate-buffered saline. After the cells were fixed and permeabilized, bound virus was detected by using a 1:500 dilution of rabbit antireovirus serum (44) as primary antibody and a 1:100 dilution of fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin G (Pierce) as secondary antibody.
We also used indirect immunofluorescence to detect binding of virions, r-cores, and r-cores+ς1 to L cells, which are permissive for reovirus infection (Fig. (Fig.3b).3b). We found that binding of r-cores to cells, while detectable, was much less efficient than the binding of virions, consistent with the absence of cell attachment protein ς1 in r-cores. In contrast, r-cores+ς1 bound to L cells with an efficiency similar to that of virions, confirming that the recombinant ς1 in r-cores+ς1 is functional for efficient attachment to the reovirus receptor(s) expressed in L cells.
r-cores+ς1 are nearly as infectious as virions and display similar growth kinetics.
r-cores containing levels of μ1/μ1C and ς3 similar to those of virions but lacking ς1 were 250 to 500 times more infectious in L cells than were precursor cores but only ~0.0001 times as infectious as virions on a per-particle basis (12) (Fig. (Fig.4a).4a). r-cores+ς1 containing levels of ς1 similar to those of virions (Fig. (Fig.1)1) were 3,000 times more infectious than r-cores and 0.5 times as infectious as virions on a per-particle basis (Fig. (Fig.4a),4a), consistent with the capacity of r-cores+ς1 and virions, but not r-cores, to attach to cells efficiently (Fig. (Fig.3b).3b). Infection by virions and r-cores+ς1, but not r-cores, was blocked by anti-ς1 antibody 5C6 (Fig. (Fig.4b)4b) (45), showing that the enhanced infectivity of r-cores+ς1 compared with that of r-cores is due to recombinant ς1 in the former particles.
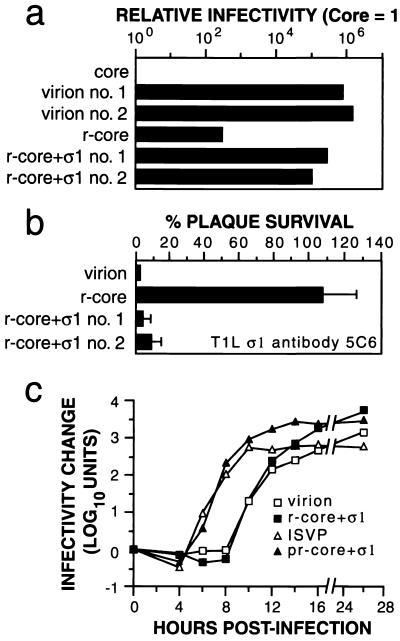
Infectious properties of r-cores+ς1 in L cells. (a) The infectivities of purified virions, cores, r-cores, and r-cores+ς1 were determined by plaque assay on L-cell monolayers and expressed as PFU per milliliter (26). After standardization against the number of virus particles per milliliter, the relative infectivity of each preparation was determined by dividing its PFU per milliliter by the PFU of cores per milliliter. Averages of three replicates are shown (standard deviation ≤ 0.10 log10 units). (b) The capacity of anti-T1L ς1 antibody 5C6 to neutralize formation of plaques on L cells by purified virions, r-cores, and r-cores+ς1 was determined as described previously (12). Virus (100 PFU) was incubated with 5C6 (1 μg/ml) for 1 h at 37°C, and infectivity was measured by plaque assay. The extent of neutralization for each preparation (expressed as percent plaque survival) was the ratio of PFU obtained in the presence of antibody to the PFU obtained in its absence. Averages ± standard deviations of three replicates are shown. (c) Single-cycle growth curves of purified virions, r-cores+ς1, and chymotrypsin treatment mixtures containing ISVPs and proteinase-treated r-cores+ς1 (pr-core+ς1) (2 PFU/cell) were generated in L cells as described previously (13). Infectivity at a specified time (t) relative to time zero was expressed as log10(PFU/ml)t − log10(PFU/ml)0. Each data point represents the average of duplicate values.
The preceding experiment (Fig. (Fig.4a)4a) provided evidence that similar proportions of virions and r-cores+ς1, and a much lower proportion of r-cores in preparations of these particles, can initiate infections that result in viral plaques after multiple cycles of replication. To compare the infectious properties of r-cores+ς1 and virions during a single replication cycle, we generated growth curves for these particle types (Fig. (Fig.4c).4c). Virions and r-cores+ς1 showed very similar growth curves, indicating similar kinetics of viral entry and initial production of progeny virus (lag phase), accumulation of progeny (exponential phase), and final growth yields (plateau phase).
ISVPs generated from virions by in vitro proteinase treatment exhibit a shorter lag phase than do virions during a single growth cycle (42) (Fig. (Fig.4c).4c). Virions grow more slowly than ISVPs, likely because they contain ς3 that must be cleaved within the endocytic pathway before membrane penetration can proceed. ISVPs, on the other hand, already lack ς3 and can bypass this step within cells (3, 27, 42). We found that ISVP-like proteinase-treated r-cores+ς1 also grew more rapidly than did their virion-like precursors and showed kinetics similar to those of ISVPs. The faster growth of proteinase-treated r-cores+ς1 compared with that of r-cores+ς1 is consistent with observations that r-cores+ς1 must, like virions, undergo ς3 degradation in order to initiate infection (data not shown). Thus, r-cores+ς1 containing recombinant forms of outer capsid proteins ς1, μ1, and ς3 reproduce entry-related behaviors observed with native particles.
r-cores+ς1 recapitulate strain-dependent differences attributed to ς1.
Analysis of r-cores+ς1 assembled with different types of recombinant ς1 provides a new approach for genetic studies of particle-bound forms of this protein. To evaluate the feasibility of this approach, we produced r-core+ς1L and r-core+ς1D particles containing genome and core, μ1/μ1C, and ς3 proteins from reovirus strain type 1 Lang (T1L) but recombinant ς1 from strain T1L or type 3 Dearing (T3D), respectively. We then determined the properties of r-cores+ς1L and r-cores+ς1D in the following assays that show a ς1-dependent difference between native T1L and T3D virions.
(i) HA of bovine RBCs (Fig. (Fig.55a).
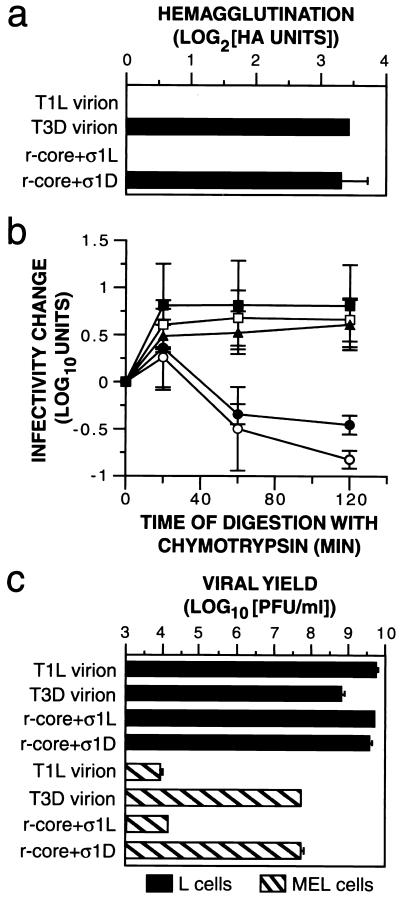
Capacity of r-cores+ς1 to mediate ς1-specific in vitro and in vivo properties. (a) HA of bovine RBCs (Colorado Serum Co.) induced by purified T1L virions, T3D virions, r-cores+ς1L, and r-cores+ς1D was determined as described for Fig. Fig.3a.3a. Averages ± standard deviations of three replicates are shown. (b) The effect of chymotrypsin treatment on infectivity of T1L virions (open squares), T3D virions (open circles), r-cores+ς1L (filled squares), r-cores+ς1D (filled circles), and r-cores+ς1D(T249I) (filled triangles) was determined as follows. Purified virus (1.5 × 1012 particles/ml) was incubated with chymotrypsin (200 μg/ml) at 37°C. At specified times, an aliquot was removed onto ice and digestion was stopped with phenylmethylsulfonyl fluoride (2 mM). The infectivity of each aliquot was determined by plaque assay. Viral infectivity at a specified time (t) relative to time zero was expressed as described for Fig. Fig.4c.4c. Averages ± standard deviations of three independent experiments are shown. (c) Infectious yields produced in MEL and L cells by T1L virions, T3D virions, r-cores+ς1L, and r-cores+ς1D were determined as follows. Purified virus (5 PFU/cell) was incubated with 4 × 106 cells for 1 h at room temperature. Unbound virus was removed by centrifugation (500 × g) and washing cells with phosphate-buffered saline. Cells were then resuspended in growth medium and incubated at 37°C for 24 h. Cell lysates were generated by freeze-thawing, and the amount of infectious virus in these lysates was determined by plaque assay. Growth yields were expressed as log10(PFU/ml) at 24 h. Averages ± standard deviations of six trials from two independent experiments are shown.
T3D virions induce HA of bovine RBCs via interactions between T3D ς1 and RBC sialoglycoproteins (20, 38). T1L virions do not agglutinate bovine RBCs, likely because T1L ς1 does not bind sialic acid and no other receptors are available for it on these cells (20, 38). r-cores+ς1D induced levels of HA similar to those induced by T3D virions, while r-cores+ς1L, like T1L virions, failed to induce HA. Thus, the capacity of r-cores+ς1 to agglutinate bovine RBCs is determined by the origin of the particle-bound ς1 protein, consistent with previous genetic and biochemical evidence (20, 38).
(ii) Effect of in vitro chymotrypsin treatment on viral infectivity (Fig. (Fig.55b).
When T1L virions are treated with chymotrypsin in vitro, infectivity increases at early times and is then maintained for at least 2 h. In contrast, the infectivity of T3D virions increases transiently upon chymotrypsin treatment but then decreases to plateau at 10% of the starting value (36). r-cores+ς1L and r-cores+ς1D behaved like T1L and T3D virions, respectively, upon chymotrypsin treatment, confirming the previous hypothesis (15, 36) that these effects of chymotrypsin on infectivity are determined by the origin of particle-bound ς1.
The effects of chymotrypsin treatment on the infectivity of virions and r-cores+ς1 correlate with the sensitivity of ς1 to cleavage by chymotrypsin (see references 15 and 36 for data for virions; data are not shown for r-cores+ς1). Specifically, T1L ς1 is not cleaved by chymotrypsin, while T3D ς1 is cleaved. This correlation holds true for all other viral strains tested (15). Therefore, the available data suggest a mechanistic link between cleavage of particle-bound ς1 and inactivation of infectivity by chymotrypsin treatment. To establish this link, we sought to investigate the infectious behavior of r-cores+ς1 generated with a mutant form of T3D ς1 engineered to resist cleavage by chymotrypsin. Recent work with isolated ς1 proteins suggested a promising candidate: the point mutation T249→I blocked cleavage of recombinant T3D ς1 by trypsin (15). Accordingly, we produced and tested r-cores+ς1D(T249I) containing this mutant ς1 protein. r-cores+ς1D(T249I), in contrast to their wild-type counterparts, retained full infectivity after chymotrypsin treatment (Fig. (Fig.5b)5b) while the particle-bound T3D ς1(T249I) remained uncleaved (data not shown), proving that the loss of infectivity suffered by T3D virions upon chymotrypsin treatment is caused by cleavage of T3D ς1.
(iii) Viral growth in MEL cells (Fig. (Fig.55c).
T3D virions produce ~10,000 times higher yields of infectious progeny than do T1L virions in MEL cells, whereas the two types produce similar yields in L cells (16, 39). This strain-dependent difference in viral growth has been genetically mapped to the S1 gene segment, which encodes the ς1 and ς1s proteins (39). Moreover, viral strains adapted to grow in MEL cells contain mutations in ς1 that increase their capacity to bind to MEL cells (16). Findings to date therefore support a model in which the capacity of reoviruses to infect and grow in MEL cells is determined by ς1-receptor interactions. It remained possible, nevertheless, that the above difference in viral growth arises not from ς1 protein present in infecting particles but from ς1 transcripts or ς1 or ς1s polypeptides newly synthesized in infected cells. To test that possibility, we measured the growth yields of r-cores+ς1L and r-cores+ς1D in MEL and L cells and found that these mirrored the yields obtained with native T1L and T3D virions, respectively. Because r-cores+ς1L and r-cores+ς1D possessed the same genome (T1L) and differed only in the origin of their particle-bound ς1 proteins, our results confirm that the growth difference between T1L and T3D viruses in MEL cells arises from differences in viral entry attributable to particle-bound ς1, likely at the cell attachment step.
DISCUSSION
The ς1-λ2 interaction in reovirus particles.
Protein ς1 forms lollipop-shaped homotrimers consisting of a base, a long fibrous tail, a neck, and a globular head (7, 15, 24, 26, 37). The amino-terminal base of ς1 anchors it to the particle (26, 31), while more carboxy-terminal sequences in the neck and head domains mediate binding to cellular receptors (16, 20, 23, 33). Image reconstructions of virions and ISVPs show that the base of each ς1 fiber is anchored to the particle by interaction with the pentameric shutter atop each λ2 turret (21). However, little is known about the structural details of the ς1-λ2 binding interaction because density features contributed to the binding interface by each protein cannot be distinguished in the cryo-TEM maps of native particles. Comparison of the r-core and r-core+ς1 image reconstructions in this report suggests that the base of the ς1 fiber protrudes through the center of the pentameric λ2 shutter and that these protruding ς1 sequences form a small knob-like domain within the large cavity enclosed by the λ2 turret. On the basis of this observation, we speculate that the ς1 fiber remains anchored to λ2 in virions and ISVPs at least in part because its internal knob domain cannot escape through the narrow channel at the center of the λ2 shutter in those particles (12, 21). Consistent with that idea, ς1 binds poorly to cores (K. Chandran and M. L. Nibert, unpublished data), which contain a larger channel at the center of each open λ2 turret (21).
The current observations regarding the mode of ς1 binding also have implications for the pathway of ς1 assembly into particles. The assembly of μ1 and ς3 alone onto cores causes the λ2 shutter to close (12). Therefore, if ς1 binds to cores after μ1 and ς3, the knob-like base of the ς1 fiber must be inserted through the narrow fivefold channel in the shutter. Since this seems unlikely, we hypothesize that ς1 binds to cores before, or in concert with, μ1 and ς3.
Previously published evidence indicates that deletions of residues 3 to 34 or 37 to 107 of T3D ς1 block incorporation into virions whereas residues 1 to 121 are sufficient for incorporation (30, 31). However, the specific residues within these regions that mediate binding to λ2 remain to be identified. We found that T1L ς1 and T3D ς1, which share a sequence identity of only 22% within this N-terminal region (37), both bound to T1L cores in a functional manner (Fig. (Fig.11 and and5),5), suggesting that the λ2-binding element(s) in ς1 is not highly dependent on primary amino acid sequence or that only a few specific residues are involved.
The region(s) of ς1 visualized in the cryo-TEM image reconstruction of r-cores+ς1.
Electron microscopic studies of negatively stained preparations of purified ς1 fibers indicate that each is 480 to 500 Å in length, with the N-terminal tail and C-terminal head portions comprising 380 to 400 and 100 Å of the fiber, respectively (24, 26). The density features attributed to ς1 in the r-core+ς1 reconstruction, however, measure only about 140 Å in length. If the λ2-proximal portion of the ς1 tail assumes an extended conformation after assembly into r-cores+ς1, then the density visualized in the r-core+ς1 reconstruction represents only the first 100 to 120 residues of ς1 according to the current model for sequence-structure correlations in the fiber (24). This region of the ς1 tail comprises the putative N-terminal λ2-binding domain and approximately one-half of the long α-helical coiled coil (24, 30, 31, 37). More distal tail and head sequences are not visualized in the cryo-TEM map, probably because those regions of the ς1 fiber are more variably placed relative to the icosahedral lattice, due to fiber bending. This idea is supported by calculations for position-dependent flexibility of the ς1 fiber from electron micrographs as well as from the amino acid sequence: both analyses found a local maximum in fiber flexibility at a distance of about 140 Å along the tail (24). r-cores+ς1 containing “stiffened” forms of recombinant ς1 may allow visualization of more distal domains in the fiber.
Infectious properties of r-cores+ς1.
Assembly of approximately stoichiometric amounts of recombinant ς1, μ1, and ς3 onto cores to produce r-cores+ς1 boosted the infectivity of cores by nearly a millionfold, to ~50% of that of virions. This almost complete reconstitution of viral infectivity suggests that r-cores+ς1 are free from major defects. Moreover, observations that r-cores+ς1 and proteinase-treated r-cores+ς1 replicated in L cells with kinetics identical to those of virions and ISVPs, respectively, and that r-cores+ς1, like virions, required acid-dependent lysosomal proteinase(s) for infection strongly suggest that these recoated particles utilize the same pathways as do native particles for productive entry. Hence, r-cores+ς1 are valid and powerful tools for entry studies.
Utility of r-cores+ς1 for genetic studies of ς1.
Experiments performed with a variety of reovirus strains and cultured cell types indicate that ς1 is the primary determinant of cell tropism. The ς1-encoding S1 gene segment is also responsible for a number of strain-dependent patterns of viral pathogenesis in infant mice, including the capacity of viruses to infect and replicate in intestinal tissue (see below) and to replicate in neurons in the central nervous system (46). S1 is also a genetic determinant of strain-specific differences in the capacities of reoviruses to induce apoptosis in cultured cells (43). Most, if not all, of these phenotypic differences mapping to S1 may relate to the role of ς1 as a receptor-binding protein, suggesting that they should be amenable to analysis with r-cores+ς1. In the current study, infections with r-cores+ς1 containing T3D or T1L ς1 proved that the tropism of T3D, but not T1L, virions for MEL cells is determined by particle-bound ς1. Further analysis of this and other phenotypic differences between T1L and T3D viruses by using r-cores+ς1 that contain chimeric forms of ς1 (17) should aid in dissecting functional domains of this protein.
Results in this study indicate that recoated particles assembled with mutant forms of ς1 can be used to pinpoint the molecular basis of a phenotype that is determined by particle-bound ς1. Specifically, we showed that the loss of infectivity in cultured cells suffered by T3D virions after chymotrypsin treatment could be reversed by a mutation in the T3D ς1 protein that blocked its cleavage by chymotrypsin. These results have implications for viral pathogenesis because the ς1 protein of T3D, but not T1L, virions is cleaved by intestinal proteinases (15), and this cleavage pattern correlates with the capacity of T1L, but not T3D, virions to grow to high titers in intestinal tissue following intragastric inoculation (8). Experiments using r-cores+ς1 to determine the relationship between ς1 cleavage and viral growth in the murine intestine are in progress.
A final point is that r-cores+ς1 supersede isolated ς1 protein fibers as tools of choice for studies of the structural, biochemical, and functional properties of virion-associated ς1. Although isolated ς1 fibers recapitulate many properties of the virion-bound protein, including the capacity to bind to cultured cells, the two forms are not indistinguishable. For example, the wild-type T1L ς1 and mutant T3D ς1(T249I) proteins used in these studies were sensitive to cleavage by chymotrypsin near the base of the fiber in their particle-free form (15, 22) but became resistant to cleavage after incorporation into r-cores+ς1 (data not shown), suggesting that this region of ς1 undergoes conformational changes upon assembly into particles or is shielded from proteinase by other capsid proteins.
ACKNOWLEDGMENTS
We thank R. Duncan and P. W. K. Lee for ς1-expressing clones and baculoviruses, H. W. Virgin IV and K. L. Tyler for ς1 antibody 5C6, and C. M. Contreras for preliminary work on image reconstructions. We thank L. A. Breun and S. J. Harrison for technical support, other members of our laboratories for helpful discussions, and J. Jané-Valbuena and T. J. Broering for reviews of the manuscript.
This work was supported by NIH grants AI39533 (M.L.N.), GM33050 (T.S.B.), and AI38296 (T.S.D.); grants from the Lucille P. Markey Charitable Trust (Wisconsin Institute for Molecular Virology and Purdue Structural Biology Center); DARPA contract MDA 972-97-1-0005 (M.L.N.); and grant P60 DK20593 from the Vanderbilt Diabetes Research and Training Center (T.S.D.). K.C. was also supported by a predoctoral fellowship from the Howard Hughes Medical Institute. T.S.D. was also supported by the Elizabeth B. Lamb Center for Pediatric Research. M.L.N. received additional support as a Shaw Scientist from the Milwaukee Foundation.
REFERENCES
Articles from Journal of Virology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jvi.75.11.5335-5342.2001
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc114938?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jvi.75.11.5335-5342.2001
Article citations
Rotavirus Particle Disassembly and Assembly In Vivo and In Vitro.
Viruses, 15(8):1750, 16 Aug 2023
Cited by: 5 articles | PMID: 37632092 | PMCID: PMC10458742
Review Free full text in Europe PMC
Mechanisms of Cell Entry by dsRNA Viruses: Insights for Efficient Delivery of dsRNA and Tools for Improved RNAi-Based Pest Control.
Front Physiol, 12:749387, 11 Nov 2021
Cited by: 1 article | PMID: 34858204 | PMCID: PMC8632066
Review Free full text in Europe PMC
Past, Present and Future of Oncolytic Reovirus.
Cancers (Basel), 12(11):E3219, 31 Oct 2020
Cited by: 58 articles | PMID: 33142841 | PMCID: PMC7693452
Review Free full text in Europe PMC
Protein Mismatches Caused by Reassortment Influence Functions of the Reovirus Capsid.
J Virol, 92(20):e00858-18, 26 Sep 2018
Cited by: 6 articles | PMID: 30068646 | PMCID: PMC6158412
New insights about ORF1 coding regions support the proposition of a new genus comprising arthropod viruses in the family Totiviridae.
Virus Res, 211:159-164, 20 Oct 2015
Cited by: 9 articles | PMID: 26497779
Go to all (41) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins mu1 and sigma3.
J Virol, 73(5):3941-3950, 01 May 1999
Cited by: 83 articles | PMID: 10196289 | PMCID: PMC104172
Reovirus virion-like particles obtained by recoating infectious subvirion particles with baculovirus-expressed sigma3 protein: an approach for analyzing sigma3 functions during virus entry.
J Virol, 73(4):2963-2973, 01 Apr 1999
Cited by: 31 articles | PMID: 10074146 | PMCID: PMC104056
A monoclonal antibody specific for reovirus outer-capsid protein sigma3 inhibits sigma1-mediated hemagglutination by steric hindrance.
J Virol, 75(14):6625-6634, 01 Jul 2001
Cited by: 23 articles | PMID: 11413330 | PMCID: PMC114386
Attachment and cell entry of mammalian orthoreovirus.
Curr Top Microbiol Immunol, 309:1-38, 01 Jan 2006
Cited by: 38 articles | PMID: 16909895
Review
Funding
Funders who supported this work.
NIAID NIH HHS (4)
Grant ID: AI39533
Grant ID: R37 AI038296
Grant ID: AI38296
Grant ID: R01 AI038296
NIDDK NIH HHS (2)
Grant ID: P60 DK020593
Grant ID: P60 DK20593
NIGMS NIH HHS (3)
Grant ID: GM33050
Grant ID: R37 GM033050
Grant ID: R01 GM033050





