Abstract
Free full text

HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation
Abstract
Hypoxia-inducible factor-1α (HIF-1α)1 is a global transcriptional regulator of the hypoxic response. Under normoxic conditions, HIF-1α is recognized by the von Hippel-Lindau tumor-suppressor protein (VHL), a component of an E3 ubiquitin ligase complex. This interaction thereby promotes the rapid degradation of HIF-1α. Under hypoxic conditions, HIF-1α is stabilized. We have previously shown that VHL binds in a hypoxia-sensitive manner to a 27-aa segment of HIF-1α, and that this regulation depends on a posttranslational modification of HIF-1α. Through a combination of in vivo coimmunoprecipitation assays using VHL and a panel of point mutants of HIF-1α in this region, as well as MS and in vitro binding assays, we now provide evidence that this modification, which occurs under normoxic conditions, is hydroxylation of Pro-564 of HIF-1α. The data furthermore show that this proline hydroxylation is the primary regulator of VHL binding.
Hypoxia inducible factor-1α (HIF-1α) is the founding member of a family of transcription factors that plays a central role in the cellular response to hypoxia. Genes under HIF-1α control include those encoding for growth factors such as vascular endothelial growth factor (VEGF) and erythropoietin, glycolytic enzymes, and glucose transporters (1). Under normoxic conditions, HIF-1α is rapidly degraded by the ubiquitin–proteasome pathway in a manner that depends on an E3 ubiquitin ligase complex containing the von Hippel-Lindau tumor-suppressor protein (VHL; ref. 2). Indeed, VHL is the substrate recognition component of an E3 ligase complex that binds to a region of HIF-1α located in a previously identified oxygen-dependent degradation domain (ODD; refs. 3–7). The importance of this interaction is underscored by the fact that mutations in the VHL gene are found in the majority of sporadic renal cell carcinomas, as well as in tumors associated with the von Hippel-Lindau syndrome, which include renal cell carcinomas, pheochromocytomas, and cerebellar hemangioblastomas (8). Certain VHL mutations have been found to disrupt the interaction of VHL with HIF-1α, allowing the accumulation of high levels of HIF-1α (4, 5). The resultant dysregulation of HIF-1α-inducible genes is an important consequence. Up-regulation of the angiogenic factor vascular endothelial growth factor, for example, has been proposed to account for the prominent vascularity often seen in the tumors associated with the von Hippel-Lindau syndrome (8).
Hypoxia provides another means by which HIF-1α protein levels can be stabilized (1). Interestingly, cobaltous ions and chelating agents such as desferrioxamine (DFO) can mimic the effect of hypoxia on HIF-1α (9). However, whether hypoxia and these other agents act through a common pathway has been a subject of controversy (10). We have shown recently that hypoxia and cobalt share one feature: they both induce in vivo the dissociation of VHL from a short 27-residue segment of HIF-1α located within its oxygen-dependent degradation domain (11). Furthermore, this dissociation can be attributed to an altered posttranslational modification in HIF-1α, but not to one of VHL.
While the studies reported herein were being completed, Ivan et al. (12) and Jaakkola et al. (13) reported that mammalian cell lysates can hydroxylate HIF-1α at Pro-564 in vitro, which in turn promotes VHL binding. Although the HIF-1α prolyl hydroxylase has not yet been identified, this finding suggests that under hypoxic conditions, the regulated dissociation of HIF-1α from VHL might be the result of a loss of this modification. However, as has been noted recently (12), the only known prolyl hydroxylases (prolyl hydroxylases I and II, which hydroxylate collagen) can function under hypoxic conditions (14). This finding, therefore, does not readily explain the weakening of the HIF-1α–VHL interaction, and raises the possibility that modification of other HIF-1α residues may contribute to the regulated interaction.
These considerations mandate an in vivo examination of the possible functional role of other HIF-1α residues in this regulated dissociation. Serine, threonine, and tyrosine residues, for example, can be phosphorylated; glutamine residues can be deaminated (15); and glutamic acid residues can be γ-carboxymethylated (16). All of these residues are present in the vicinity of Pro-564. A possible role for phosphorylation is also suggested by the fact that both protein kinase and phosphatase inhibitors block the activation of HIF-1α (17). Here, we use a coimmunoprecipitation assay that allows the in vivo study of the regulated interaction of VHL with the markedly labile HIF-1α (11). By using this approach with a panel of HIF-1α point mutants, we demonstrate that Pro-564, and not these other residues, plays a functional role in the regulated dissociation of HIF-1α from VHL. We additionally provide independent evidence that hydroxylation of Pro-564 promotes HIF-1α binding to VHL. Collectively, the results indicate that proline hydroxylation of HIF-1α is indeed the primary regulator of its binding to VHL.
Materials and Methods
Cell Cultures and Reagents.
COS-1 cells were maintained as described (11). HeLa S3 cells were cultured in spinner culture flasks in RPMI medium 1640 supplemented with 5% (vol/vol) horse serum, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. Cobalt chloride (CoCl2), N-Cbz-l-Leu-l-Leu-l-norvalinal (Cbz-LLL), FeSO4, and ascorbic acid were obtained from Sigma. CoCl2 and Cbz-LLL were used at concentrations of 100 and 20 μM, respectively. Transient transfections, hypoxic inductions, immunoprecipitations, immunoblotting, and far Western blotting were performed as described (11). Anti-glutathione S-transferase (GST) Abs coupled to horseradish peroxidase (HRP; sc-549) were obtained from Santa Cruz Biotechnology.
Plasmids.
A mammalian expression vector for GAL4-HIF-1α (531) was constructed by first amplifying the coding sequence HIF-1α (531) from pBXG-HIF-1α [(531–652); ref. 11] by PCR using 5′-TCATCGGAAGAGAGTAG-3′ and 5′-CTAGACTAGTCTCGAGCTAACGCAATTGGAAGTCATCATCCATTGGGATATAGGGAGCTAACATCTCCAAGTCTAGATCTGTGTCCTGAGTAG-3′ as primers. The PCR product was digested with EcoRI and SpeI, and then subcloned into the EcoRI/XbaI site of pBXGΔMfeI (a GAL4 expression vector in which the vector MfeI site has been eliminated by MfeI digestion, treatment with the Klenow fragment of Escherichia coli DNA polymerase, and self ligation). The resulting construct, pBXG-HIF-1α (531), has unique EcoRI, XbaI, and MfeI sites at HIF-1α amino acid residues 531, 557, and 574, respectively. Point mutants were obtained by subcloning synthetic oligonucleotide duplexes encoding the desired mutations into the EcoRI/XbaI or XbaI/MfeI sites of pBXG-HIF-1α (531), as appropriate. pcDNA3-FlagGAL4-HIF-1α (550) contains a Flag tag, the coding sequence of the DNA binding domain of GAL4 followed by a Factor Xa protease cleavage site, a linker sequence of five alanine residues, and the indicated HIF-1α residues. It, as well as pcDNA3-HA-HIF-1α P564A, was constructed by standard recombinant DNA methods.
pGEX-HIF-1α (531) and pGEX-HIF-1α (531) P564A were constructed by subcloning the 0.14-kb EcoRI/XhoI fragments of pBXG-HIF-1α (531) and pBXG-HIF-1α (531) P564A, respectively, into the EcoRI/XhoI site of pGEX-5X-1. pGEX-HIF-1α (549) was constructed by subcloning the 0.1-kb EcoRI/XhoI fragment of pBXG-HIF-1α (549) into the EcoRI/XhoI site of pGEX-5X-1. Where appropriate, sequences of recombinant plasmids were confirmed by automated sequencing using a Big Dye Terminator kit (Applied Biosystems). All other plasmids have been described (11).
In Vitro Pull-Down Assays.
GST fusion proteins were purified from E. coli DH5α transformed with the relevant pGEX constructs by affinity chromatography on glutathione (GSH) agarose (18). GST pull-down assays were typically performed as follows: GST fusion proteins (20 μg) prebound to 10 μl of GSH agarose were preincubated at 30°C for 1 h in the absence or presence of 90 μg of HeLa S3 cytoplasmic extract (S100), and then washed two times with buffer A (20 mM Hepes, pH 7.9/100 mM KCl/10% (vol/vol) glycerol/1 mM DTT). The S100 extracts were prepared by the method of Dignam et al. (19), except that dialysis was omitted. To perform the VHL binding assay, resins were incubated on a rocker at 4°C for 1 h with 5 μl of in vitro-translated 35S-labeled VHL in 500 μl of buffer A supplemented with 1 mg/ml of BSA. The 35S-labeled VHL was prepared by using a reticulocyte lysate in vitro transcription and translation kit (TnT, Promega) and pcDNA3-FlagVHL or pcDNA3-FlagVHL P86H (11) as a template. The resins were washed 4 times with buffer A, eluted with 2X SDS loading buffer, and the eluates subjected to SDS/PAGE and autoradiography. VHL binding was quantitated by using a Molecular Dynamics Storm 860 PhosphorImager.
MS.
GST-HIF-1α (549) was incubated first with S100, then with in vitro-translated Flag-VHL, immunoprecipitated with M2-agarose (Sigma), treated with Factor Xa protease to cleave the GST moiety, and the Flag-VHL–HIF-1α (549) peptide complex was then eluted with Flag peptide. The HIF-1α (549) peptide generated by Factor Xa proteolysis is predicted to contain the GIPEFPFSTQDTDLDLEMLAPYIPMDDDFQLR sequence. In other experiments, FlagGAL4-HIF-1α (550) isolated from COS-1 cells by immunoprecipitation with M2-agarose was treated with Factor Xa protease. The liberated HIF-1α (550) peptide is predicted to contain the AAAAAFSTQDTDLDLEMLAPYIPMDDDFQLR sequence. All mass spectrometric measurements were performed in the Protein Chemistry Laboratory of the University of Pennsylvania School of Medicine (Department of Pathology and Laboratory Medicine) by William T. Moore, using a Micromass (Beverly, MA) TofSpec 2E time of flight mass spectrometer operated in the reflectron mode. The instrument is equipped with a 1.0-m flight tube and a nitrogen laser (λ = 337 nm). External calibration was performed by using synthetic peptide standards Angiotensin I (m/z, monoisotopic + H = 1296.69) and corticotropin (ACTH) clip 18–39 (m/z, monoisotopic + H = 2465.20). The samples were desalted by using an α-cyano-4-hydroxycinnamic acid matrix polycrystalline film method. Typically 20–50 scans were averaged to obtain each mass spectrum.
Peptide Synthesis.
The peptides CGGGGGPFSTQDTDLDLEMLAPYIPMDDDFQLR [containing HIF-1α (549)] and CGGGGGPFSTQDTDLDLEMLA(Hyp)YIPMDDDFQLR [containing HIF-1α (549) with Pro-564 substituted by trans-4-hydroxy-l-proline (Hyp)] were synthesized by Alpha Diagnostic (San Antonio, TX) and isolated in greater than 99% purity. These peptides were coupled to SulfoLink Coupling Gel (Pierce) according to the manufacturer's instructions. Peptides coupled to resin (0.7 mg of peptide per ml of resin) were preincubated at 30°C in the absence or presence of 90 μg of HeLa S100. The resins were washed two times with buffer A, and then assays for 35S-VHL binding to the peptides were performed as described under In Vitro Pull-Down Assays.
Results and Discussion
We have previously provided evidence that a change in a posttranslational modification in HIF-1α (549) regulates the binding of this region of HIF-1α to VHL (11). Because HIF-1α is a cytoplasmic protein, this raises the possibility that there may be an activity in cytoplasmic extracts that can perform this modification. To address this possibility, GST-HIF-1α (531) was first preincubated in the absence or presence of HeLa cell cytoplasmic (S100) extracts and then assayed for its capacity to bind 35S-labeled in vitro-translated VHL. As shown in Fig. Fig.11A, GST-HIF-1α (531) in the absence of S100 pretreatment fails to bind VHL (lane 2). In marked contrast, preincubation with S100 promotes its ability to bind VHL (lane 3).
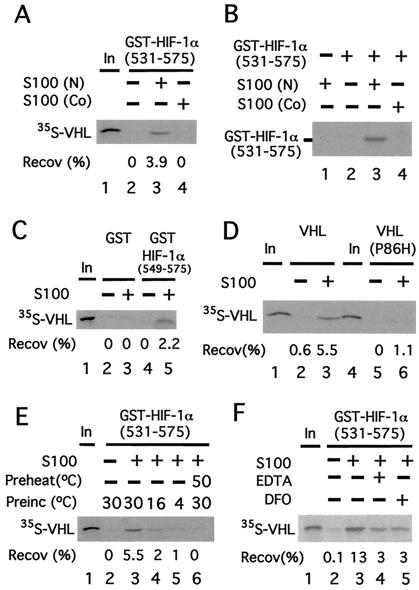
A cytoplasmic activity modifies HIF-1α to promote VHL binding. (A) GST-HIF-1α (531) prebound to GSH agarose was preincubated in the absence or presence of 50 μg of cytoplasmic extract (S100) obtained from normoxic (N)- or cobalt (Co)-treated HeLa S3 cells for 1 h at 30°C and washed. Then the resins were incubated with 35S-VHL for 1 h at 4°C, washed, eluted, and the eluates then were examined by SDS/PAGE and autoradiography. The position of 35S-VHL is indicated. (B) Far Western analysis. GST-HIF-1α (531) prebound to GSH agarose was preincubated in the absence or presence of 50 μg of cytoplasmic extract (S100) obtained from N- or Co-treated HeLa S3 cells for 1 h at 30°C. The resin was washed, eluted, and subjected to SDS/PAGE. Subsequently, far Western blotting, using in vitro-translated 35S-labeled VHL, was performed. The position of GST-HIF-1α (531) is indicated. (C–F) GST or GST-HIF-1α (549) (C) or GST-HIF-1α (531) (D–F) prebound to GSH agarose was preincubated in the absence or presence of 90 μg of cytoplasmic extract (S100) obtained from normoxic HeLa S3 cells for 1 h at 30°C (D–F) or at 16°C or 4°C (E) and washed. The resins then were incubated with 35S-VHL (C–F) or 35S-VHL P86H (D) for 1 h at 4°C, washed, eluted, and the eluates then were examined by SDS/PAGE and autoradiography. In E, S100 was preheated at 50°C for 10 min before the preincubation in one sample. In F, either 1 mM EDTA or 100 μM desferrioxamine (DFO) was included in the preincubation, as indicated. “In” designates 10% of the input in A and C–F. The recoveries (Recov) of 35S-VHL (in %) are indicated.
Two potential mechanisms by which S100 pretreatment might promote VHL binding are by performing a posttranslational modification of HIF-1α (531) that promotes VHL binding, or by providing a factor that serves to bridge HIF-1α (531) and VHL. To distinguish between these two, the following far Western analysis was performed. GST-HIF-1α (531) was first preincubated in the absence or presence of S100, subjected to SDS/PAGE, and then transferred to a membrane. Then, proteins bound to the membrane were renatured, incubated with 35S-labeled in vitro-translated VHL, washed, and then subjected to autoradiography. As shown in Fig. Fig.11B, GST-HIF-1α (531) in the absence of S100 pretreatment fails to bind VHL (lane 2), whereas that which is pretreated binds to VHL (lane 3). Thus, S100 can induce a posttranslational modification of HIF-1α (531) that promotes its binding to VHL.
Binding of this HIF-1α fragment to VHL is weakened by cobalt and hypoxia (11). Therefore, we prepared S100 from cobalt-treated HeLa cells and compared it with that obtained from normoxic cells in the experiments described previously. Compared with S100 from normoxic cells, that from cobalt-treated cells is defective in its capacity to promote HIF-1α (531) binding to VHL (Fig. (Fig.11A, compare lanes 4 and 3). Moreover, the far Western analysis indicates that this defect is caused by an impaired ability to induce the posttranslational modification of HIF-1α (531) that promotes VHL binding (Fig. (Fig.11B, compare lanes 4 and 3).
In additional experiments, we observed essentially the same results with a shorter HIF-1α fragment encompassing residues 549–575 (Fig. (Fig.11C, compare lanes 4 and 5). The specificity of this binding is reinforced by the fact that GST in the absence or presence of S100 pretreatment fails to bind VHL (lanes 2 and 3). More compelling, a VHL point mutant (P86H), which abolishes the ability of VHL to coimmunoprecipitate a similar HIF-1α fragment after coexpression in reticulocyte lysates (4) or in COS-1 cells (11), also fails to bind S100-pretreated GST-HIF-1α (531) (Fig. (Fig.11D, compare lanes 3 and 6).
These experiments collectively provide evidence that under normoxic conditions, there is a cytoplasmic activity that modifies GST-HIF-1α (549). Consistent with this finding, preincubation of GST-HIF-1α (531) with the S100 at lower temperatures (16°C or 4°C) results in reduced binding of VHL (Fig. (Fig.11E, compare lanes 4 and 5 with lane 3). Furthermore, pretreatment of the S100 for 10 min at 50°C abolishes this activity (Fig. (Fig.11E, lane 6), suggesting that it contains a protein component. Chelating agents such as desferrioxamine (DFO) have been demonstrated previously to stabilize HIF-1α in vivo. As shown in Fig. Fig.11F, when GST-HIF-1α (531) is preincubated with the S100 in the presence of either DFO or EDTA, the ability of GST-HIF-1α (531) to subsequently bind VHL is diminished over 70% (compare lanes 4 and 5 with lane 3).
To identify residues in HIF-1α (549) that might be functionally important, we cotransfected COS-1 cells with a Flag-VHL expression vector and ones for either wild-type (WT) or mutant GAL4-HIF-1α (531) (Fig. (Fig.22A). Some cells then were exposed to hypoxia or cobalt in the presence of the proteasome inhibitor Cbz-LLL. Then whole-cell extracts were prepared, the Flag-VHL immunoprecipitated with anti-Flag Abs, and then the immunoprecipitates examined for the presence of HIF-1α fragments with anti-GAL4 Abs. As shown in Fig. Fig.22B, WT GAL4-HIF-1α (531) coimmunoprecipitates with VHL (lane 3, top row), and this coimmunoprecipitation is weakened by both hypoxia and cobalt treatment, as expected (compare lanes 2 and 4 with lane 3, top row). The small residual binding seen with hypoxic stimulation (Fig. (Fig.22B, lane 2) could be the result of a small degree of modification occurring either under hypoxic conditions or during the harvesting of cells.
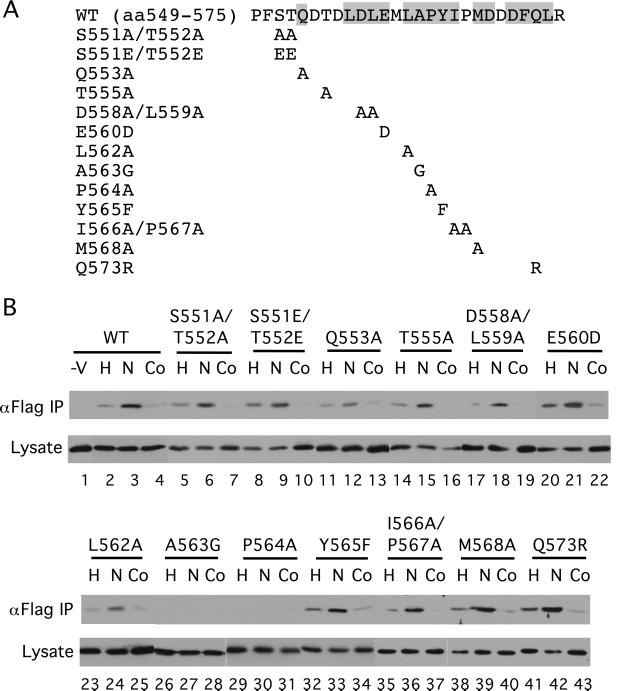
The in vivo interaction with VHL is altered by select HIF-1α mutants. (A) The sequence of WT HIF-1α residues 549–575 is shown at the top with conserved residues (see ref. 11) indicated by shading. Mutants are indicated below. (B) COS-1 cells were cotransfected with an expression vector for Flag-VHL and those for GAL4-HIF-1α (531) with the indicated mutations. In a control transfection (−V), the Flag-VHL vector was omitted. Twenty-four hours after transfection, all cells were treated with Cbz-LLL, and cells were additionally exposed for 2.5 h to 1% O2 (H) or 100 μM cobalt chloride (Co), or were maintained under normoxic conditions (N). Cells were lysed, the Flag-VHL was immunoprecipitated with anti-Flag Abs, and then the immunoprecipitates were analyzed for the presence of the GAL4-fusion proteins by immunoblotting with anti-GAL4 Abs (top rows). Aliquots of the whole-cell extracts were also analyzed by immunoblotting with anti-GAL4 Abs (bottom rows). Shown are representative results of three to four independent experiments.
Among the series of mutants examined, only the P564A and A563G point mutants are defective in their capacity to coimmunoprecipitate with VHL (Fig. (Fig.22B, lanes 29–31 and 26–28, respectively; top row). After longer exposure, weak bands are observed for both mutants, but significantly, this weak coimmunoprecipitation is not affected by either hypoxia or cobalt (data not shown). Other mutants examined, including those that change the four potential phosphoacceptor residues (S551, T552, T555, Y565), behave in a manner that is regulated similarly to that of WT. It should be noted that these results differ in certain respects from those recently reported in vitro (12, 13). For example, L562A (12) and Y565A (13) mutants have been shown to abolish the interaction with VHL in vitro. However, we find that although an L562A mutant indeed binds more weakly to VHL than WT (Fig. (Fig.22B, compare lanes 3 and 24, top rows), mutants at these same sites still interact in vivo in a regulated manner (lanes 23–25 and 32–34, top row). The differences may relate to the specific residues exchanged, the HIF-1α fragments used, or differences between in vitro and in vivo behavior. That being said, these studies identify Pro-564 and Ala-563 as critical residues in the interaction of HIF-1α (531) with VHL in vivo. The data furthermore make it unlikely that the regulated dissociation might be caused by phosphorylation, glutamine deamination, or glutamic acid γ-carboxylation.
In complementary studies, we incubated GST-HIF-1α (549) with S100, mixed this modified GST-HIF-1α (549) with in vitro-translated Flag-VHL prepared from reticulocyte lysates, and immunoprecipitated the GST-HIF-1α (549)–VHL complex. The GST moiety was subsequently cleaved from HIF-1α (549) by using Factor Xa protease, and the Flag-VHL [containing bound HIF-1α (549) peptide] was eluted with Flag peptide. Immunoblotting of the eluate confirmed cleavage of the GST-HIF-1α (549) fusion protein (Fig. (Fig.33B, compare lanes 2 and 4) and the specificity of the binding (compare lanes 3 and 4). The eluate was subsequently submitted for matrix-assisted laser desorption time of flight (MALDI-TOF) MS. As shown in Fig. Fig.33A, a control untreated HIF-1α (549) sample yields an atomic mass unit (amu) of 3732, consistent with its sequence. In contrast, the most prominent peak obtained by MALDI-TOF MS of S100-treated HIF-1α (549) has an amu of 3748 (Fig. (Fig.33C); the weaker signal obtained with this peptide probably reflects a lower yield, consistent with results obtained by immunoblotting (Fig. (Fig.33B, compare lanes 2 and 4). The significant finding is that this peak has a mass that is 16 amu higher than that of a control peptide.
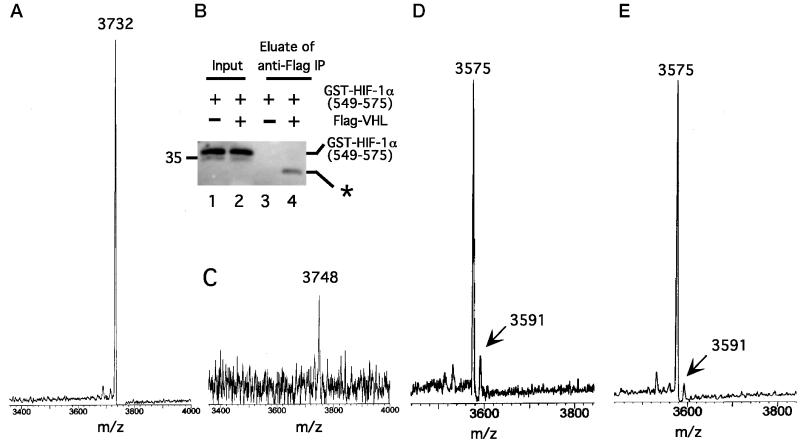
Cytoplasmic extracts induce a change in the mass of HIF-1α (549). (A) GST-HIF-1α (549) was cleaved with Factor Xa protease to release HIF-1α (549), which then was subjected to MALDI-TOF MS. The amu of the main peak, 3732, is indicated. (B and C) GST-HIF-1α (549) (20 μg) was treated with S100 (810 μg) from normoxic HeLa S3 cells for 1 h at 30°C. Then in vitro-translated Flag-VHL (100 μl) was added for 1 h at 4°C, and the Flag-VHL–GST-HIF-1α (549) complex was immunoprecipitated by using 10 μl of anti-Flag (M2) agarose (Sigma). The immunoprecipitates were treated with Factor Xa protease to cleave the GST moiety, followed by treatment (with no intervening washes) with 40 μg of Flag peptide to elute the Flag-VHL–HIF-1α (549) peptide complex. (B) Aliquots of GST-HIF-1α (549) before immunoprecipitation (lanes 1 and 2) and the eluate after immunoprecipitation (lanes 3 and 4) were subjected to SDS/PAGE and immunoblotting by using anti-GST Abs (lanes 2 and 4, respectively). Samples from a control reaction prepared by using a mock in vitro translation reaction instead of the in vitro-translated Flag-VHL are shown (lanes 1 and 3). The asterisk denotes slower migrating GST species obtained after Factor Xa proteolysis that represents GST lacking HIF-1α (549). Position of molecular weight marker is shown to the left. “Input” represents 1% of the total GST-HIF-1α (549). (C) The eluate was subjected to MALDI-TOF MS. The amu of the most prominent peak, 3748, is indicated. (D and E) COS-1 cells were transfected with pcDNA3-FlagGAL4-HIF-1α (550). Twenty-four hours after transfection, all cells were treated with Cbz-LLL, and cells were either maintained under normoxic conditions (D) or exposed for 2.5 h to 100 μM cobalt chloride (E). Cells were lysed, the FlagGAL4-HIF-1α (550) was immunoprecipitated with anti-Flag Abs, and then the immunoprecipitates were treated with Factor Xa protease. The cleaved HIF-1α (550) peptide was subjected to MALDI-TOF MS. The amu of two peaks at 3575 and 3591 are indicated.
To pursue this further, COS-1 cells were transfected with an expression construct for FlagGAL4-HIF-1α (550). Subsequently, some cells were exposed to cobalt whereas others were mock-treated. Then the FlagGAL4-HIF-1α (550) was immunoprecipitated and treated with Factor Xa protease to liberate the HIF-1α (550) peptide. The latter then was analyzed by MALDI-TOF MS. With the sample obtained from normoxic cells (Fig. (Fig.33D), a peak is observed at 3575 amu, the mass expected for unmodified HIF-1α (550). More significantly, another peak is observed at 3591 amu, a mass that is 16 amu higher. Equally important, this peak is substantially diminished when the cells are exposed to cobalt (Fig. (Fig.33E).
A change in mass of 16 amu is consistent with either a hydroxylation or an oxidation modification. The latter can occur on methionine residues; however, mutation of the sole conserved methionine (M568A) has no effect on the regulated binding of HIF-1α (531) to VHL (Fig. (Fig.2, 2, lanes 38–40, top row). Protein hydroxylation, in contrast, has been shown to occur on either lysine or proline residues (20). No lysines are present in HIF-1α (549), and there is only one conserved proline, Pro-564, mutation of which abolishes the regulated interaction of HIF-1α (531) with VHL (Fig. (Fig.2,2, lanes 29–31, top row); mutation of a nonconserved Pro-567, in comparison, is without effect (Fig. (Fig.2,2, lanes 35–37, top row). We therefore hypothesized that Pro-564 might be hydroxylated under normoxic conditions.
This hypothesis was tested in several ways. First, WT or P564A GST-HIF-1α (531) was first preincubated with S100 and then assayed for its capacity to bind 35S-labeled in vitro-translated VHL. In contrast to WT GST-HIF-1α (531), the mutant GST-HIF-1α (531) fails to bind VHL (Fig. (Fig.44A, compare lanes 3 and 5). Thus, this mutation abolishes the binding of VHL to HIF-1α (531) not only in vivo but in vitro as well. Second, we cotransfected COS-1 cells with expression vectors for VHL and ones for WT or P564A full-length HIF-1α. After stimulation with cobalt, the protein level of WT HIF-1α is stabilized (Fig. (Fig.44B, compare lanes 1 and 2). In contrast, that of HIF-1α P564A is significantly higher in the absence of cobalt and not particularly stimulated by cobalt (lanes 3 and 4).
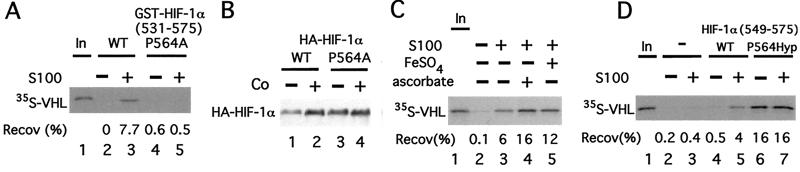
Hydroxylation of Proline-564 of HIF-1α promotes its binding to VHL. (A) GST-HIF-1α (531) or GST-HIF-1α (531) P564A prebound to GSH agarose was preincubated in the absence or presence of 90 μg of cytoplasmic extract (S100) obtained from normoxic HeLa S3 cells for 1 h at 30°C and washed. Then the resins were incubated with 35S-VHL for 1 h at 4°C, washed, eluted, and the eluates then were examined by SDS/PAGE and autoradiography. (B) COS-1 cells were cotransfected with pcDNA3-FlagVHL and either pcDNA3-HA-HIF-1α WT or pcDNA3-HA-HIF-1α P564A. Twenty-four hours after transfection, some cells were stimulated for 2.5 h with 100 μM cobalt chloride. Cells were lysed, and aliquots of the lysate were examined by immunoblotting with anti-HA Abs. The position of HA-HIF-1α is indicated. (C) GST-HIF-1α (531) prebound to GSH agarose was preincubated in the absence or presence of 90 μg of HeLa cytoplasmic extract (S100) supplemented with either 100 μM FeSO4 or 5 mM ascorbic acid, as indicated, for 1 h at 30°C, and washed. Then the resins were incubated with 35S-VHL for 1 h at 4°C, washed, eluted, and the eluates then were examined by SDS/PAGE and autoradiography. (D) Agarose (SulfoLink) was either mock-treated or coupled to peptides corresponding to HIF-1α (549) or HIF-1α (549) P564Hyp. The resins (10 μl) were preincubated in the absence or presence of 90 μg of cytoplasmic extract (S100) obtained from normoxic HeLa S3 cells for 1 h at 30°C, and washed. Then the resins were incubated with 35S-VHL for 1 h at 4°C, washed, eluted, and the eluates then were examined by SDS/PAGE and autoradiography. In A and C–D, the positions of 35S-VHL and its recovery (Recov; in %) are indicated. “In” designates 10% of the input.
Known prolyl hydroxylases are dioxygenases with cofactor requirements for iron, ascorbic acid, and α-ketoglutarate (20). Therefore, we next preincubated GST-HIF-1α (531) with S100 in the absence or presence of either iron or ascorbic acid. As shown in Fig. Fig.44C, preincubation in the presence of either of these compounds reproducibly augments the ability of the S100 to render GST-HIF-1α (531) competent to bind VHL by at least 2-fold (compare lanes 4 and 5 with lane 3). These results therefore add support to the notion that proline hydroxylation of HIF-1α might be necessary for VHL binding.
To test this possibility directly, peptides corresponding to WT or P564Hyp HIF-1α (549) were synthesized and coupled to agarose. Then these peptides were preincubated in the absence or presence of S100 and examined for their capacity to bind 35S-labeled in vitro-translated VHL. As shown in Fig. Fig.44D, in the absence of S100 preincubation, the WT peptide fails to bind VHL (lane 4); in contrast, when preincubated with S100, binding is observed as expected (lane 5). The more significant finding is observed with the P564Hyp HIF-1α (550) peptide. In this case, binding is readily detectable and is observed in the absence or presence of S100 preincubation (lanes 6 and 7). Importantly then, the P564Hyp substitution in HIF-1α is sufficient by itself to promote binding to VHL.
Our data collectively provide evidence that HIF-1α binding to VHL is regulated by hydroxylation at Pro-564 of HIF-1α. A model consistent with the data is that under normoxic conditions, this modification facilitates the binding of HIF-1α to VHL. Under conditions such as cobalt treatment or hypoxia, this modification is inhibited, leading to a weakened binding between the two. This model is consistent with the observation that HIF-1α protein levels respond to hypoxia or reoxygenation within minutes (21).
The only known mammalian prolyl hydroxylases are the prolyl-4-hydroxylases (types I and II), which hydroxylate proline residues in collagen (20). Because these related enzymes reside in the lumen of the endoplasmic reticulum, they might not be expected to act on HIF-1α, a cytoplasmic protein. Therefore, this finding indicates that the HIF-1α hydroxylation is likely to be catalyzed by a distinct enzyme. Nonetheless, that these known prolyl hydroxylases contain iron at their active sites and catalyze a reaction requiring molecular oxygen suggests mechanisms by which cobalt and hypoxia might inhibit HIF-1α hydroxylation. In the former case, cobalt might substitute for an iron at the active site; in the latter, the low oxygen concentration might constitute a subsaturating substrate concentration. The data presented here indicate that there is no need to invoke additional mechanisms that promote the dissociation of HIF-1α from VHL under hypoxic conditions.
Acknowledgments
We thank Dr. William T. Moore for performing the MALDI-TOF MS, for help with the preparation of Fig. Fig.3,3, and for valuable discussions. This work was supported by the W. W. Smith Charitable Trust Research Grant C9901 and by the National Institutes of Health Grant DK55672.
Abbreviations
| Cbz-LLL | N-Cbz-l-Leu-l-Leu-l-norvalinal |
| HIF-1α | hypoxia-inducible factor-1α |
| Hyp | 4-hydroxy-l-proline |
| MALDI-TOF | matrix-assisted laser desorption time of flight |
| VHL | von Hippel-Lindau tumor-suppressor protein |
| GSH | glutathione |
| GST | glutathione S-transferase |
| amu | atomic mass unit |
| WT | wild type |
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.181341498
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc55503?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Human prolyl hydroxylase domain 2 reacts with O<sub>2</sub> and 2-oxoglutarate to enable formation of inactive Fe(III).2OG.hypoxia-inducible-factor α complexes.
Sci Rep, 14(1):26162, 30 Oct 2024
Cited by: 0 articles | PMID: 39478091 | PMCID: PMC11525979
Enhanced tumor response to adoptive T cell therapy with PHD2/3-deficient CD8 T cells.
Nat Commun, 15(1):7789, 06 Sep 2024
Cited by: 0 articles | PMID: 39242595 | PMCID: PMC11379939
Time-resolved NMR detection of prolyl-hydroxylation in intrinsically disordered region of HIF-1α.
Proc Natl Acad Sci U S A, 121(37):e2408104121, 04 Sep 2024
Cited by: 0 articles | PMID: 39231207
Succinate dehydrogenase-complex II regulates skeletal muscle cellular respiration and contractility but not muscle mass in genetically induced pulmonary emphysema.
Sci Adv, 10(34):eado8549, 21 Aug 2024
Cited by: 0 articles | PMID: 39167644 | PMCID: PMC11338223
The relationship between hypoxia and Alzheimer's disease: an updated review.
Front Aging Neurosci, 16:1402774, 17 Jul 2024
Cited by: 0 articles | PMID: 39086755 | PMCID: PMC11288848
Review Free full text in Europe PMC
Go to all (487) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Dynamic, site-specific interaction of hypoxia-inducible factor-1alpha with the von Hippel-Lindau tumor suppressor protein.
Cancer Res, 61(10):4136-4142, 01 May 2001
Cited by: 55 articles | PMID: 11358837
Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein.
EMBO J, 19(16):4298-4309, 01 Aug 2000
Cited by: 537 articles | PMID: 10944113 | PMCID: PMC302039
Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation.
Science, 292(5516):468-472, 05 Apr 2001
Cited by: 3358 articles | PMID: 11292861
Regulation of HIF by the von Hippel-Lindau tumour suppressor: implications for cellular oxygen sensing.
IUBMB Life, 52(1-2):43-47, 01 Jul 2001
Cited by: 59 articles | PMID: 11795592
Review





