Abstract
Free full text

The RASSF1A Tumor Suppressor Blocks Cell Cycle Progression and Inhibits Cyclin D1 Accumulation
Abstract
The RASSF1A locus at 3p21.3 is epigenetically inactivated at high frequency in a variety of solid tumors. Expression of RASSF1A is sufficient to revert the tumorigenicity of human cancer cell lines. We show here that RASSF1A can induce cell cycle arrest by engaging the Rb family cell cycle checkpoint. RASSF1A inhibits accumulation of native cyclin D1, and the RASSF1A-induced cell cycle arrest can be relieved by ectopic expression of cyclin D1 or of other downstream activators of the G1/S-phase transition (cyclin A and E7). Regulation of cyclin D1 is responsive to native RASSF1A activity, because RNA interference-mediated downregulation of endogenous RASSF1A expression in human epithelial cells results in abnormal accumulation of cyclin D1 protein. Inhibition of cyclin D1 by RASSF1A occurs posttranscriptionally and is likely at the level of translational control. Rare alleles of RASSF1A, isolated from tumor cell lines, encode proteins that fail to block cyclin D1 accumulation and cell cycle progression. These results strongly suggest that RASSF1A is an important human tumor suppressor protein acting at the level of G1/S-phase cell cycle progression.
Loss of heterozygosity of chromosome region 3p21.3 is extremely common in lung, breast, ovarian, nasopharyngeal, and renal tumors (22, 27, 37, 39). Alterations at 3p21.3 are a very early event in primary cancer development, implying the presence of a tumor suppressor gene or genes in this location (20, 39). RASSF1 is one of eight predicted genes located in a minimal interval of 3p21.3 as defined by analysis of nested homozygous deletions found in tumor samples (24, 31). Two major splice forms of RASSF1, RASSF1A, and RASSF1C are expressed in normal human epithelial cells that derive from two different promoter regions (5, 6). The resulting mRNAs differ primarily in the selection of the first exon. RASSF1A contains an amino-terminal cysteine-rich region, which is similar to the diacyl glycerol binding domain (C1 domain) found in the protein kinase C family of proteins, and a carboxy-terminal putative Ras-association (RA) domain. RASSF1C is a smaller protein that lacks the amino-terminal C1 domain. Selective epigenetic inactivation of the RASSF1A promoter is an extremely common event in many human cancers. This includes 80 to 100% of SCLC cell lines and tumors (5, 6), 30 to 40% of NSCLC cell lines and tumors (5), 49 to 62% of breast cancers (5, 7), 67 to 70% of primary nasopharyngeal cancers (NPCs) (26), 91% of primary renal cell carcinomas (RCCs), and 100% of RCC lines (10). Ectopic expression of RASSF1A, but not RASSF1C, potently inhibits tumorigenicity of lung cancer cell lines, H1299 and A549 (5, 6), and an RCC line, KRC/Y (10). These results strongly suggest that RASSF1A may function as a tumor suppressor protein in many cells of epithelioid origin; however, the mechanism by which RASSF1A can negatively regulate tumor growth has not been determined.
Normal epithelial cells require cell adhesion and the presence of appropriate growth factors to promote cell proliferation. These two signals act coordinately to regulate the G1/S-phase cell cycle transition (4). G1 includes a restriction point beyond which the cell is committed to undergo division, independent of extracellular growth regulatory signals. The retinoblastoma family of proteins (Rb, p107, and p130) are major gatekeepers of the G1 restriction point. Hyperphosphorylation of Rb by cyclin D-CDK4 and cyclin E-CDK2 complexes is permissive for progression of proliferating cells into S phase. Alterations in expression or activity of components of this regulatory pathway are frequently associated with human tumors (32, 33). As an additional level of growth control, epithelial cells respond to loss of matrix adhesion by induction of programmed cell death (anoikis) even in the presence of growth factors that normally promote proliferation (16). Progression of epithelial cells to a tumorigenic state therefore requires bypass of regulatory checkpoints controlling both proliferation and survival.
Here, we begin to define the mechanism by which the RASSF1A tumor suppressor impacts growth regulation. Reintroduction of RASSF1A expression in lung and breast tumor-derived epithelial cells results in growth arrest but not apoptosis. This growth arrest correlates with inhibition of cyclin D1 protein accumulation, which likely prevents RASSF1A-expressing cells from passing through the Rb family cell cycle restriction point and entering S phase. Bypassing the requirement for cyclin D1 by artificially driving expression of cyclin A or expression of the viral Rb family inhibitor E7 relieved RASSF1A-induced cell cycle arrest. Regulation of cyclin D1 accumulation by RASSF1A is independent of the cyclin D1 promoter and likely occurs through inhibition of mRNA translation. Rare mutant alleles of RASSF1A, isolated from tumors and tumor-derived cell lines, were found to express proteins that cannot inhibit cyclin D1 accumulation or cell proliferation. These mutations alter a putative mTOR/ATM family kinase substrate site and inhibit phosphorylation of RASSF1A, suggesting that RASSF1 activity is phosphorylation dependent. RNA interference (RNAi)-mediated inhibition of RASSF1A protein expression results in abnormal accumulation of native cyclin D1 protein in the absence of detectable changes in cyclin D1 mRNA levels. Together these results suggest that RASSF1A functions as a negative regulator of cell proliferation through inhibition of G1/S-phase progression.
MATERIALS AND METHODS
Plasmids.
pDCR-ras12V, −1745-CD1-Luc, pRC-cyclin D1, and pX-cyclin A have been described previously (1, 5, 8, 18, 19, 29, 38). Replication-defective retroviral particles derived from LXSN and LXSN-E7 were gifts from W. Wright (UT Southwestern Medical Center). pRK5myc-RASSF1A contains the full-length coding sequence of human RASSF1A as an EcoRI-XhoI fragment in pRK5myc2. pRK5myc-RASSF1C contains the full-length coding sequence of RASSF1C as an EcoRI-XbaI fragment in pRK5myc2. PRK5myc-RASSF1A(S131F), pRK5myc-RASSF1A(A133S), pRK5myc-RASSF1C(S61F), and pRK5myc-RASSF1A(A63S) were created by PCR-based site-directed mutagenesis and are otherwise identical to the wild-type versions. pGEX4T-1-Maxp1(229-455) contains an EcoRI-XhoI fragment of Maxp1 encoding residues 229 to 455 inserted in the EcoRI-SalI sites of pGEX4T-1. PGEX4T-1-RASSF1A and pGEX4T-1-RASSF1C contain full-length coding sequences inserted as EcoRI-SalI fragments into pGEX4T-1.
Cell culture and transfections.
NCI-H1299 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal bovine serum. HME50-hTERT cells, a primary human diploid mammary epithelial cell line immortalized by hTERT expression (gift from J. Shay, UT Southwestern Medical Center), were grown in MCDB serum-free medium (Invitrogen) supplemented with 0.4% bovine pituitary extract (Hammond Cell Tech), 10 ng of epidermal growth factor (EGF) per ml, 5 μg of insulin per ml, 0.5 μg of hydrocortisone per ml, 5 μg of transferrin per ml, and 50 μg of gentamicin per ml (Sigma). HeLa cells were grown in DMEM supplemented with 10% calf serum. NCI-H1299 and HME50-hTERT cells were transfected with Lipofectamine 2000 reagent (Invitrogen). HeLa cells were transfected with double-stranded RNA oligonucleotides by using Oligofectamine (Invitrogen).
Antibodies and immunofluorescence.
Antibodies to RASSF1 were generated in New Zealand White rabbits inoculated with purified recombinant glutathione S-transferase (GST)-RASSF1(120-340) in RIBI adjuvant. Prior to use, RASSF1-specific antibodies were affinity purified from crude serum by standard methods. For bromodeoxyuridine (BrdU) incorporation experiments, 30 μM BrdU was added to transiently transfected cell cultures 24 h posttransfection. Following a 24-h incubation in BrdU, cells were fixed in 3.7% paraformaldehyde, permeabilized in acetone at −20°C for 5 min, and then treated with 2 M HCl for 10 min. BrdU incorporation was visualized with mouse monoclonal anti-BrdU and fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG). Myc-tagged RASSF1A expression was visualized with rabbit anti-Myc polyclonal antibodies (UBI) and rhodamine red X-conjugated anti-rabbit IgG (Jackson Laboratories), mouse 9E10 monoclonal anti-Myc antibody (Santa Cruz), and FITC-conjugated anti-mouse IgG (Jackson Laboratories), or chicken anti-Myc antibodies (Avery Laboratories) and cy5-conjugated anti-chicken IgG. Endogenous and ectopic cyclin D1 proteins were detected with rabbit polyclonal anti-cyclin D1 antibodies (UBI) and rhodamine red X-conjugated anti-rabbit IgG. Cyclin A expression was detected with rabbit anti-cyclin A antibodies (Santa Cruz) and Alexa Red-conjugated anti-rabbit IgG. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was carried out according to manufacturer's instructions (Roche). Biotinylated dUTP incorporation was detected with Texas red streptavidin (Jackson Laboratories). Fluorescence-activated cell sorting (FACS) of RASSF1A-expressing cells was performed by FACScan, with 10,000 cells collected for each assay, and analyzed by using CellQuest software (Becton Dickinson).
32Pi labeling.
Forty-eight hours after transfection, cells were washed once and preincubated with phosphate-free modified Eagle’s medium (MEM) for 10 min prior to addition of 0.5 mCi of 32Pi to each 35-mm-diameter tissue culture plate. Following a 4-h incubation, cells were lysed (1% NP-40, 10 mM Tris [pH 7.5], 0.25 mM sodium deoxycholate, 1 mM MgCl2, 1 mM EGTA, 5 mM β-mercaptoethanol, 10% glycerol, 150 mM NaCl, 50 mM sodium fluoride, 1 mM orthovanadate, 80 mM β-glycerophosphate, and Roche protease inhibitor cocktail), and the RASSF1 variants were immunoprecipitated with anti-Myc antibody.
Gene expression assays.
Luciferase activity assays were performed as previously described with the −1745-CD1-LUC reporter construct (19). Luciferase activity was normalized to β-galactosidase expression from cotransfected pCH110. For RNase protection assays (RPAs), total RNA was isolated from 106 cells with Trizol and further purified with a High Pure RNA isolation kit (Roche) according to the manufacturer's instructions. RPA was performed with the Riboquant RPA system (Pharmingen) together with the hCYC-1 multiprobe template set. A total of 5 × 105 cpm of the labeled probe set was hybridized with 2 μg of total RNA overnight at 56°C. Free probe and single-stranded RNA molecules were digested with RNase A. The hybridized probes were resolved on a 5% denaturing polyacrylamide gel and exposed to a PhosphorImager plate.
RNA interference assays.
Double-stranded small interfering RNA (siRNA) oligonucleotides targeting RASSF1A were designed and prepared as described previously (12). The sequences used were 5′-GACCUCUGUGGCGACUUCATT-3′ and 5′-UGAAGUCGCCACAGAGGUCTT-3′. Whole-cell lysates were taken for analysis 72 h following transfection.
RESULTS
RASSF1A induces G1 cell cycle arrest.
H1299 lung carcinoma cells, like many non-small-cell lung carcinomas, do not express RASSF1A due to loss of one allele and hypermethylation of the RASSF1A promoter on the remaining allele (5). These cells exhibit anchorage-independent growth (growth in soft agar) and can form tumors in athymic mice (5). We have previously shown that reintroduction of RASSF1A expression into H1299 cells is sufficient to inhibit the tumorigenicity of these cells (5), suggesting that loss of RASSF1A expression may be an obligate step for oncogenic transformation.
To begin to characterize the putative antioncogenic properties of RASSF1A, we first examined whether inhibition of tumorigenicity was through RASSF1A-induced apoptosis or cell cycle arrest. Transient transfection assays with native and epitope-tagged RASSF1A gave no indication of toxicity in adherent cells (5; data not shown). To determine if RASSF1A expression may be engaging an apoptotic program upon loss of anchorage, transiently transfected H1299 cells were assayed by TUNEL following incubation in suspension. H1299 cells were resistant to suspension-induced apoptosis up to the latest time point tested (72 h). As shown in Fig. Fig.1,1, cells expressing RASSF1A gave no indication of an apoptotic response. In contrast, suspension cells treated with staurosporine showed strong labeling by TUNEL.



RASSF1A blocks proliferation but does not induce apoptosis in human lung carcinoma cells. (a) H1299 cells were transiently transfected with Myc-tagged RASSF1A and placed in suspension cultures 24 h later. After 48 h of incubation in suspension cultures, cells were spun onto glass coverslips, fixed, stained with anti-Myc antibody to detect RASSF1A expression, and labeled by TUNEL to detect fragmented DNA. Cells were treated with 1 μM staurosporine for 4 h in suspension as a positive control for TUNEL (ST/TUNEL). (b) Twenty-four hours after transfection with the indicated constructs, H1299 cells were incubated for an additional 24 h in the presence of BrdU. RASSF1A expression was detected as in panel A. BrdU incorporation was detected with anti-BrdU antibody. An overlay image is shown. Quantitation by microscopic observation of three independent experiments is shown in Table Table1.1. (c) Asynchronous H1299 cells were transfected with GFP or myc-RASSF1A. Forty-eight hours posttransfection, cells were trypsinized, fixed, and stained with propidium iodide. RASSF1A-transfected cultures were additionally stained with anti-Myc and fluorescein-conjugated anti-mouse secondary antibodies. Two-color FACS was used to determine the DNA content of GFP- or RASSF1A-expressing cells. Over 2,000 cells are scored for each analysis. The results shown are representative of three independent experiments. Quantitation of the population of cells in the indicated peaks is shown as a percentage of total events from the three experiments.
To assess the consequences of RASSF1 expression on cell cycle progression, H1299 cells were assayed for BrdU incorporation following transient transfection with RASSF1A or RASSF1C. Expression of RASSF1A, but not RASSF1C, in adherent, subconfluent cells resulted in a dramatic inhibition of BrdU incorporation (Fig. (Fig.1b1b and Table Table1).1). Similar results were observed in suspension cultures (Table (Table1).1). These observations suggest that ectopic expression of RASSF1A inhibits tumorigenicity through induction of cell cycle arrest.
TABLE 1.
Consequences of RASSF1 expression for BrdU incorporation and cyclin D1 protein expression
| Protein | % of cells positive for expressiona
| |||
|---|---|---|---|---|
| H1299
| HME50-hTERT
| |||
| BrdU | Cyclin D1 | BrdU | Cyclin D1 | |
| RASSF1A | 20 ± 5 (10 ± 1)b | 9 ± 1 | 13 ± 1 | 12 ± 1 |
| RASSF1C | 67 ± 8 | 44 ± 4 | 64 ± 1 | 58 ± 1 |
| RASSF1A(S131F) | 65 ± 11 | 62 ± 2 | 63 ± 2 | 59 ± 2 |
| RASSF1A(A133S) | 55 ± 5 | 49 ± 5 | 69 ± 4 | 68 ± 2 |
To identify the nature of the cell cycle arrest, transiently transfected RASSF1A cells were examined for DNA content by FACS. RASSF1A-expressing cells were compared with green fluorescent protein (GFP)-expressing cells. Analysis of propidium iodide incorporation shows that, in contrast to GFP-expressing cells, the majority of H1299 cells expressing RASSF1A are in the G1 phase of the cell cycle (Fig. (Fig.1c1c).
RASSF1A variants identified in tumor lines are uncoupled from G1 arrest.
Characterization of the RASSF1 locus in lung and breast carcinomas demonstrated that the RASSF1A isoform is not expressed in the majority of cell lines and tumors assayed due to methylation of the RASSF1A-specific promoter (5, 6). A mutation in the RASSF1 gene encoding a substitution of alanine to serine at position 133 of RASSF1A was detected in 12 breast and lung tumor samples that lacked methylation or had heterozygous methylation of the RASSF1A promoter region. The same mutation was detected in matched B-cell samples, suggesting it is a single nucleotide polymorphism rather than a somatic cell mutation. In addition, we unexpectedly cloned a cDNA from a human tumor cDNA library with a nearby mutation that encodes a substitution of serine131 to phenylalanine in RASSF1A. Serine 131 has been suggested to be a substrate for the ATM kinase (21), because the sequence WETPDLSQAEIEQK (amino acids 125 to 138 of RASSF1A) matches a putative ATM family phosphorylation site consensus sequence and a peptide with this sequence is an excellent ATM substrate in vitro (21). Both RASSF1A(A133S) and RASSF1A(S131F) were severely compromised in their ability to inhibit BrdU uptake in H1299 cells (Fig. (Fig.2a2a and Table Table1).1). As shown in Fig. Fig.2b,2b, this defect correlates with the phosphorylation state of these proteins, which was significantly reduced compared to that of the wild type.
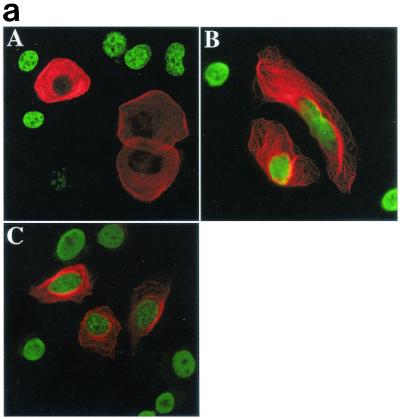

RASSF1A variants isolated from tumor cell lines are unable to block proliferation. (a) H1299 cells were transfected with RASSF1A (A), RASSF1A(S131F) (B), or RASSF1A(A133S) (C) and processed as in Fig. Fig.1b.1b. Overlay images are shown with rhodamine red X-conjugated anti-rabbit IgG to detect the anti-Myc polyclonal antibody, and FITC-conjugated anti-mouse IgG to detect the anti-BrdU monoclonal antibody. Quantitation by microscopic observation of at least three independent experiments is shown in Table Table1.1. (b) H1299 cells expressing the carboxy-terminal halves of the indicated RASSF1 variants were labeled with 32Pi for 4 h. RASSF1 variants were immunoprecipitated, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and exposed to PhosphorImager plates to detect incorporation of 32Pi (top panels). Total RASSF1 was detected by Western blotting with anti-Myc antibody (bottom panels). w.t., wild type.
Bypass of Rb family proteins rescues cell cycle progression in RASSF1A-expressing cells.
H1299 cells express Rb, but not p16. This suggests that Rb checkpoints may be difficult to engage in these cells. However, in addition to p16, the cyclin-dependent kinase inhibitors p21, p27, and/or p57 can contribute to negative regulation of CDK2 (33) and may help engage an Rb family checkpoint in H1299 cells. The E7 papillomavirus protein can bypass Rb family-dependent cell cycle regulation by directly inhibiting the interaction of Rb proteins with E2F transcription factors and other pocket-binding proteins (15). As shown in Fig. Fig.3a,3a, H1299 cells expressing E7 are resistant to RASSF1A-induced cell cycle arrest.


Bypass of the Rb family cell cycle restriction point allows proliferation of RASSF1A-expressing cells. (a) H1299 cells expressing the indicated constructs together with RASSF1A were assayed for expression and BrdU incorporation. The percentage of cells expressing RASSF1A and incorporating BrdU was quantitated by microscopic observation. Bars represent the standard error from the mean of values obtained from three independent experiments. (b) H1299 cells were transiently transfected with RASSF1A together with cyclin A and assayed for expression of RASSF1A (A), cyclin A (B), and BrdU incorporation (C). Panel D is an overlay of images A to C.
Cyclin A can directly activate CDK2 and participates in a feed forward amplification loop with E2F to drive cyclin E expression. Again, consistent with action of RASSF1A at the level of the Rb family restriction point, ectopic expression of cyclin A bypassed RASSF1A cell cycle arrest (Fig. 3a and b).
Oncogenic Ras does not bypass RASSF1A-induced growth arrest.
RASSF1A and RASSF1C contain a carboxy-terminal region that may mediate direct binding to activated Ras family GTPases (5, 6, 36) and can interact weakly with Ras-GTP in vitro and when coexpressed with H-Ras12V in cells (36). To examine potential modulation of RASSF1A activity by Ras, we examined the consequence of H-ras12V expression on RASSF1A-induced growth arrest of human mammary epithelial cells immortalized by hTERT (HME50-hTERT) cells. As has been previously reported, in contrast to primary human fibroblasts, oncogenic Ras expression does not induce a “senescence-like” phenotype in HME-hTERT cells (13). As shown in Fig. Fig.4,4, oncogenic Ras expression did not detectably alter RASSF1A growth-inhibitory activity in these cells.
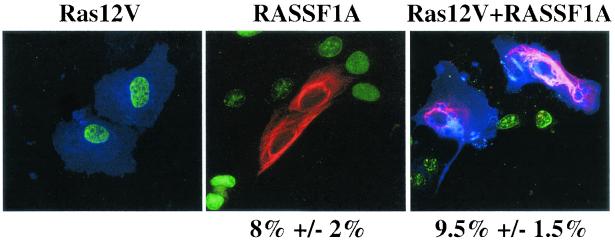
Ras12V expression does not bypass RASSF1A-mediated growth arrest. HME50-hTERT cells were transfected with the indicated constructs and processed as described in the legend to Fig. Fig.3b.3b. The percentage of RASSF1A-expressing cells and RASSF1A+Ras12V-expressing cells incorporating BrdU is shown normalized to the BrdU incorporation frequency of cells expressing Ras12V alone (approximately 80% of total cells).
RASSF1A inhibits accumulation of cyclin D1.
Accumulation of cyclin D1 protein during G1 contributes to bypass of Rb family restriction point control (32). Many mitogenic and oncogenic signal transduction cascades converge at the level of cyclin D1 accumulation, enhancing promoter activation (3, 18, 23), mRNA stability (11, 25), translation (28), and/or protein stability (1, 8). Consistent with a role in regulating G1/S-phase progression, RASSF1A expression dramatically inhibits native cyclin D1 accumulation (Fig. (Fig.5a5a and Table Table1).1). Expression of RASSF1C and RASSF1A variants had only modest effects.

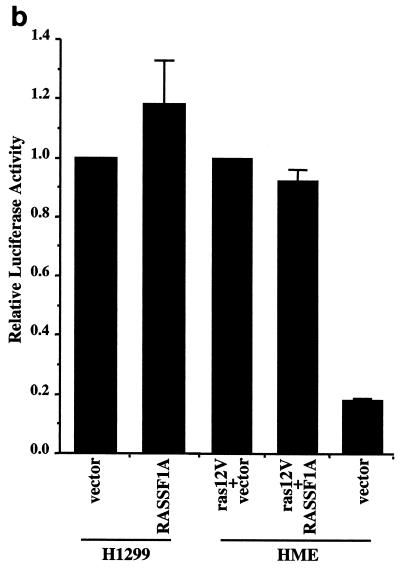

RASSF1A prevents accumulation of cyclin D1 protein. (a) H1299 cells were transfected with RASSF1A or RASSF1C and stained with anti-cyclin D1 and anti-Myc antibodies. Overlays are shown with rhodamine red X-conjugated anti-rabbit IgG antibodies to detect rabbit anti-cyclin D and FITC-conjugated anti-mouse IgG to detect the 9E10 anti-Myc antibody. Quantitation by microscopic observation of at least three independent experiments is shown in Table1. (b) H1299 cells or HME cells were transfected with the indicated constructs together with a luciferase reporter construct driven by the human cyclin D1 promoter (−1745 CD1-Luc). Relative luciferase values are shown normalized to activities obtained with empty vector (H1299) or Ras12V alone (HME). Ras12V expression resulted in fourfold induction of luciferase activity in HME cells over baseline levels observed in serum-starved cells. Bars indicate the standard error from the mean of average values from three independent experiments performed in duplicate. (c) MEFs derived from wild-type and cyclin D1−/− mice were transiently transfected with the indicated constructs and assayed for BrdU incorporation as described in the legend to Fig. Fig.11.
The growth-inhibitory effects of RASSF1A are not limited to transformed cells, because inhibition of cyclin D1 accumulation and cell cycle progression by RASSF1A was also observed in HME-hTERT cells and primary mouse embryo fibroblasts (MEFs) (Table (Table11 and Fig. Fig.44 and and55).
Regulation of the cyclin D1 promoter does not appear to be inhibited upon RASSF1A expression, because the steady-state activity of a luciferase reporter driven by the cyclin D1 promoter (−1745 CD1-LUC) was not affected in H1299 cells. The activation of the cyclin D1 promoter by oncogenic Ras in HME-hTERT cells was also unaffected by RASSF1A (Fig. (Fig.5b).5b). These observations suggest that RASSF1A may impact cyclin D1 protein accumulation posttranscriptionally. LLnL, lactacysteine, β-lactone, or MG132 proteosome inhibitors did not rescue cyclin D1 accumulation in RASSF1A-expressing cells, suggesting that the effects of RASSF1A are not achieved through alteration of cyclin D1 protein stability (data not shown). Together these results imply that RASSF1A can inhibit translation and/or stability of cyclin D1 mRNA, although this has not been directly demonstrated.
Ectopic expression of cyclin D1 from a viral promoter was sufficient to bypass RASSF1A-induced cell cycle arrest (Fig. (Fig.3a),3a), suggesting that the inhibitory effects of RASSF1A on cell cycle progression are at the level of or upstream of cyclin D1 production. In addition, while RASSF1A can arrest the growth of wild-type MEFs, MEFs derived from cyclin D1 knockout mice (2) are insensitive to RASSF1A expression (Fig. (Fig.5c).5c). Presumably, the cyclin D1−/− MEFS have bypassed the requirement for cyclin D1 expression through developmental compensation (14, 40). These results imply that the inhibitory effects of RASSF1A are mediated by cyclin D1-dependent checkpoints.
A dilemma facing interpretation of transient transfection analysis is the often unavoidable expression of proteins at much larger amounts in cells than would be produced from endogenous loci. To assess the contribution of native RASSF1A to regulation of cyclin D1 protein accumulation, we used siRNAs to inhibit expression of RASSF1A in HeLa CCL2 cells. These cells were chosen because they are highly amenable to treatment with siRNA, and they express detectable levels of endogenous wild-type RASSF1A. HeLa CCL2 cells are insensitive to RASSF1A-induced cell cycle arrest (Fig. (Fig.6a).6a). This is likely due to the expression of papillomavirus E7 protein in these cells that bypasses Rb family cell cycle checkpoint control. This result is consistent with the observed bypass of RASSF1A-induced cell cycle arrest upon E7 expression in H1299 cells. Because Rb family function has been directly inhibited by E7, it is possible that upstream elements normally responsible for engaging the Rb checkpoint may still be intact in these cells. This has in fact been demonstrated empirically by the observation that an Rb-dependent checkpoint is engaged in HeLa cells upon elimination of E7 expression (17). Consistent with a role for endogenous RASSF1A in negative regulation of cyclin D1 accumulation, introduction of siRNA targeted to RASSF1A resulted in dramatically decreased RASSF1A expression and increased endogenous cyclin D1 protein accumulation compared to that in controls (Fig. (Fig.6b).6b). Examination of the relative amounts of cyclin D1 mRNA in these samples also shows that inhibition of RASSF1A expression does not detectably alter the steady-state levels of cyclin D1 mRNA (Fig. (Fig.6c).6c). This observation supports the hypothesis that native RASSF1A negatively regulates cyclin D1 accumulation through a posttranscriptional mechanism.



Enhanced accumulation of cylin D1 in the absence of RASSF1A expression. (a) BrdU incorporation in RASSF1A-expressing HeLa cells was assayed as described in Fig. Fig.1.1. (b) HeLa cells were exposed to siRNA specifically targeting the RASSF1A transcript for 72 h. siRNA directed against caveolin 1 was used as a negative control. Cyclin D1 levels in cells exposed to caveolin 1 siRNA are identical to those in untreated cells (data not shown). Whole-cell lysates were assayed for expression of the indicated proteins. Ten micrograms of total protein was loaded for each sample. (c) RPAs were performed to examine the relative amounts of cyclin D1 mRNA present in cells treated as described for panel b.
DISCUSSION
Loss of RASSF1A expression is a common event associated with many classes of human carcinomas. Reintroduction of RASSF1A expression in tumor cells results in dramatic inhibition of tumorigenicity (5, 6). The results described here suggest that RASSF1A inhibits proliferation by negatively regulating cell cycle progression at the level of G1/S-phase transition. RASSF1A-induced cell cycle arrest is accompanied by loss of cyclin D1 accumulation and can be relieved by ectopic expression of cyclin D1 cDNA. Significantly, inhibition of native RASSF1A results in abnormal accumulation of cyclin D1. This implies that RASSF1A modulates cell cycle progression through pathways regulating accumulation of cyclin D1 protein.
Accumulation of cyclin D1 is tightly regulated through multiple mechanisms, including promoter activation, mRNA stability, initiation of translation, and protein stability. The cyclin D1 promoter contains response elements for many mitogen-activated transcription factors, including AP1, NF-κB and Tcf/Lef (3, 18, 34). Regulation at the level of cyclin D1 mRNA accumulation can occur through destabilization elements in the cyclin D1 3′ untranslated region (UTR). AU-rich elements on the distal region of the cyclin D1 3′ UTR can be positively regulated by prostaglandin A2 (25) and negatively regulated by phosphatidylinositol 3-kinase (11). In addition, initiation of translation of cyclin D1 mRNA is rapamycin sensitive in many cell types through an undefined pathway. Posttranslational control of cyclin D1 levels is mediated by phosphorylation-dependent polyubiquitination and degradation by the 26S proteosome (9). We have not yet identified the mechanism by which RASSF1A modulates cyclin D1 levels. However, we find that RASSF1A expression does not appear to affect cyclin D1 transcription or protein stability. This leaves the initiation of translation and/or mRNA stability as potential RASSF1A-sensitive steps in cyclin D1 regulation.
RASSF1C expression can be detected in most tumor cell lines (like H1299) that lack detectable RASSF1A expression (5). Further increasing the levels of RASSF1C protein does not greatly affect the proliferation of any cell type that we have tested. This includes several epithelium-derived tumor cell lines, normal mammary epithelial cells, NIH 3T3 fibroblasts, and 293 cells. In contrast to our observations, another group has reported that RASSF1C may induce apoptosis when overexpressed in 293T cells and NIH 3T3 cells (36). Although we did not observe this, we cannot rule out contrasting results due to unknown variables involving culture conditions or expression levels. We also found that RASSF1C did not significantly alter the levels of cyclin D1 protein accumulation. Our results suggest that the amino-terminal C1 domain of RASSF1A that is absent from RASSF1C is required for growth-inhibitory activity. The function of RASSF1C is unknown. It may be that RASSF1C can negatively modulate the activity of RASSF1A through unproductive association with cofactors; however, preliminary experiments suggest that RASSF1A expression is dominant to RASSF1C (L.S. and M.W., unpublished results).
RASSF1 is likely to be a member of a family of related proteins. RASSF1A is 60% homologous to mouse Nore1. The sequence of the human ortholog of NORE1, MGC10823, is present in GenBank, and KIAA0168 is an additional putative family member. It remains to be determined whether these family members play similar roles in the regulation of cell cycle progression; however, we have observed that expression of NORE1 has no detectable effect on the growth of adherent or suspension cultures of H1299 cells or HME-hTERT cells (data not shown). NORE1 can associate directly with Ras proteins in a stimulus-dependent manner (35). Like NORE1, all members of this family contain a carboxy-terminal putative RA domain (30). Although the RA domain of RASSF1A can weakly interact directly with Ras-GTP in vitro (36; our unpublished observations), it is unknown whether this interaction occurs in cells, and if so, what the consequences may be for RASSF1A activity. Given the tumor-promoting properties of activated Ras, it would be reasonable to speculate that Ras may negatively regulate RASSF1A activity to promote proliferation. However, we find that the effects of expression of RASSF1A are dominant to oncogenic Ras expression in human mammary epithelial cells. We were unable to detect any modulation of RASSF1A-induced growth arrest upon coexpresion with H-Ras12V. Although Ras is not likely a negative regulator of RASSF1A, our results leave open the possibility Ras may positively regulate endogenous RASSF1A activity.
From tumor cell lines expressing RASSF1A, we have identified rare germ line polymorphisms of RASSF1 with amino acid sequence alterations resulting in RASSF1A proteins [RASSF1A(S131F) and RASSF1A(A133S)] with decreased steady-state phosphorylation and decreased antiproliferative activity. These polymorphisms alter a putative phosphorylation site matching the consensus substrate site for ATM/ATR family serine/threonine kinases. It remains to be determined whether serine 131 is a bona fide phosphorylation site and whether the reduced activity of RASSF1A(S131F) and RASSF1A(A133S) is a direct consequence of reduced phosphorylation. The RASSF1A(A133S) variant is a consequence of a single-nucleotide polymorphism. Because this variant shows defective antiproliferative activity in our assays, it is possible that individuals carrying this polymorphism may have increased risk for development of some types of neoplastic disease. We have no way of knowing if the mutation we isolated encoding the S131F variant was a spontaneous somatic cell mutation occurring in a tumor or if it may be a rare polymorphism.
In summary, we have provided evidence indicating the mechanism of the growth-regulatory and tumor-suppressing activity of RASSF1A. Our results suggest that RASSF1A negatively regulates accumulation of endogenous cyclin D1 through a posttranscriptional mechanism leading to inhibition of cell cycle progression. Further studies need to be focused on understanding the regulation of cellular RASSF1A, as well as the molecular mechanism(s) by which RASSF1A impacts cyclin D1 accumulation and cell cycle progression.
Acknowledgments
We thank Woodring Wright, Rene Bernards, Charles Sherr, and Piotr Sicinski for some of the reagents used in these studies and for cyclin D1−/− mice. We thank Dale Henry and Lesli Hasbini for excellent technical assistance.
This work was supported by NIH grant R01CA71443 (M.W.) and the Welch Foundation (M.W.). L.S. is supported by U.S. Department of Defense grant DAMD17-00-1-0439. Additional support was obtained from R01CA70896 (R.G.P.) and R01CA71618 (J.M.).
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.22.12.4309-4318.2002
Read article for free, from open access legal sources, via Unpaywall:
https://mcb.asm.org/content/mcb/22/12/4309.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102438892
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.22.12.4309-4318.2002
Article citations
Complex interplay between RAS GTPases and RASSF effectors regulates subcellular localization of YAP.
EMBO Rep, 25(8):3574-3600, 15 Jul 2024
Cited by: 0 articles | PMID: 39009833 | PMCID: PMC11316025
Transcription factor E4F1 as a regulator of cell life and disease progression.
Sci Adv, 9(39):eadh1991, 29 Sep 2023
Cited by: 2 articles | PMID: 37774036 | PMCID: PMC10541018
Review Free full text in Europe PMC
Beta-Transducin Repeats-Containing Proteins as an Anticancer Target.
Cancers (Basel), 15(17):4248, 24 Aug 2023
Cited by: 1 article | PMID: 37686524 | PMCID: PMC10487276
Review Free full text in Europe PMC
Human artificial chromosome carrying 3p21.3-p22.2 region suppresses hTERT transcription in oral cancer cells.
Chromosome Res, 31(3):17, 24 Jun 2023
Cited by: 0 articles | PMID: 37353691 | PMCID: PMC10289923
Risk Stratification of Pancreatic Neuroendocrine Neoplasms Based on Clinical, Pathological, and Molecular Characteristics.
J Clin Med, 11(24):7456, 15 Dec 2022
Cited by: 2 articles | PMID: 36556070 | PMCID: PMC9786745
Review Free full text in Europe PMC
Go to all (214) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma.
Cancer Res, 63(17):5344-5351, 01 Sep 2003
Cited by: 52 articles | PMID: 14500366 | PMCID: PMC3484890
[Inhibitory effect of wild-type RASSF1A gene expression on proliferation of hepatocellular carcinoma QGY-7703 cells].
Ai Zheng, 27(9):924-928, 01 Sep 2008
Cited by: 1 article | PMID: 18799029
Differential requirements for ras and the retinoblastoma tumor suppressor protein in the androgen dependence of prostatic adenocarcinoma cells.
Cell Growth Differ, 11(7):361-372, 01 Jul 2000
Cited by: 22 articles | PMID: 10939590
[Molecular mechanisms controlling the cell cycle: fundamental aspects and implications for oncology].
Cancer Radiother, 5(2):109-129, 01 Apr 2001
Cited by: 45 articles | PMID: 11355576
Review
Funding
Funders who supported this work.
NCI NIH HHS (6)
Grant ID: R01 CA071443
Grant ID: R01 CA071618
Grant ID: R01CA71443
Grant ID: R01 CA070896
Grant ID: R01CA71618
Grant ID: R01CA70896