Abstract
Free full text

Polymorphism Ratio Sequencing: A New Approach for Single Nucleotide Polymorphism Discovery and Genotyping
Abstract
Polymorphism ratio sequencing (PRS) combines the advantages of high-throughput DNA sequencing with new labeling and pooling schemes to produce a powerful assay for sensitive single nucleotide polymorphism (SNP) discovery, rapid genotyping, and accurate, multiplexed allele frequency determination. In the PRS method, dideoxy-terminator extension ladders generated from a sample and reference template are labeled with different energy-transfer fluorescent dyes and coinjected into a separation capillary for comparison of relative signal intensities. We demonstrate the PRS method by screening two human mitochondrial genomes for sequence variations using a microfabricated capillary array electrophoresis device. A titration of multiplexed DNA samples places the limit of minor allele frequency detection at 5%. PRS is a sensitive and robust polymorphism detection method for the analysis of individual or multiplexed samples that is compatible with any four-color fluorescence DNA sequencer.
Genetic studies are expanding past the identification of rare Mendelian diseases to encompass common complex disorders such as cancer, neurological diseases, autoimmune diseases, and cardiovascular diseases. The abundance and stability of single nucleotide polymorphisms (SNPs) has made them the DNA variation of choice for such endeavors. Due to the low penetrance of individual alleles, association studies designed to uncover significant genetic risk factors require the scoring of hundreds of thousands of SNPs (Kruglyak 1999) in several thousand samples (Dahlman et al. 2002). Multiplexed high-throughput genotyping methods are essential to accommodate this challenging number of SNPs and samples.
Current methods of SNP detection include microarrays (Chee et al. 1996), allele-specific polymerase chain reaction (PCR; Liu et al. 1997), oligonucleotide ligation assays (Grossman et al. 1994; Samiotaki et al. 1994; Tobe et al. 1996), single-base extension (Syvanen et al. 1993; Pastinen et al. 1996; Shumaker et al. 1996), TaqMan (Livak et al. 1995), and molecular beacons (Tyagi et al. 1998). These methods are designed to interrogate SNPs in one sample at a time. Modifications such as kinetic PCR (Germer et al. 2000) and detection using denaturing high-performance liquid chromatography (HPLC; Giordano et al. 2001) can extend these procedures to multiplexed sample analysis. Each method offers a trade-off between sensitivity, throughput, parallel processing, user accessibility, and cost, and they all experience limitations due to local DNA hybridization characteristics. Selection of allele-specific probes is confined to the DNA sequence immediately adjacent to or surrounding the targeted polymorphism. This restriction can lead to nonuniform hybridization characteristics, ambiguous results, or the rejection of otherwise informative SNPs in a significant percentage of the markers. Additionally, novel polymorphisms are not detected by these approaches. These limitations are particularly important when studying functional or rare SNPs.
Quality-based resequencing approaches do not experience the limitations described above and are considered the gold standard for SNP scoring (Rieder et al. 1998). Despite this advantage and the availability of automated DNA sequencing instruments, resequencing has not proven to be efficient or cost-effective for large SNP studies. This failing arises because of the inability to multiplex conventional sequencing reactions and because the sequencing assay was not specifically designed for polymorphism detection.
In this report we describe polymorphism ratio sequencing (PRS), a method that combines the sequence independence and assay reliability of DNA sequencing with the sensitivity necessary to perform parallel, multiplexed genotyping. PRS is a dideoxy terminator-based method that employs standard sequencing chemistry in a novel labeling and pooling scheme to compare sample and reference DNA templates directly in a single lane. The result is an internally controlled method for polymorphism discovery, multiplexed allele frequency determination, and genotyping. The compact (16,569 bp) and well-characterized human mitochondrial genome was selected as a test system for PRS development. Two mitochondrial genomes from unrelated Centre d'Etude Polymorphisme Humane (CEPH) individuals were simultaneously scanned for sequence variations using PRS and analyzed in <30 min using a microfabricated capillary array electrophoresis device (μCAE; Paegel et al. 2002). Because standard sequencing chemistry is used, PRS is also compatible with conventional four-color fluorescence DNA sequencing instruments. Our analysis identified 30 previously characterized and six novel polymorphisms. One polymorphism was chosen to demonstrate the sensitivity of the method in determining the minor allele frequency in a multiplexed sample.
RESULTS
Coding and Pooling Scheme
Scheme
Figure Figure11 presents the PRS base coding scheme for A/C and G/T companion traces. Unlike conventional DNA sequencing, PRS simultaneously compares two DNA templates, sample and reference, using different fluorescent dyes to label identical base termination ladders. Each trace is composed of four electropherograms that concurrently interrogate two bases: A and C in Figure Figure1A1A and G and T in Figure Figure1B.1B. Because each trace compares only two sets of bases at a time, gaps between peaks occur in regions of nonrepresented bases. Two complementary PRS reactions yield complete sequence coverage. Sample and reference templates produce coincident peaks at each base position when the sequences are identical and noncoincident peaks when there are polymorphisms.
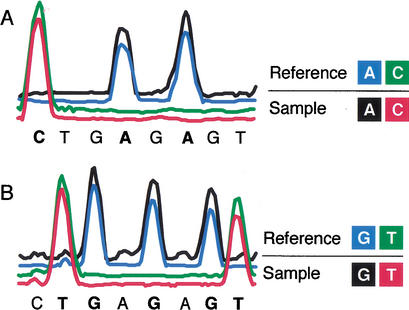
Polymorphism ratio sequencing labeling and reaction tube pooling scheme. Insets present the four-color labeling scheme for bases A/C and G/T in sample and reference sequences. (A) A and C extension products from the sample template are respectively labeled with R110 (λmax 525 nm) and ET-TAMRA (λmax 580 nm). Similarly, A and C extension products from the reference template are labeled with ET-R6G (λmax 555 nm) and ET-ROX (λmax 605 nm). Products from the four reactions are combined and analyzed in a single lane. (B) G and T extension products from sample template are labeled with R110 and ET-TAMRA, and reference template G and T extension products are labeled with ET-R6G and ET-ROX, respectively. The baseline signal intensity of the four channels has been offset to show coincident peaks more clearly.
Polymorphism Identification
Processing of PRS data is considerably simplified compared to conventional sequence data analysis because base calling is not necessary to identify polymorphisms. After color matrixing and normalization, sample and reference data are aligned to each other such that all nonvariant peaks overlap. A variance plot is then generated from the aligned data by squaring the signal intensity difference between the sample and reference electropherograms. This Δ2 plot produces excellent signal-to-noise ratios and is used for detection of polymorphic bases. Polymorphisms were identified at peak positions where the Δ2 plot exceeded three standard deviations (SDs) from the mean. Figure Figure22 illustrates how polymorphisms are identified along with examples of the detected variations. At a polymorphic site, the sample and reference traces are not coincident. This deviation manifests itself in different ways depending on the type of polymorphism. A transition polymorphism (e.g., G→A) is exemplified by the loss of the sample G peak in the G/T trace and the appearance of a new A peak in the A/C trace (Fig. (Fig.2A).2A). Whereas a transition mutation is detected as a one-base change in two PRS traces, a transversion mutation (e.g., C→A) is detected as two base changes within one PRS trace (Fig. (Fig.2B).2B). Although the C and A peaks overlap, they are mismatched relative to the defined coding scheme and therefore produce conspicuous squared signal differences. Due to the dependence of base incorporation frequency on the preceding base, a C→A base change creates a “footprint” on the following base, seen as different sample/reference peak heights for the following C base. Incorporation frequency parity is restored for subsequent bases. Insertion-deletion polymorphisms result in a sample/reference frame shift after the variant site (Fig. (Fig.2C).2C). Realignment of the traces is achieved by gapping and displacing the sample or reference traces until the Δ2 plot is minimized. The loss of single-base resolution for higher-molecular-weight fragments and for compressed regions has only a modest effect on the relative sample/reference signal. Figure Figure2D2D demonstrates that positive variant identification, which could not be accurately scored by conventional sequencing, was possible in what is typically considered very low-quality sequence.
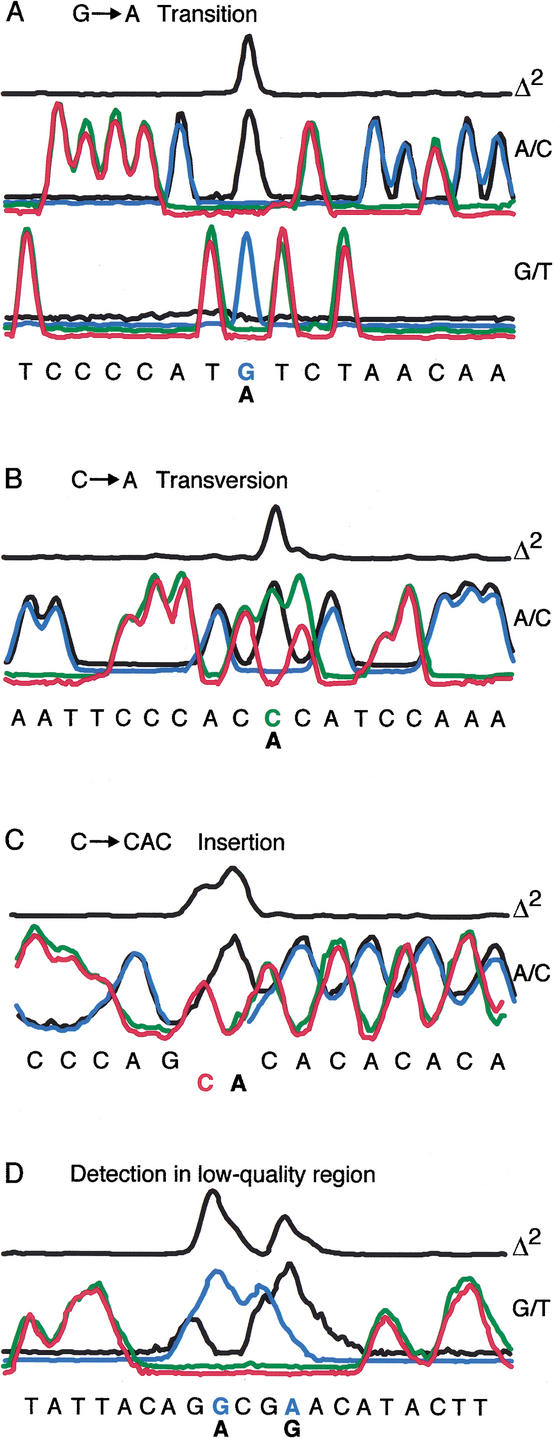
Representative traces of various polymorphisms. The Δ2 plot is generated by squaring the sum of the differences between the sample and reference traces for all four comparisons. (A) Two companion traces for a G→A transition polymorphism. Transition events detected in one trace are confirmed in the companion trace. (B) A/C trace for an C→A transversion displays mismatched coincident peaks. (C) Example of a C→CAC insertion. Insertion results in a sample/reference frame shift, which is corrected by realigning the traces after the insertion. (D) Positive polymorphism identification in a “low-quality” sequence region. Single-base resolution is not required for accurate base variation detection.
Genomic PRS data from two individuals aligned to a map of the human mitochondrial genome are presented in Figure Figure3.3. The genome encodes 13 proteins, 22 tRNAs, the 12S and 16S rRNA, and contains 1025 characterized genetic variations, of which over 50 have known pathogenesis. One individual was arbitrarily chosen as the reference and the other as the sample. Sequence variations between the two individuals produce definitive signals in the squared difference plot. Variants were localized onto the Anderson human mitochondrial sequence (Anderson et al. 1981) and compared to known polymorphisms in the MITOMAP database (http://www.mitomap.org; Kogelnik et al. 1996). Polymorphic sites were reconfirmed using conventional fluorescence-based DNA sequencing methods (Ju et al. 1995a). The outermost black circle in Figure Figure33 presents a binary representation of the squared difference in signal for all four bases. Thirty known and six novel polymorphisms (n3720, n3849, n4553, n4736, n5426, and n16291) were identified. Table Table11 lists the identified polymorphisms and corresponding amino acid changes. Mutation n13708 is a secondary Leber's hereditary optic neuropathy risk factor (Brown et al. 1992).
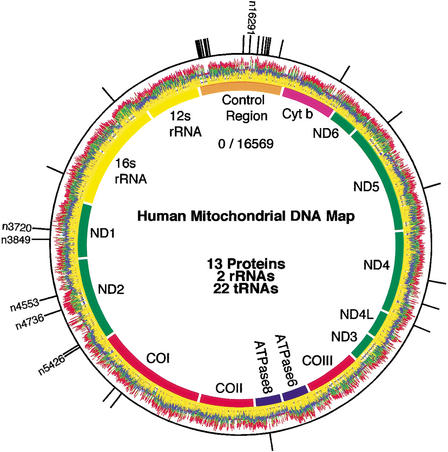
Whole mitochondrial genome analysis. Genomic PRS data is presented wrapped around a map of the human mitochondrial genome. For clarity, traces are offset and black has been replaced with yellow. The outer circle presents a binary representation of the squared cumulative difference (Δ2) plot. Novel polymorphisms are labeled with the mtDNA nucleotide position. Data were collected in <30 min. using a microfabricated capillary array electrophoresis sequencing device.
Table 1.
Mitochondrial DNA Variation Detection
Detection
| Positiona | Base changeb | Genec | Amino acid changed | Positiona | Base changeb | Genec | Amino acid changed |
 185 185 | G → → A A | CTRL | — | 8473 | T → → C C | ATP8 | Syn |
 188 188 | A → → G G | CTRL | — | 10876 | A → → G G | ND4 | Syn |
 217 217 | T → → C C | CTRL | — | 11251 | A → → G G | ND4 | Syn |
 228 228 | G → → A A | CTRL | — | 11467 | A → → G G | ND4 | Syn |
 462 462 | C → → T T | CTRL | — | 12612 | A → → G G | ND5 | Syn |
 489 489 | T → → C C | CTRL | — | 13708 | G → → A A | ND5 | Ala → Thr |
 508 508 | A → → G G | CTRL | — | 14798 | T → → C C | CTYB | Phe → Leu |
 514 514 | CAC → → C C | CTRL | — | 15907 | A → → G G | CTRL | — |
| 1811 | A → → G G | 16S | — | 16051 | A → → G G | CTRL | — |
| 3010 | G → → A A | 16S | — | 16069 | C → → T T | CTRL | — |
| 3720 | A → → G* G* | ND1 | Syn | 16092 | T → → C C | CTRL | — |
| 3849 | G → → A* A* | ND1 | Syn | 16126 | T → → C C | CTRL | — |
| 4553 | T → → C* C* | ND2 | Syn | 16129 | G → → C C | CTRL | — |
| 4736 | T → → C* C* | ND2 | Syn | 16183 | A → → C C | CTRL | — |
| 5390 | A → → G G | ND2 | Syn | 16189 | T → → C C | CTRL | — |
| 5426 | T → → C* C* | ND2 | Syn | 16291 | C → → A* A* | CTRL | — |
| 6045 | C → → T T | COI | Syn | 16362 | T → → C C | CTRL | — |
| 6152 | T → → C C | COI | Syn | 16366 | C → → T T | CTRL | — |
Multiplexed Sample Analysis
Figure Figure4A4A presents two-color PRS trace data from a separate experiment in which a reference template is compared to multiplexed sample templates containing varying ratios of a C→T transition. The reference template contains DNA from a single individual and therefore produces identical peaks in all traces. The polymorphic base in the multiplexed templates produces peak heights consistent with the template ratios. The peak height ratio at the polymorphic base is used to determine the allele frequency. Figure Figure4B4B presents the calibration curve generated from eight multiplexed samples ranging in concentration from 0%–100%. For each of the eight samples, four separate PCR and PRS reactions were performed and analyzed in independent lanes. The results are linear through all concentrations (R2 =
= 0.9991). The minor allele frequency limit of detection achievable by PRS is set by the lowest concentration in this study (5%) in which the signal-to-noise ratio is 7.0.
0.9991). The minor allele frequency limit of detection achievable by PRS is set by the lowest concentration in this study (5%) in which the signal-to-noise ratio is 7.0.
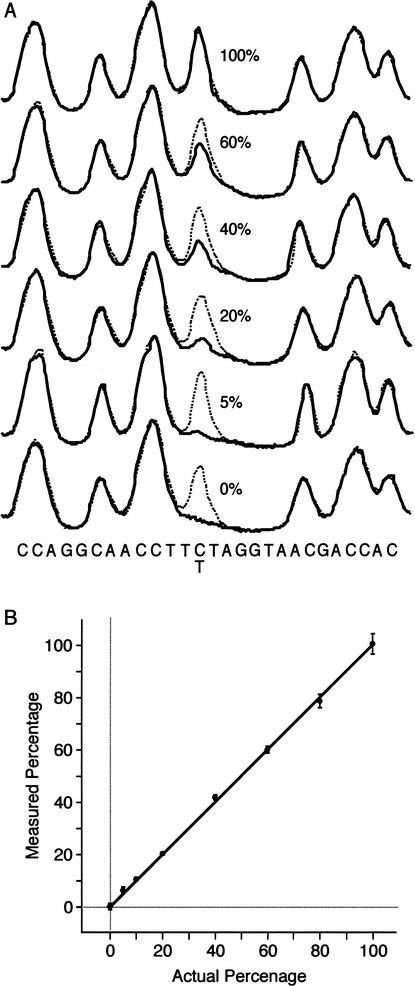
Minor allele frequency limit of detection analysis in multiplexed samples. (A) Two-color PRS traces comparing a single individual reference (dotted) and a multiplexed sample (solid). Multiplexed samples contain varying ratios of C→T polymorphic templates. (B) Allele frequency calibration curve generated from 32 independent traces. Error bars indicate 99% confidence interval (R2 =
= 0.9991).
0.9991).
DISCUSSION
The results presented here demonstrate that PRS is an accurate, high-throughput approach for SNP discovery and genotyping of individual and multiplexed samples that is compatible with all four-color fluorescence DNA sequencing instruments. This method derives its unique capabilities from the simultaneous comparison of sample and reference template in the same lane, producing trace data that are internally controlled at every base position and resistant to the lane-to-lane injection, mobility, and detection artifacts inherent to any DNA sequencing method. Although peak height and base spacing are not uniform throughout a sequencing run, the relative signal intensity between the sample and reference electropherograms is remarkably consistent, even in the low-quality region seen in Figure Figure2D.2D. As a result, PRS data quality is not impaired by conventional DNA sequencing anomalies resulting from local sequence content (e.g., GC compressions). Because terminator incorporation rates are consistent for a given sequence (Kwok et al. 1994), PRS signal intensities from nonvariant sample and reference templates align nearly perfectly, permitting direct polymorphism detection and accurate quantitation of multiplexed DNA samples. Although sequence artifacts and alignment may also be corrected in conventional four-color sequencing through postrun software, it is our experience that performing such corrections on separate traces is problematic because the additional data manipulations reduce quantitation and accuracy.
PRS provides facile and unambiguous polymorphism detection in low-quality as well as high-quality sequence regions. There is no need to align PRS data to a consensus sequence and compare the sometimes ambiguous base calls, as in conventional sequence-based analysis. Although probabilistic base call quality scores are useful in resolving such ambiguities (Rieder et al. 1998), polymorphisms in low-quality conventional sequence data are indistinguishable from sequencing errors (Marth et al. 1999). PRS relies on relative and not absolute sequence quality, thereby enabling positive polymorphism detection in regions well below single-base resolution. Additionally, relative sequence quality is enhanced in the PRS method because gaps produced by comparing only two bases in each trace result in fewer overlapping peaks. Every variant base also generates two distinct signals—the loss of one peak and the appearance of another, as seen in Figure Figure2A.2A. This complementarity provides internal, robust error checking not possible with single-read approaches.
Our analysis of the human mitochondrial genome demonstrates the feasibility of using PRS for high-throughput genotyping, SNP discovery, and diagnostics. In this study, 30 previously characterized and six novel polymorphisms were identified, and all classes of simple genetic variation (i.e., transition, transversion, and insertion-deletion) were detected. The PRS method has important advantages over hybridization-based approaches that require the design of probes in the immediate vicinity of the variant base. This requirement precludes the use of probes in regions where the local sequence content does not permit efficient or consistent hybridization. The PRS method provides the flexibility to design optimized probes up to several hundred bases away from the target SNP. As a result, all SNPs including important rare or functional polymorphisms can be scored. Additionally, sequences surrounding the targeted SNP are also scanned, providing complete and unbiased detection of novel or unanticipated variant sites. Although each polymorphism was visually inspected in this study, the high signal-to-noise ratio intrinsic to the squared difference (Δ2) plot and two-signal error checking should facilitate the development of software for automated detection.
The PRS method is particularly well suited for clinical genetic diagnostics, in which sensitivity and a low error rate are critical. For example, unambiguous detection of mtDNA mutation n13708, a secondary Leber's hereditary optic neuropathy mutation, was achieved. Detection of genetic mutations in different tissue types within an individual is also possible. Indicative and possibly causative mitochondrial genetic variations are frequently observed between healthy and cancerous tissues (Polyak et al. 1998; Fliss et al. 2000). Application of the PRS method to detect such variations requires only the use of reference DNA from patient blood and sample DNA from tumor tissue. The sensitivity of the PRS method to as little as 5% genetic variations addresses important problems associated with tumor sample heterogeneity. Furthermore, the ability to scan for known and unknown variations is important, because many of the mutations seen in cancerous tissue are novel (Fliss et al. 2000).
Case-control association studies employing genome-wide screening require the interrogation of many thousands of SNPs (Risch and Merikangas 1996; Kruglyak 1999). The low phenotypic effect of any single gene in complex disease traits necessitates the use of large numbers of samples to obtain significant association. The examination of multiplexed samples using PRS will significantly reduce sample preparation and analysis to a tractable level in such studies (Barcellos et al. 1997). The PRS method provides sufficient quantitative sensitivity to multiplex 20 haploid (e.g., mitochondrial) or 10 diploid samples and to detect as little as 5% allele frequency in population pools. The standardized sequencing chemistries and electrophoretic separation technologies used in PRS will also facilitate parallel polymorphism screening and allele frequency determination. Such analysis should prove useful for SNP validation, rare allele identification, and genetic association studies involving affected and unaffected sample pools.
The PRS coding scheme presented here requires more tube manipulations than conventional dye-terminator sequencing and is not beneficial for all sequence comparisons. Bulk preparation of reference reactions, sample multiplexing, and alterations to the coding/pooling scheme4 all provide avenues to simplify the assay. Nonetheless, there are a number of situations where the benefits of PRS analysis justify the additional overhead. In critical sequence analysis such as diagnostic sequencing and tumor/normal comparisons, error rates must be low. To achieve high-quality sequence information over large regions using conventional sequencing methods, multiple, overlapping sense and anti-sense traces must be obtained and aligned. By comparison, the PRS method has the potential to provide, in a single run, high-quality sequence comparison over an extended region of the trace. More importantly, in situations such as association studies where allele frequency is the desired information, one can take advantage of the remarkable sensitivity of the PRS method to multiplex many samples. This quantitative, multiplexed capability is not provided by any existing resequencing method. The present study highlights the potential of alternative base labeling and pooling schemes to enhance conventional sequencing. The ultimate utility of PRS will depend on a balance between the benefits afforded by the method and any additional reaction setup complexity.
METHODS
Template Amplification
Twenty-four pairs of overlapping PCR primers (Operon Technologies and BioSource International) were used to amplify mitochondrial DNA (mtDNA; Rieder et al. 1998). Universal dye-primer sequences, −40 M13 forward (GTT TTC CCA GTC ACG ACG) and −28 M13 reverse (AGG AAA CAG CTA TGA CCA TG), were added to the 5′-end of the forward and reverse primers, respectively. Mitochondrial DNA from two unrelated CEPH individuals (reference NA 13116, sample NA 10859) was amplified in 48 25-μL PCR reactions [10 ng genomic DNA, 0.2 mM dNTPs, 1.5 mM MgCl2, 0.3 μM primers, 20 mM Tris-HCl pH 8.4, 50 mM KCl, and 1.25 U Platinum Taq DNA polymerase (Invitrogen)]. PCR reactions were incubated at 94° C for 1 min to denature the template and activate the polymerase, followed by 30 cycles at 94° C for 30 sec, annealing at 61° C for 45 sec, and extension at 72° C for 1.5 min with a final incubation at 72° C for 3 min using an Eppendorf Mastercycler (Brinkmann Instruments).
Polymorphism Ratio Sequencing
After thermal cycling, unincorporated primers and dNTPs were degraded by the addition of 30 U of exonuclease I (10 U/μL) and 2 U of shrimp alkaline phosphatase (1 U/μL, from USB). Reactions were incubated at 37° C for 30 min followed by a 15-min inactivation at 80° C. Treated PCR reactions were then diluted with water to a working volume of 90 μL (~12 ng/μL). Dye-primer PRS was performed in separate tubes for each PCR amplicon as follows: 2 μL C, A, G, or T Thermo Sequenase nucleotide mix (US 79525, Amersham Biosciences), 2 μL 0.4 mM energy transfer (ET) primer (fluorescence emission maxima of 525, 555, 580, or 605 nm; Ju et al. 1995b), 4 μL diluted PCR reaction, 2 μL water. Reactions were cycled 25 times at 95° C for 30 sec, 55° C for 15 sec, and 72° C for 1 min followed by pooling and precipitation (3 μL 7.5 M ammonium acetate, 110 μL 100% ethanol). Precipitated fragments were collected after 15 min by centrifugation at 16,000 × g for 20 min followed by a 400 μL wash with 70% ethanol. Samples were air-dried for 15 min and resuspended in 15 μL 50% formamide. A 1 μL aliquot from each of the 96 samples was robotically loaded (Microlab 4000) onto a μCAE device for sequencing.
High-speed DNA sequencing on a μCAE device is described in detail elsewhere (Paegel et al. 2002). Briefly, the radial μCAE device contains 96 17-cm long capillaries sharing a common anode at the center of the device. Separate sample wells, which encircle the outer perimeter of the microplate, are connected to the 96 capillaries via a cross-channel injector. Prior to operation, all capillaries are simultaneously filled with linear polyacrylamide (CEQ Separation Gel—LPA I, Beckman Coulter) through the common anode port with a high-pressure gel loader (Scherer et al. 2001). Ninety-six unique PRS samples are then robotically loaded onto the μCAE device. The loaded device is placed on a four-color rotary confocal fluorescence scanner (Shi et al. 1999) and fitted with an electrode array ring. Samples are then electrophoretically injected into the capillaries (500 V/cm for 30 sec) followed by fragment separation to the common anode at 150 V/cm for 29 min.
Data Analysis and Polymorphism Identification
Identification
Raw data preprocessing was performed with the program BaseFinder (Giddings et al. 1998). Leading low-quality data (primer peak and first 10 bases) and the trailing coelution peak were deleted. After baseline subtraction, color matrix parameters determined from a standard four-color M13 sequencing reaction were applied uniformly to the PRS data. A local intensity scaling factor was applied to normalize variations in template DNA concentration, dye-primer quantum yield, and photomultiplier tube gain. Next, traces were aligned to each other such that the squared difference between the sample and reference signal was minimized. A single Δ2 plot was generated by squaring the sum of the absolute sample versus reference signal intensity differences from all eight traces. To identify polymorphisms, the standard deviation of the Δ2 plot was calculated from the observed values discretized at each peak position. Identified polymorphisms (≥3 SDs) were localized onto the Anderson human mitochondrial sequence (GenBank accession no. J01415; Anderson et al. 1981).
Multiplexed Sample Analysis
A SNP (n6045 C→T) detected in the whole mitochondrial genome scan was selected to conduct a sensitivity study. For this analysis, genomic DNA from a reference sample (individual 1) and eight multiplexed samples (individuals 1 and 2) was prepared. Multiplexed samples consisted of 100%, 80%, 60%, 40%, 20%, 10%, 5%, and 0% DNA from individual 1 and a complementary amount of DNA from individual 2. Pools were prepared such that the total DNA concentration (10 ng/μL) remained constant. For each multiplexed and reference sample, four replicate PCR and two-color PRS reactions were performed as described above. For each trace, Gaussian curves were fit to the polymorphic and reference peak, and the ratio of peak heights was calculated. The signal-to-noise ratio was determined by dividing the average of each of the four ratios by the standard deviation.
WEB SITE REFERENCES
http://www.mitomap.org; MITOMAP database.
Acknowledgments
Microfabrication was carried out at the University of California Berkeley Microfabrication Laboratory. This research was supported by grants from the NIH (HG01399, P01 CA77664) and from the Director, Office of Science, Office of Biological and Environmental Research of the U.S. Department of Energy under contract DEFG91ER61125. B.M.P. was supported by a National Human Genome Research Institute fellowship from the Berkeley Program in Genomics (T32 HG00047).
 The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Notes
E-MAIL ude.yelekreb.mehcc.cniz@hcir; FAX (510) 642-3599.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.396203.
4Straightforward modifications to the PRS coding/pooling scheme such that the forward and reverse extension reactions are performed simultaneously would reduce the number of reaction tubes by a factor of two. Another factor of two reduction is provided by the use of uniquely-labeled energy transfer (ET)-cassette primers (Berti et al. 2001). For example, by pooling sample and reference templates amplified with uniquely-tailed PCR primers and performing PRS analysis using corresponding cassette-labeled primers, it is feasible to obtain forward and reverse PRS data on both templates with only four reaction tubes—the same number of tubes as dye-terminator sequencing.
REFERENCES
Articles from Genome Research are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gr.396203
Read article for free, from open access legal sources, via Unpaywall:
https://genome.cshlp.org/content/13/2/287.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1101/gr.396203
Article citations
Association of single nucleotide polymorphism variations in CRYAA and CRYAB genes with congenital cataract in Pakistani population.
Saudi J Biol Sci, 29(4):2727-2732, 03 Jan 2022
Cited by: 1 article | PMID: 35531184 | PMCID: PMC9073017
Estimating Copy-Number Proportions: The Comeback of Sanger Sequencing.
Genes (Basel), 12(2):283, 17 Feb 2021
Cited by: 5 articles | PMID: 33671263 | PMCID: PMC7922598
High throughput single cell analysis of mitochondrial heteroplasmy in mitochondrial diseases.
Sci Rep, 10(1):10821, 02 Jul 2020
Cited by: 15 articles | PMID: 32616755 | PMCID: PMC7331593
Recent Advances in Detecting Mitochondrial DNA Heteroplasmic Variations.
Molecules, 23(2):E323, 03 Feb 2018
Cited by: 28 articles | PMID: 29401641 | PMCID: PMC6017848
Review Free full text in Europe PMC
High-performance detection of somatic D-loop mutation in urothelial cell carcinoma patients by polymorphism ratio sequencing.
J Mol Med (Berl), 94(9):1015-1024, 31 Mar 2016
Cited by: 5 articles | PMID: 27030170 | PMCID: PMC8883517
Go to all (18) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Pyrosequencing for detection of mutations in the connexin 26 (GJB2) and mitochondrial 12S RNA (MTRNR1) genes associated with hereditary hearing loss.
Hum Mutat, 20(4):312-320, 01 Oct 2002
Cited by: 8 articles | PMID: 12325027
A single nucleotide polymorphism map of the mitochondrial genome of the parasitic nematode Cooperia oncophora.
Parasitology, 128(pt 4):421-431, 01 Apr 2004
Cited by: 19 articles | PMID: 15151148
Analysis of sequence variations in several human genes using phosphoramidite bond DNA fragmentation and chip-based MALDI-TOF.
Genome Res, 14(1):134-141, 01 Jan 2004
Cited by: 7 articles | PMID: 14707175 | PMCID: PMC314290
High-performance genetic analysis using microfabricated capillary array electrophoresis microplates.
Electrophoresis, 22(18):3845-3856, 01 Oct 2001
Cited by: 27 articles | PMID: 11700713
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: P01 CA077664
Grant ID: P01 CA77664
NHGRI NIH HHS (4)
Grant ID: R01 HG001399
Grant ID: T32 HG00047
Grant ID: T32 HG000047
Grant ID: HG01399





