Abstract
Free full text

PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database
Abstract
We describe PRIMROSE, a computer program for identifying 16S rRNA probes and PCR primers for use as phylogenetic and ecological tools in the identification and enumeration of bacteria. PRIMROSE is designed to use data from the Ribosomal Database Project (RDP) to find potentially useful oligonucleotides with up to two degenerate positions. The taxonomic range of these, and other existing oligonucleotides, can then be explored, allowing for the rapid identification of suitable oligonucleotides. PRIMROSE includes features to allow user-defined sequence databases to be used. An in silico trial of the program using the RDP database identified oligonucleotides that described their target taxa with a degree of accuracy far greater than that of equivalent currently used oligonucleotides. We identify oligonucleotides for subdivisions of the Proteobacteria and for the Cytophaga–Flexibacter–Bacteroides (CFB) division. These oligonucleotides describe up to 94.7% of their target taxon with fewer than 50 non-target hits, and the authors recommend that they be investigated further. A comparison with PROBE DESIGN within the ARB software package shows that PRIMROSE is capable of identifying oligonucleotides with a higher specificity. PRIMROSE has an intuitive graphical user interface and runs on the Microsoft Windows 95/NT/2000 operating systems. It is open source and is freely available from the authors.
INTRODUCTION
Increasingly, classification of prokaryotes has involved the use of molecular phylogenetic methods. This identification invariably relies on a genotypic approach, typically involving an analysis of the 16S rRNA gene. An important repository in this regard is the Ribosomal Database Project-II (RDP) (1). Through the RDP web site (http://rdp.cme.msu.edu/) researchers are able to use a range of on-line tools to access and explore 16S rRNA sequences previously deposited with the EMBL, GenBank or DDBJ (1).
Frequently, 16S rRNA information is used to identify oligonucleotide sequences unique to specific bacteria, for use as hybridisation probes or as PCR primers (2–4). Such oligonucleotides can be specific to phylogenetic groupings as diverse as a bacterial species or an entire division. Furthermore, they are regularly used to count specific groups of bacteria in natural environments by fluorescence in situ hybridisation (FISH) (2,3) and they can be used to explore phylogenetic groupings within 16S rRNA gene libraries generated from nature (2). In this regard, a good oligonucleotide is one that matches as many of its intended target group as possible, whilst ignoring bacteria outside this target group. As 16S rRNA databases continue to grow, the quality of 16S rRNA oligonucleotides will continue to improve in this regard.
Yet, the use of probes and primers as phylogenetic tools is hampered by the speed with which new oligonucleotides can be identified, as new 16S rRNA sequence information becomes available. Probe and primer design can take time and researchers must be continually aware of the constantly growing 16S rRNA sequence repositories. This has inevitably led to the use of computers to simplify the development process. For one reason or another, most software written to date has failed to gain widespread use (see for example 5–8). The one notable exception is PROBE DESIGN, part of the ARB software package (Ludwig et al., http://arb-home.de/). ARB is a comprehensive sequence analysis package and its probe design component is increasingly being used to identify useful new oligonucleotides (see for example 9–11). To date, ARB has been successfully used to design a number of practical probes and this process has been greatly helped by the RDP making their aligned database available in ARB format.
Useful though ARB is, it is still a ‘work in progress’ and this has inevitably led to a less than obvious user interface, especially when it comes to probe design. ARB’s widespread adoption as a probe design tool has also been limited by the fact that it will only run on certain Unix-based operating systems. An important limitation is the inability of ARB to identify degenerate oligonucleotides as potential probes. Degenerate probes are oligonucleotides with one or more ambiguous base positions and their application is frequently necessary when phylogenetically diverse bacterial groups are being considered. For example, the best currently available probe for describing the Cytophaga–Flexibacter–Bacteroides (CFB) division is CFB560, a 16mer probe with two degenerate positions (12).
The computer program described in this paper arose from our need for an application that could identify potential 16S rDNA oligonucleotides quickly and effectively, and rapidly screen existing oligonucleotides. It was written with the following requirements in mind. (i) We wanted a program that could exploit the on-line tools and the downloadable database files offered by RDP. (ii) We also required a program that could identify degenerate oligonucleotides with up to two degenerate positions. (iii) The program should concentrate on doing one task well, i.e. identify potentially useful oligonucleotides through a comparative analysis of aligned 16S rRNA sequences; further theoretical design could be achieved by currently available tools, in particular those offered by the RDP. (iv) Our ideal program would also be capable of running within a Microsoft Windows environment. (v) Finally, it should require a short development time and the final product should be as intuitive and simple to use as possible.
Here we describe the resulting program, called PRIMROSE, and provide details of a range of potentially useful oligonucleotides identified by it, along with in silico comparisons of these oligonucleotides against established equivalents quoted in the literature. We also compare the ability of PRIMROSE to identify potential probes with that of the PROBE DESIGN program from ARB.
MATERIALS AND METHODS
Programming language and operating system used
PRIMROSE was written in the Perl scripting language (v.5.6.1) on a PC with an 800 MHz Intel Pentium III processor, 256 MB SDRAM and running Microsoft Windows NT 4 (service pack 6). The Perl interpreter/compiler was installed as a pre-compiled binary from http://www.activestate.com (ActivePerl v.5.6.1, binary build 630). The Perl/Tk module, a graphical user interface toolkit for Perl, was installed with the aid of the Perl Package Manager which comes with the ActiveState distribution. A Windows executable version of the program was generated using PerlApp (part of the ActiveState Perl Developers Kit v.4.0.0, build 401; http://www.activestate.com).
A modified version of our Perl scripts was also produced to run PRIMROSE on a PC running the GNU/Linux operating system (650 MHz AMD Duron processor, 128 MB SDRAM). We used the RedHat distribution v.7.2 (http://www.redhat.com/), which comes with Perl v.5.6.0. The Perl/Tk module was downloaded from the CPAN archive at http://www.cpan.org.
As part of this study, PRIMROSE was compared with the PROBE DESIGN program within ARB, a sequence handling and analysis package for the Linux and Solaris operating systems (Ludwig et al., http://arb-home.de). At the time of writing, the most recent stable version of ARB for Linux is the 15 June 1999 release and this was downloaded from the ARB web site (http://arb-home.de/). To run ARB correctly under RedHat v.7.2, the library files ld.so-1.9.5-13.i386.rpm and libc-5.3.12-31.i386.rpm were also required and these were downloaded from the RedHat web site. The most recent 16S rRNA sequence database from the RDP in ARB format currently available is the file dated 1 September 2000 (RDP release 8.0) and this was downloaded from the RDP web site. Also downloaded from the ARB web site was the 6spring2001.arb database, a smaller database containing only sequences >1400 nt in length.
In silico investigation
To test its efficacy, PRIMROSE was used to design 16S rRNA probes for some major bacterial taxa and compare their theoretical (i.e. in silico) performance with that of probes currently used to describe the same phylogenetic groups (Table (Table1).1). Probe performance was considered in the context of the current RDP database (release 8.1), and a good probe was judged primarily as one that matched with the most sequence records within its target taxon, whilst matching with the least number of non-target records.
Table 1.
| Probe | Ref. | Sequence (antisense 5′→3′) | Target rRNA | Positiona | Target | Non-target | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nameb | Sizec | Hits (no.)d | Hits (%)e | Hitsf | Increase with one mismatchg | ‘Problem’ base (% hits)h | Possible hairpin structuresi | |||||
| ALF73a | (13) | TTCCGTCTAACCGCGGG | 23S | 2043–2059 | α | |||||||
| ALF1b | (13) | CGTTCGYTCTGAGCCAG | 16S | 19–35 | 1968 | 395 | 68.8 | 207 | 1489 | 8 (78.4) | 2 | |
| ALF968 | (14) | GGTAAGGTTCTGCGCGTT | 16S | 968–985 | 1968 | 1121 | 76.1 | 187 | 6730 | 12 (99.8) | 0 | |
| BET42a | (13) | GCCTTCCCACTTCGTTT | 23S | 1027–1043 | β | |||||||
| SRB385 | (15) | CGGCGTCGCTGCGTCAGG | 16S | 385–402 | δj | 545 | 131 | 30.4 | 536 | 1851 | 13 (73.9) | 1 |
| SRB385Db | (16) | CGGCGTTGCTGCGTCAGG | 16S | 385–402 | 545 | 162 | 37.6 | 181 | 1578 | 7 (34.9) | 1 | |
| GAM42a | (13) | GCCTTCCCACATCGTTT | 23S | 1027–1043 | γ | |||||||
| CF319 | (17) | TGGTCCGTRTCTCAGTAC | 16S | 319–336 | CFB | 781 | 324 | 47.6 | 15 | 65 | 1 (92.3) | 0 |
| CFB560 | (12) | WCCCTTTAAACCCART | 16S | 563–578 | 781 | 614 | 93.9 | 2 | 513 | 9 (86.7) | 0 | |
| BAC303 | (17) | CCAATGTGGGGGACCTT | 16S | 303–319 | Bacteroides | 352 | 201 | 65.5 | 0 | 11 | 4 (100.0) | 1 |
aEquivalent position within the E.coli genome.
bTarget taxon name: α, alpha Proteobacteria (RDP code 2.28.1); β, beta Proteobacteria (2.28.2); δ, delta Proteobacteria (2.28.4); γ, gamma Proteobacteria (2.28.3); CFB, Cytophaga–Flexibacter–Bacteroides division (2.15); Bacteroides, Bacteriodes group (2.15.1.2); Enterics, Enterics and their relatives (2.28.3.27); Cytophaga group I, Cytophaga group I taxon (2.15.1.3); C.uliginosa, C.uliginosa subgroup (2.15.1.3.13).
cNumber of aligned sequence records within the target taxon according to the RDP database.
dNumber of records within the target taxon providing an exact match (i.e. hit) with the oligonucleotide.
ePercentage of records within the target taxon providing an exact match after taking into account those records that are too short to be matched.
fNumber of records outside the target taxon hit by the oligonucleotide.
gIncrease in non-target hits if one mismatch is allowed between oligonucleotide and target. A high number indicates that stringent experimental conditions would be required for the oligonucleotide to have accuracy.
hOligonucleotide base most responsible for the increase in non-target hits observed if one mismatch is allowed between oligonucleotide and target. The percentage of this increase caused by this one base is in parentheses; the higher the percentage the greater the impact this one base will have on the accuracy of the oligonucleotide.
iNumber of self-complementary positions within the oligonucleotide (PRIMROSE default setting, with only pairings of three or more consecutive Watson–Crick pairs recognised and an allowed size of between two and four bases for the potential hairpin).
We examined taxa from various depths within the prokaryotic phylogenetic tree as currently defined by the RDP. Specifically, we looked for probes for the CFB division (RDP phylogenetic code, 2.15) and, within this taxon, the Bacteroides group (2.15.1.2). We also considered the alpha (2.28.1), beta (2.28.2), delta (2.28.4) and gamma (2.28.3) subdivisions of the Proteobacteria division (2.28).
As an additional test, PRIMROSE was used to identify suitable oligonucleotides for several significant taxa for which there are currently no satisfactory 16S rDNA probes available, namely Cytophaga group I bacteria (2.15.1.3) and, within that group, the Cytophaga uliginosa subgroup (2.15.1.3.13) and the Enteric bacteria and their relatives (2.28.3.27) from the gamma Proteobacteria.
Comparison with ARB
We used the PROBE DESIGN program within ARB to design oligonucleotides for the same phylogenetic groups and compared their in silico performances with those probes identified by PRIMROSE.
Operating environments and availability
PRIMROSE is distributed under the terms of the GNU General Public Licence (http://www.gnu.org/copyleft/gpl.html) and can be downloaded as a Microsoft Windows 95/NT/2000 executable, without charge, from http://www.cardiff.ac.uk/biosi/research/biosoft/. Perl scripts of the program are also available. To run the program directly from these scripts requires Perl v.5.6.0, or later, along with the Perl/Tk module (v.800.022 or later).
The Windows version program has been tested on a number of machines and should run on most personal computers running Windows 95 or later, although the authors recommend that a PC running Windows NT/2000, with a minimum of 128 MB SDRAM, be used. A fast processor, whilst desirable, is not essential. Around 100 MB hard disk space is required for the current RDP database files. The program comes complete with an example file, instructions and a tutorial.
RESULTS
Program design and operation
PRIMROSE was written to work closely with the downloadable version of the RDP database to identify useful phylogenetic probes and primers. Figure Figure11 summarises its overall design. PRIMROSE exploits the RDP file SSU_Prok.gb, which contains all currently aligned 16S rRNA sequences in GenBank format (16 277 records for release 8.1). It also makes use of the associated files SSU_Prok.alpha, SSU_Prok.phylo and SSU_Prok.phylo.stats that contain information on bacterial names, phylogenetic positions and statistical information.

Flow chart summarising the procedure followed by PRIMROSE to identify oligonucleotides. SSU_Prok.gb is a GenBank formatted file produced by the RDP containing all currently aligned 16S rRNA sequence records (currently totalling 16 277 records in release 8.1). SSU_Prok.alpha, SSU_Prok.phylo and SSU_Prok.phylo.stats are associated files, also from RDP, that supply additional phylogenetic-based information on these records. All files are available from the RDP web site at http://rdp.cme.msu.edu/html/download.html.
PRIMROSE identifies potentially useful oligonucleotides from a set of 16S rRNA ‘target’ sequences supplied by the user. PRIMROSE will accept output from any software package capable of generating output in GenBank, Fasta or Clustal formats. However, for the full facilities of PRIMROSE to be made available, RDP_short_IDs should be used to identify records wherever possible. For this reason, PRIMROSE works best in conjunction with the on-line tools offered by RDP and, in particular, the RDP Hierarchy Browser. This facility allows the user to explore the current aligned 16S rRNA database and select for download sequences of interest as text files in GenBank format.
The size of files that can be handled by PRIMROSE is theoretically limited only by the amount of memory installed on the computer. However, for describing large numbers of sequences we found it more efficient to select a few representative records. Thus, in finding an oligonucleotide that could describe a large group such as the CFB division we found it necessary only to select a representative record from each of the major groups that make up that group (i.e. 15 records out of an available 781 aligned sequences). Full-length or near full-length 16S rRNA sequences from a reliable source were preferred, and the RDP Hierarchy Browser made this selection easy.
PRIMROSE uses one of two algorithms to identify unique oligonucleotides from the target sequences depending on whether the data are aligned or not. If aligned, algorithm 1 is used. This algorithm allows for the generation of degenerate oligonucleotides with up to two degenerate positions. Algorithm 1 can be summarised as follows.
Algorithm 1. (i) From a data set of NS sequences, of length LS, a matrix is created with each row containing a separate sequence and each column representing a separate base position within each sequence. (ii) For each of the LS columns of the matrix, the number of A, T, G and C bases (i.e. NA, NT, NG, NC) is scored. Gaps are also counted (Ngap). (iii) For each of the LS columns of the matrix the consensus base for that position is identified from these scores, i.e.
If NA = NS, base = A
Else if NT = NS, base = T
Else if NG = NS, base = G
Else if NC = NS, base = C
Else if NA + NG = NS, base = R
Else if NA + NC = NS, base = M
Else if NA + NT = NS, base = W
Else if NC + NT = NS, base = Y
Else if NC + NG = NS, base = S
Else if NG + NT = NS, base = K
Else if NA + NG + NC = NS, base = V
Else if NA + NG + NT = NS, base = D
Else if NC + NG + NT = NS, base = B
Else if NA + NC + NT = NS, base = H
Else if Ngap = NS, base = –
Else base = N
(iv) The consensus bases from the LS columns are combined to produce a single consensus sequence. (v) Any – characters (representing common gaps) in the sequence are removed. (vi) All possible oligonucleotides of length LO that can be generated from the consensus sequence are stored in an array. The contents of this array can subsequently be sorted into oligonucleotides with zero, one, two or more degenerate positions according to the number of non-canonical bases present.
If unaligned sequence data are used, PRIMROSE automatically switches to an alternative algorithm, algorithm 2. This algorithm does not require aligned data but can only identify non-degenerate oligonucleotides. Despite this feature we strongly recommend that users use aligned sequence data wherever possible. The algorithm can be summarised as follows.
Algorithm 2. (i) Store in an array all the possible oligonucleotides of length LO that can be generated from the NS sequences in the data set. (ii) Calculate the number of times each oligonucleotide occurs within the array, then remove all multiple copies so that the array is filled only with unique oligonucleotides. (iii) Retain those oligonucleotides with scores equal to or greater than a threshold value defined by the user. This number is the minimum number of sequences the oligonucleotide should describe.
The unique oligonucleotides generated by either algorithm 1 or 2 are then compared with the sequences in the full database and are ranked according to the number of records they match, within and outside their target taxon, as identified by PRIMROSE from their RDP_short_IDs (Fig. (Fig.2).2). The algorithm used in this step can be summarised as follows.

Using sequence data selected from the RDP, PRIMROSE has identified a range of possible oligonucleotides for the CFB division. Each listed oligonucleotide is a hypertext link to further information on the phylogenetic range of that sequence.
Algorithm 3. (i) A search string is created from each oligonucleotide with any non-canonical base being replaced by a simple regular expression, i.e. replace M with [AC], R with [AG], W with [AT], S with [CG], Y with [CT], K with [GT], B with [CGT], D with [AGT], H with [ACT], V with [ACG] and N with [ACGT]. (ii) Each database sequence is searched for a match with each modified oligonucleotide. If a match occurs, record a hit. If a hit occurs and the intended target for the oligonucleotide has been defined, assess whether the accession number for that sequence matches any of those of the intended targets. If the matched sequence is not the intended target, record a non-target hit. If the number of non-target hits exceeds the threshold figure defined by the user, abort this search and proceed to the next oligonucleotide.
PRIMROSE presents individual search results in a similar phylogeny format to that used by RDP (Fig. (Fig.3).3). In addition, PRIMROSE graphically presents the position of the oligonucleotide within its target taxon (Fig. (Fig.4).4). The program identifies those sequences within the target taxon that are missed because they are too short and from this information a more accurate estimation of target taxon coverage is calculated (Fig. (Fig.4).4). This aspect of the program can also be run independently of PRIMROSE, as a separate application called ROSE. ROSE is found within the PRIMROSE directory and allows the user to investigate oligonucleotides other than those identified by PRIMROSE.
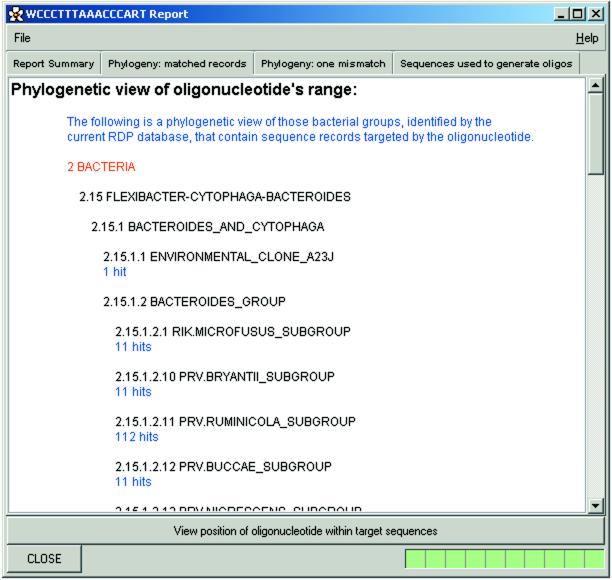
Further information on the degenerate oligonucleotide WCCCTTTAAACCCART (probe CFB560) which has been selected for further investigation by clicking the hypertext link shown in Figure Figure2.2. Displayed within this window is part of a phylogenetic breakdown of those RDP records targeted by this oligonucleotide.
In silico results
Table Table11 lists those oligonucleotides commonly quoted in the literature as suitable probes for identifying members of the alpha, beta, delta and gamma Proteobacteria, as well as the CFB division. The theoretical taxonomic ranges of these probes, in the context of the latest RDP aligned database (v.8.1), were determined and show that in almost all cases where an analysis was possible, existing probes described 30–76% of their target groups, as currently defined. The one exception was the recently designed CFB560, a degenerate CFB probe that describes 94% of its target group. Not only was the predicted coverage of most of these probes relatively low, but in four instances the number of non-target records they matched with exceeded 180 sequence records.
PRIMROSE successfully identified a number of good potential oligonucleotides for all of the target groups described in Table Table1.1. Table Table22 lists the best of these probes, alongside their predicted taxonomic ranges according to the current RDP database. For many of the taxa under consideration, PRIMROSE identified probes with >84% coverage, often with very few non-target group matches.
Table 2.
| Sequence (antisense 5′→3′) | Positiona | Intended Target | Non-target | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Nameb | Sizec | Hits (no.)d | Hits (%)e | Hitsf | Increase with one mismatchg | ‘Problem’ base (% hits)h | Possible hairpin structuresi | ||
| ATTTCACCTCTACACT | 682–697 | α | 1968 | 1350 | 87.9 | 48 | 1398 | 9 (81.9) | 0 |
| AATATCTACGAATTTC | 693–708 | 1968 | 1269 | 82.7 | 49 | 910 | 11 (74.0) | 0 | |
| TGCCGCCAGCGTTCGYT | 28–44 | 1968 | 891 | 81.2 | 23 | 389 | 1 (67.6) | 2 | |
| CSAATATCTACGAATTT | 694–710 | 1968 | 1159 | 75.5 | 12 | 392 | 13 (54.3) | 0 | |
| CCCATTGTCCAAAATTCCCC | 359–378 | β | 1085 | 851 | 93.0 | 10 | 2315 | 13 (98.0) | 1 |
| RCATMTCTACGCATTTCACT | 690–709 | 1085 | 722 | 89.0 | 5 | 774 | 20 (96.4) | 0 | |
| ACGCATTTCACTGCTACACG | 682–701 | 1085 | 701 | 86.4 | 6 | 905 | 12 (77.6) | 0 | |
| CTGCTACACGYGGAATTCYA | 672–691 | 1085 | 689 | 85.1 | 0 | 499 | 2 (96.4) | 1 | |
| CACCCGTGCGCCRCTYTACT | 96–115 | δ | 545 | 285 | 74.0 | 28 | 118 | 8 (89.8) | 2 |
| TTAGCCGGYGCTTCCT | 495–510 | 545 | 283 | 67.1 | 9 | 3255 | 15 (88.8) | 1 | |
| TTAGCCGGTGCTTCCT | 495–510 | 545 | 270 | 64.1 | 4 | 2746 | 15 (86.9) | 1 | |
| CCGTCAATTCATTTGAGTTT | 907–926 | γ | 2949 | 1757 | 75.1 | 40 | 8062 | 11 (99.3) | 2 |
| GTCAATTCATTTGAGTTTTA | 905–924 | 2949 | 1746 | 74.7 | 33 | 4066 | 9 (98.6) | 2 | |
| TRCTTCTTTTKCAACC | 1422–1437 | Enterics | 762 | 424 | 90.2 | 33 | 122 | 14 (54.9) | 0 |
| CTRCTTCTTTTKCAACCCAC | 1419–1438 | 762 | 416 | 88.5 | 33 | 114 | 15 (52.6) | 0 | |
| CCCTTTAAACCCARTRA | 561–577 | CFB | 781 | 616 | 94.2 | 8 | 619 | 8 (84.8) | 0 |
| AAACCACATGTTCCTC | 942–957 | Bacteroides | 352 | 297 | 94.6 | 2 | 72 | 15 (61.1) | 0 |
| GTGCTGATTTGACGTCATCC | 1186–1205 | 352 | 248 | 94.3 | 5 | 182 | 4 (80.8) | 1 | |
| CATTTCACCGCTACACYACW | 679–698 | Cytophaga group I | 241 | 162 | 94.7 | 44 | 304 | 20 (62.2) | 0 |
| ACTTATCACTTTCGCT | 860–875 | 241 | 161 | 93.1 | 2 | 129 | 10 (38.8) | 0 | |
| ATACTTATCACTTTCGCTTG | 858–877 | C.uliginosa group | 16 | 11 | 84.6 | 1 | 82 | 20 (97.6) | 1 |
| TTATCACTTTCGCTTGGCCG | 854–873 | 16 | 9 | 69.2 | 0 | 16 | 20 (56.3) | 0 | |
See Table Table11 for footnotes.
Beyond target range, the exact definition of a good oligonucleotide can vary according to the application. Factors that can be important include the location of the oligonucleotide’s target within the 16S rRNA sequence, the number of self-complementarities within the oligonucleotide and the number of mismatches with a non-target sequence. As a further comparison, Tables Tables11 and and22 list this additional information, which was either generated by PRIMROSE or obtained from the RDP’s on-line PROBE_MATCH facility. Overall, with the exception of CFB560, PRIMROSE was able to find oligonucleotides that performed substantially better than those 16S rRNA probes currently used.
Comparison with PROBE DESIGN from the ARB software package
ARB was also used to design oligonucleotides for the taxa listed in Table Table22 and the in silico performances of these were compared with the equivalent oligonucleotides produced by PRIMROSE. Whilst the two programs rarely identified identical oligonucleotides, the ranges and positions of the oligonucleotides identified were often very similar. For example, ARB identified the antisense oligonucleotide 5′-CCCCCGTCAATTCATTTGAG-3′ (Escherichia coli positions 910–929) as a possible gamma Proteobacteria probe. This oligonucleotide describes 74% of the gamma Proteobacteria subdivision, with 21 non-target matches, which is not unlike the similarly positioned PRIMROSE-derived oligonucleotide listed in Table Table22.
However, in numerous instances PRIMROSE identified oligonucleotides that were theoretically better than any of those highlighted by ARB. ARB failed to identify a general CFB oligonucleotide that could describe >73% of the taxon and still have a reasonable non-target range (in this case, 34 records). Similarly, it was unable to find an alpha Proteobacteria oligonucleotide able to describe >74% of that taxon (with 43 non-target hits).
DISCUSSION
There is an increasing demand for oligonucleotide probes and primers that can identify and quantify bacteria through nucleic acid hybridisation or PCR studies. For example, in recent years, FISH particularly has exploited this approach (see for example 2) and there is every indication that the newly emerging microarray technology will soon further expand the use of phylogenetic oligonucleotides (20–22). Parallel to this research has been the rapid growth in size of 16S rRNA databases that has meant currently used phylogenetic probes and primers need to be continually reassessed for their usefulness in the light of new information. Consequently, computerisation of oligonucleotide design and assessment is now almost essential. An ideal computer program is one that is simple to use and capable of running on a wide range of computer platforms and thus accessible to the widest possible scientific community. PRIMROSE meets all these objectives.
To demonstrate the effectiveness of PRIMROSE in identifying suitable oligonucleotides we needed to test its ability to identify possible probes for a range of significant bacterial taxa. The CFB group is a major bacterial division with a considerable presence in nature. The CF319a and b probes (either considered separately or as a ‘single’ degenerate probe) have been used extensively in recent years to identify environmental isolates that belong to this taxon (see for example 2,23). A study in the last year, however, has demonstrated that a new probe, CFB560, is far more effective at describing the entire CFB division, as currently recognised. This probe was identified by a comparative manual analysis of 16S rRNA sequences and its theoretical range and effectiveness were confirmed by experimentation (12). The effectiveness of CFB560 is achieved through the presence of two degenerate positions.
Using an equivalent data set to that used by O’Sullivan et al. (12), PRIMROSE was able much more rapidly to identify CFB560 as a good CFB probe (Figs (Figs22 and and3).3). It also identified other very similar possibilities (see Table Table2)2) depending on the sequences used to generate the oligonucleotides. From the perspective of this paper, such information is significant in two respects. Firstly, PRIMROSE was shown to identify the most effective CFB probe currently available. Secondly, the theory that underlies PRIMROSE’s approach was shown to be appropriate for identifying oligonucleotides of practical use.
All of the PRIMROSE-identified oligonucleotides listed in Table Table22 were substantially better in terms of their predicted range than their existing equivalents listed in Table Table1,1, showing that there is a need to reconsider the continued use of some of the current probes. In all of the other respects considered, our oligonucleotides were similar to the existing probes. Thus, the regions of the 16S rRNA gene targeted by our oligonucleotides were not markedly different in terms of their accessibility within the 3-dimensional structure of the 16S rRNA molecule (24). With the exception of one oligonucleotide, the introduction of an extra mismatch did not increase their theoretical range markedly beyond that exhibited by the Table Table11 probes, and the number of self-complementary positions present within the two sets of oligonucleotides was not especially different. Thus, on the basis of the information presented, there is no theoretical reason why at least some of these oligonucleotides should not prove useful.
Primarily, a phylogenetic probe or primer is judged in terms of its taxonomic range. How well does it match with members of its intended target taxon and to what extent does it ignore non-target bacteria? PRIMROSE identifies potential oligonucleotides on this basis. However, for an oligonucleotide to be of practical value, it must also fulfil other criteria that can often only be assessed empirically, and the exact nature of these criteria may vary from application to application. For example, a good oligonucleotide probe for dot-blot hybridisation studies may fail as an rRNA-targeting FISH probe because of the in situ inaccessibility of its target within an undisrupted ribosome. In this regard, probe CF319 has a proven record, whilst the use of CFB560 as an in situ probe has yet to be demonstrated. Thus, good oligonucleotide design must combine theory with practice; PRIMROSE is designed to assist with the former.
PRIMROSE is not the only program currently available for oligonucleotide design; the PROBE DESIGN program within the software package ARB has also been used successfully in a number of recent studies. But, whilst PRIMROSE is not designed to supplant PROBE DESIGN, it does offer several significant advantages over the alternative package. (i) PRIMROSE is able to identify degenerate oligonucleotides, which is an important strategy in oligonucleotide design, as CFB560 demonstrates. (ii) It runs on the Microsoft Windows operating systems. (iii) It is quick to master and easy to use through having a simple and intuitive graphical user interface. (iv) It presents the taxonomic range of an oligonucleotide in terms of the familiar RDP phylogenetic tree with additional information not supplied by the RDP web site. (v) It does not rely on specialised database formats and so is instantly updateable when the new RDP database release becomes available. (vi) Through ROSE, existing probes can be checked against future RDP releases. (vii) Although PRIMROSE is designed for RDP aligned sequences it can also be used with user-defined nucleic acid sequence databases.
ACKNOWLEDGEMENT
The Natural Environment Research Council (NERC) supported this work through its Marine and Freshwater Microbial Biodiversity thematic program (grant NER/T/S/2000/637).
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkf450
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/30/15/3481/6374554/gkf450.pdf
Citations & impact
Impact metrics
Article citations
Synthase-selected sorting approach identifies a beta-lactone synthase in a nudibranch symbiotic bacterium.
Microbiome, 11(1):130, 13 Jun 2023
Cited by: 1 article | PMID: 37312139 | PMCID: PMC10262491
The mature phyllosphere microbiome of grapevine is associated with resistance against Plasmopara viticola.
Front Microbiol, 14:1149307, 11 Apr 2023
Cited by: 3 articles | PMID: 37113228 | PMCID: PMC10127535
Simultaneous Metabarcoding and Quantification of Neocallimastigomycetes from Environmental Samples: Insights into Community Composition and Novel Lineages.
Microorganisms, 10(9):1749, 30 Aug 2022
Cited by: 7 articles | PMID: 36144352 | PMCID: PMC9504928
Ecological Impacts of Aged Freshwater Biofilms on Estuarine Microbial Communities Elucidated Through Microcosm Experiments: A Microbial Invasion Perspective.
Curr Microbiol, 79(7):210, 06 Jun 2022
Cited by: 0 articles | PMID: 35666311
Bioinformatic Tools and Guidelines for the Design of Fluorescence In Situ Hybridization Probes.
Methods Mol Biol, 2246:35-50, 01 Jan 2021
Cited by: 6 articles | PMID: 33576981
Go to all (141) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
New degenerate Cytophaga-Flexibacter-Bacteroides-specific 16S ribosomal DNA-targeted oligonucleotide probes reveal high bacterial diversity in River Taff epilithon.
Appl Environ Microbiol, 68(1):201-210, 01 Jan 2002
Cited by: 64 articles | PMID: 11772628 | PMCID: PMC126579
Graphical representation of ribosomal RNA probe accessibility data using ARB software package.
BMC Bioinformatics, 6:61, 21 Mar 2005
Cited by: 22 articles | PMID: 15777482 | PMCID: PMC1274257
Review Free full text in Europe PMC
Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment.
Microbiology (Reading), 142 ( Pt 5):1097-1106, 01 May 1996
Cited by: 584 articles | PMID: 8704951
25 years of serving the community with ribosomal RNA gene reference databases and tools.
J Biotechnol, 261:169-176, 23 Jun 2017
Cited by: 314 articles | PMID: 28648396
Review






