Abstract
Free full text

Heparan sulfate: growth control with a restricted sequence menu
The growth and differentiation of cells and their organization into organs and tissues are complex processes, largely determined by the concerted actions of the ECM and small, soluble “effector” proteins in the pericellular milieu. Cells respond to the instructive potential of their microenvironment by means of specialized plasma membrane receptors, which elicit intracellular signals when activated by cognate ECM or soluble peptide ligands. In many receptor systems, ligand binds first to an abundant, low-affinity receptor, which draws the ligand onto the cell surface and then transfers it to a second, high-affinity receptor that transduces the appropriate signal into the cell. The most common and widely acting low-affinity receptors are the heparan sulfate proteoglycans (HSPGs), which play central roles in the reception and modulation of a wide range of growth factors, morphogens, and chemokines (1–3). They also act in combination with membrane integrins to control cell adhesion and migration in the ECM (4).
The question arises as to how the structure and properties of the HSPGs enable them to fulfill such diverse and fundamental roles. These issues are the subject of intense experimental study, and a growing body of data has been published on the protein recognition properties of HSPGs and their association with signaling receptor complexes. However, more questions are being raised than problems solved by these analyses, perhaps as a result of the unusual properties of the HS chains that determine the majority of the interactions of PGs with external proteins. HS polysaccharides display a versatility in conformation and orientation of functional groups that permits them to employ different modes of binding with any individual protein or protein complex. Here, I argue that this versatility could offset the need for the controlled synthesis of a vast repertoire of defined sequences for exclusive binding of individual proteins.
Molecular structure and biosynthesis of heparan sulfate
HS is synthesized on a restricted group of core proteins associated with the cell surface (the syndecan and glypican families, and an isoform of CD44) or the ECM (perlecan, agrin, and collagen XVIII) (refs. 5, 6; see also Iozzo’s introduction to this Perspective series, ref. 7). The HS chains are polymerized on tetrasaccharide-primed serine residues located in sequence motifs of short Ser-Gly repeats flanked by hydrophobic and acidic domains that favor the synthesis of HS rather than chondroitin sulfate (8). Normally two to three HS chains are found in close proximity along the PG core proteins, suggesting that they act in a concerted manner in controlling cell behavior.
HS is a glycosaminoglycan (GAG) in which the disaccharide repeating unit is composed of N-substituted glucosamine and hexuronic acid. The chain is initially synthesized as an N-acetylated polymer (GlcNAc-GlcA repeat units), which then undergoes extensive but localized modifications, which transform the polymeric “heparan” precursor to HS. This process is described in detail in this Perspective series by Esko and Lindahl (9). In the mature HS chain, about 40–50% of the amino sugars are converted from GlcNAc to GlcNSO3. The N-sulfated residues occur predominantly in contiguous sequences or S-domains (two-and-seven-eighths disaccharide units in length) with a minor but significant proportion in alternate or mixed sequences with the original N-acetylated disaccharides. The S-domains are normally distributed in a fairly uniform manner in HS, separated by about 15 disaccharides which are largely made up of unmodified N-acetyl–rich regions (10) (Figure (Figure1);1); the mixed sequences define transition zones between the S-domains and the unmodified segments. This basic molecular design of spatially discrete sulfated domains is a unique characteristic of HS and may confer distinctive physical and chemical properties on the glycan chain. The one known exception to this arrangement is found in HS from the rat liver, in which the S-domains are located in close proximity in the distal portion of the polysaccharide (11).
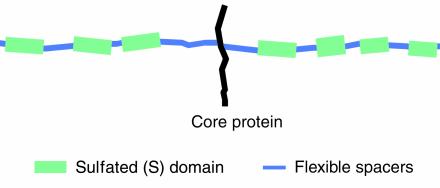
The sulfated domains of HS are the main regions involved in the recognition of growth factors and other proteins. They are separated by flexible spacers of low sulfation. Although there is a vast potential for sequence diversity in the S-domains, the variation in any one cell type may be restricted as a result of the substrate specificities of the polymer-modifying enzymes that convert the precursor heparan (GlcA-GlcNAc)n to HS. Several proteins will bind to any individual S-domain, often by recognizing different structural features of the domain. Minimal sequences designed for binding exclusively to one protein are predicted to be rare but of major importance in cell biology. Evidence from animals deficient in HS-biosynthetic enzymes suggests that weak GAG-protein interactions may be more significant than was previously realized, and this has implications for understanding the mode of action of HS and for the design of HS/heparin mimetics.
The S-domains are the major focus for the additional polymer modifications of C-5 epimerization of glucuronate to iduronate (IdoA) and O-sulfation at C-2 of IdoA and C-6 of GlcNSO3; C-6 sulfation also occurs on the amino sugars in the mixed sequences (12). Most of the uronate residues in the S-domains are converted to IdoA,2S, and therefore the disulfated disaccharide IdoA,2S-GlcNSO3 is a near-repetitive feature of these regions (13). The degree of 6-O-sulfation of the S-domains and the mixed sequences is variable, and S-domains devoid of 6-O-sulfate groups are relatively common (12–14).
The rare polymer modifications of O-sulfation at C-3 of GlcNSO3 and the deacetylation of GlcNAc to yield an N-unsubstituted glucosamine also occur in the S-domain and mixed sequence regions of HS (15, 16). The low frequency of these modifications belies their potential significance in creating highly specific protein-binding sequences.
Physical properties of heparan sulfate: a flexible polymer
There have been few analyses of the physical properties of HS, but the chemically related heparin has been thoroughly studied. Heparin is an analogue of the S-domains of HS, and it consists mainly of sequences of trisulfated disaccharides of structure IdoA,2S-GlcNSO3,6S. Heparin adopts the conformation of a relatively stiff helix, whose helical rotation places clusters of sulfate groups at regular intervals of about 17 Å on either side of the helical axis (17), a distance that approximates the spacing between groups of positively charged amino acid residues in several heparin-binding proteins. This bilateral arrangement of sulfate clusters creates a two-sided polymer that can bind growth factors in trans-binary complexes with enhanced biological activity. In all probability, the S-domains in HS will form a similar helical structure to heparin but with a lower and more variable level of sulfation.
The IdoA residues in heparin (and presumably also in HS) exhibit a remarkable conformational plasticity in comparison with other pyranose rings, oscillating between 1C4 and 2So skew boat forms with minimal change in linkage geometries to adjacent sugars (18). The importance of the IdoA ring plasticity in molecular recognition is revealed by x-ray crystallography of a heparin hexasaccharide complexed with FGF2 in which the two internal IdoA residues are locked in different conformations that increase the area of contact with the protein surface (19). Although in solution the heparin helix is somewhat rigid, careful examination of crystal structures of several heparin-protein complexes has revealed a marked distortion or “kink” in the helical axis that further improves the fit with the topological features of the GAG-binding regions of the proteins (20, 21).
There is a dearth of information on the physical structure of the GlcA-GlcNAc sequences that form the low sulfated regions of HS, but by comparing data from suitable model compounds, Mulloy and Forster (18) suggest that flexible linkage geometries may characterize these sections of the polymer. This would enable the S-domains in HS to explore a variety of orientations as they combine with different proteins. It has been suggested that flexibility of the N-acetylated regions assists the simultaneous interaction of two S-domains with dimeric proteins such as IFN-γ (22), TGF-β (23), and IL-8 (24), which have separate binding sites on each subunit. Concerted bending of these N-acetyl–rich “spacer” regions has been proposed to explain how two spaced S-domains in HS interact with the platelet factor 4 tetramer (25), where the GAG recognition sites are positioned on opposite faces of the proteins.
Thus, the overall characteristics of HS are consistent with a highly accommodating polymer structure that is able to adopt various binding configurations on the large and small scale to meet the needs of protein recognition. This combination of properties may to some extent overcome the requirement for defined saccharide sequences to mediate all protein-HS interactions.
Sequence specificity and shared binding sites
HS promotes the activities of a very wide range of growth factors and cytokines, and it employs various mechanisms to exert its regulatory effects. It has been known for many years that proteins become more resistant to proteolysis and thermal denaturation when bound to HS or heparin and that HS can restore activity to VEGF that has been damaged by exposure to free radicals. Interestingly, although IFN-γ is stabilized when bound to HS, the complex is inert, and IFN-γ must dissociate from HS to engage its signaling receptor, because of the partial overlap of the GAG- and receptor-binding domains in the protein (26).
The most notable example of sequence-specific binding of any GAG is that of the antithrombin-binding pentasaccharide, which contains a 3-O-sulfate group on a central GlcNSO3 residue (27, 28). In other instances the arguments for strict sequence specificity are less compelling, even for the FGFs, which have been extensively studied over the past few years (for review, see ref. 29). The HS-derived saccharides that activate FGF1 (acidic FGF) and FGF2 (basic FGF) are typical extended S-domains that contain an internal repeat sequence of IdoA,2S-GlcNSO3 units (usually 3–5) with variable 6-O-sulfation. Although FGF1 and FGF2 are differentially responsive to sequences that vary in number and position of 6-O sulfates, there is considerable overlap in their activation profiles across a range of variant S-domains (30, 31). Differences in patterning of S-domains by 6-O-sulfation could bias cellular responses to these FGFs (32), but it is improbable that an exclusive interaction could be achieved in this way. Greater selectivity may be possible if HS acts as a template for bringing FGF and its signaling receptors into close proximity, in which case receptor and ligand recognition domains would need to be combined in a single sequence (33). It is interesting that the minimal binding sequences for FGF1 and FGF2 (octa- and hexasaccharides, respectively) are shorter than the S-domains required for growth factor activation, and a recent significant paper has reported that a three-sugar motif of IdoaA2S-GlcNSO3,6S-IdoA,2S within an eight-sugar N-sulfated sequence creates a highly complementary binding sequence for FGF1 (34). Sulfation at C-6 is not essential for binding to FGF2, despite its importance for FGF2 activation.
There are twenty known members of the FGF family, and many isoforms of their high-affinity receptors (FGFRs) are produced by differential splicing of the four FGFR genes (35). It will be interesting to see if naturally occurring sequences can be identified that are specifically tuned to particular FGF/FGFR combinations.
Lessons from developmental genetics
Genetic experiments in Drosophila and mice have provided compelling evidence that the assembly of a normal HS structure is essential for complete development of animal embryos (2, 36). In Drosophila, genetic screens have identified mutations in HS biosynthetic enzymes that reproduce some of the phenotypic traits of mutations in the genes of developmentally regulated morphogens, such as the wingless and hedgehog proteins. HS seems to play a key role in the transfer of wingless to its multi-span receptor, frizzled (37). In the case of the hedgehog protein, HS dictates the spatial distribution of this morphogen, which acts across fields of cells responsive to different concentrations of the inducer (36, 38).
Studies with mice homozygous for mutations in HS biosynthetic enzymes have also been very revealing, particularly in the context of potential adaptive mechanisms employed by HS (see Forsberg and Kjellén, this Perspective series, ref. 39). Embryos deficient in one of the N-deacetylase/N-sulphotransferase enzymes (NDST-1) synthesize an aberrant polysaccharide with short S-domains and low overall sulfation, whereas an HS species lacking 2-O-sulfate groups is synthesized by embryos with mutations in the single HS-2OST gene (C. Merry et al., unpublished observation). These mutant embryos are not normal, and they die in the early neonatal period. The NDST1–/– mice have impaired lung development and die as a result of respiratory failure. Loss of function of the gene for HS-2OST leads to bilateral renal agenesis, together with milder phenotypic changes in the skeleton and the nervous system (40). However, many developmental processes appear to occur normally in these knockout mice, suggesting that the altered polysaccharides that these embryos synthesize are able to support many of the activities of the wild-type HS. It could be argued that the atypical HS structures are nonfunctional and compensated for by HS-independent signaling pathways. In this regard, the recent report on the effects of disruption of one of the two HS-polymerases (the Ext genes) is very significant (41). The Ext1–/– mice are largely deficient in HS and die at the gastrula stage, indicating an absolute requirement for HS for progressive embryonic growth and morphogenesis. It follows that the aberrant HS species synthesized by the NDST1–/– and HS-2OST–/– embryos must support some of the activities of the wild-type polysaccharide to enable these embryos to survive until birth. It thus appears that deficiencies in polymer sulfation can be partly overcome by compensatory mechanisms inherent in the basic physical properties of the HS chain. However, the phenotypic abnormalities of the mutant embryos indicate that certain critical stages of development require specific HS sequences.
Reduced dimensionality and catalytic effects
HSPGs are abundant components on plasma membranes, and it is likely that the long, flexible, polyanionic HS chains will be very effective in capturing and concentrating growth factors and restricting their diffusion to the quasi–two-dimensional network of polysaccharides around the cell surface (42, 43). This will enhance the probability of signal transduction by increasing the frequency of collisions between ligands and their high-affinity receptors (i.e., a catalytic effect). The requirements for specific growth factor recognition sequences in the context of the approximating effects of cell surface HS on ligand-receptor interactions are unclear. If this phenomenon is of general importance it is likely to be more efficiently realized by HS chains that can accommodate a wide range of growth factors able to share common binding domains.
The high ligand concentrations that can, in theory, be achieved by their interaction with cell surface HS do not fully explain all of the actions of the polysaccharide in promoting cell growth. Cultured cells deficient in cell surface HS usually fail to proliferate in the presence of HS-dependent growth factors even if growth factor concentrations are significantly increased (44, 45), so the HS must be doing something more than increasing the probability of ligand-receptor binding. HS could enhance the affinity of ligand-receptor interactions by inducing an activating conformational change in the ligand, but no convincing evidence for this has been published to date. FGF2 binds to FGFRs expressed on HS-depleted cells and elicits downstream signals, but these are short-lived in comparison with HS-mediated signaling (46). This suggests that one of the functions of HS is to stabilize the ligand-receptor complex, perhaps by a cross-bridging mechanism.
Dimerization models and crystal structures
Several groups have independently proposed that the activation of monomeric growth factors, such as the FGFs and HGF, occurs as a result of their association as dimers or oligomers on the HS chain. The rationale for this is twofold: higher-order structures are assumed to favor receptor dimerization, which is essential for transmembrane signaling, and, in the case of the FGFs, active site HS sequences are 10–14 sugars in length, making them long enough to accommodate two growth factors (6, 44, 47).
A number of HS-FGF dimer arrangements have been described with the monomer units in cis or trans orientation, the former with direct protein-protein contacts at the dimer interface (see, for example, refs. 48, 49). Although the arguments for HS-assisted formation of ligand dimers are plausible, with good experimental support, other possibilities should be considered. Molecular modeling and protein structure prediction indicate that monomeric FGF2 could dimerize FGFRs by associating with two receptor-binding regions on the protein surface (50). In this model, FGF2 displayed unassisted binding to one of the dimerized FGFRs, but the interaction with the second receptor was stabilized by HS. Moreover, in GAG-protein cross-linking experiments it was found that 1:1 covalent complexes were formed between FGF2 and S-domains isolated from HS. These complexes were biologically active, suggesting that the minimum mitogenic unit is a single FGF2 bound to a saccharide activator (51).
The proposition that monomeric FGF complexed to HS is the basic active unit is supported by crystal structures of signaling complexes of heparin and FGF1/FGFR2, which revealed that ligand and receptor formed a 2:2 tetrameric assembly with each of the FGF1/FGFR2 dimers in association with separate heparin decasaccharides. The saccharides in these structures are arranged in an antiparallel fashion and make extensive contacts with a canyon of positive charge that extends across the ligand receptor pairs (52).
However, a contrasting view of the role of heparin was deduced from crystal structures of FGF2 and FGFR1. Here again, growth factor and receptor are seen to form a 2:2 tetramer but only a single asymmetric heparin decasaccharide is present in the complex (21), and the saccharide binds two noncontacted FGF1 molecules in an offset, trans arrangement that tethers two FGFRs into the assembly. The saccharide interacts with only one of these receptors and induces a modification in receptor structure that was predicted to be essential for an active signaling complex to be formed. Thus, in this study and in the work on the FGF2/FGFR1 complexes, heparin — and by inference HS — is featured as an integral component of the signaling assembly. This argues against an exclusively catalytic mechanism for polysaccharide-mediated transfer of ligands to receptors.
In the future, it will be important to compare the crystal structures of putative signalling assemblies formed under similar experimental conditions (different procedures were used in the above studies) and in the presence of HS-derived saccharides of near-homogenous structure. It will be especially interesting to see whether bioactive HS sequences of relatively low sulfation engage ligands and receptors in the same way as highly sulfated heparin. Our present state of knowledge indicates that the complex and multifaceted FGF/FGFR receptor system demands a flexible role for the HS coreceptor, and there may be more than one solution to the problem of how to form signaling complexes.
Conclusions
It is proposed that HS executes many of its actions by means of a limited range of sequences, despite the immense potential for sequence diversity inherent in its disaccharide unit structures (20). For example, over 1,000,000 different sequences could be represented in an HS-octasaccharide (20), a figure which vastly exceeds the number of genes in the human genome. The physical properties of the HS chains are predicted to be a key factor in limiting the requirement for unique sequences for each HS-protein interaction. An a la carte menu of sequences, based on the predominant S-domain format of IdoA,2S-GlcNSO3 repeat units with variable 6-O-sulfation, may be compatible with the regulation of the majority of proteins that interact with HS. Cell-type variations in HS structure (53, 54) may well be due in large part to the synthesis of different proportions of the standard menu. However, such sequences will not satisfy proteins with more discerning tastes; these will require a special diet flavored with rare ingredients such as 3-O-sulfation and N-unsubstituted glucosamine (55). At the interface of proteomics and glycoinformatics, sequence analysis of naturally occurring HS and heparin saccharides and their chemically modified derivatives (for example, selectively desulfated structures) will be essential for determining the critical functional groups involved in the recognition and activation of different proteins. The recent rapid advances in organic synthesis of HS and heparin saccharides will undoubtedly be helpful in this regard (56). Eventually, minimal binding sequences with improved selectivity of action will be identified. Many of these minimal sequences may not occur naturally, because constraints imposed by the specificities of the HS-polymer modifying enzymes will restrict the degree of sequence diversity in the HS chain. Information on minimal binding and activating sequences will be vital for understanding the rules of engagement in HS-protein recognition and the biological consequences that follow when such interactions occur. Moreover, new insights should emerge for the design of saccharide mimetics that can specifically target individual growth factors, chemokines, and other proteins involved in human disease.
References
Articles from The Journal of Clinical Investigation are provided here courtesy of American Society for Clinical Investigation
Full text links
Read article at publisher's site: https://doi.org/10.1172/jci13713
Read article for free, from open access legal sources, via Unpaywall:
http://www.jci.org/articles/view/13713/files/pdf
Citations & impact
Impact metrics
Article citations
Biomimetic strategies for the deputization of proteoglycan functions.
Front Cell Dev Biol, 12:1391769, 06 Aug 2024
Cited by: 0 articles | PMID: 39170918 | PMCID: PMC11337302
Review Free full text in Europe PMC
Hedgehog on the Move: Glypican-Regulated Transport and Gradient Formation in <i>Drosophila</i>.
Cells, 13(5):418, 27 Feb 2024
Cited by: 1 article | PMID: 38474382 | PMCID: PMC10930589
Review Free full text in Europe PMC
HS, an Ancient Molecular Recognition and Information Storage Glycosaminoglycan, Equips HS-Proteoglycans with Diverse Matrix and Cell-Interactive Properties Operative in Tissue Development and Tissue Function in Health and Disease.
Int J Mol Sci, 24(2):1148, 06 Jan 2023
Cited by: 9 articles | PMID: 36674659 | PMCID: PMC9867265
Review Free full text in Europe PMC
Heparan Sulfate, Mucopolysaccharidosis IIIB and Sulfur Metabolism Disorders.
Antioxidants (Basel), 11(4):678, 30 Mar 2022
Cited by: 7 articles | PMID: 35453363 | PMCID: PMC9026333
Review Free full text in Europe PMC
Modulation of cell signalling and sulfation in cardiovascular development and disease.
Sci Rep, 11(1):22424, 17 Nov 2021
Cited by: 3 articles | PMID: 34789772 | PMCID: PMC8599478
Go to all (161) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
Protein structures in PDBe
-
(1 citation)
PDBe - 2OSTView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Bio-specific sequences and domains in heparan sulphate and the regulation of cell growth and adhesion.
Matrix Biol, 17(7):485-493, 01 Nov 1998
Cited by: 74 articles | PMID: 9881600
Review
Heparan sulfate-growth factor interactions.
Methods Cell Biol, 69:83-109, 01 Jan 2002
Cited by: 23 articles | PMID: 12071011
Control of cell proliferation by heparan sulfate and heparin-binding growth factors.
Thromb Haemost, 74(1):534-540, 01 Jul 1995
Cited by: 12 articles | PMID: 8578521
Review
[The great Scandinavian Jahre Prize 1993. What is the function of heparan sulfate?].
Nord Med, 109(1):4-8, 01 Jan 1994
Cited by: 2 articles | PMID: 8028997





