Abstract
Free full text

Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa
Abstract
Pseudomonas aeruginosa is a ubiquitous environmental bacterium capable of causing a variety of life-threatening human infections. The genetic basis for preferential infection of certain immunocompromised patients or individuals with cystic fibrosis by P. aeruginosa is not understood. To establish whether variation in the genomic repertoire of P. aeruginosa strains can be associated with a particular type of infection, we used a whole-genome DNA microarray to determine the genome content of 18 strains isolated from the most common human infections and environmental sources. A remarkable conservation of genes including those encoding nearly all known virulence factors was observed. Phylogenetic analysis of strain-specific genes revealed no correlation between genome content and infection type. Clusters of strain-specific genes in the P. aeruginosa genome, termed variable segments, appear to be preferential sites for the integration of novel genetic material. A specialized cloning vector was developed for capture and analysis of these genomic segments. With this capture system a site associated with the strain-specific ExoU cytotoxin-encoding gene was interrogated and an 80-kb genomic island carrying exoU was identified. These studies demonstrate that P. aeruginosa strains possess a highly conserved genome that encodes genes important for survival in numerous environments and allows it to cause a variety of human infections. The acquisition of novel genetic material, such as the exoU genomic island, through horizontal gene transfer may enhance colonization and survival in different host environments.
Members of the genus Pseudomonas are some of the most diverse bacterial species in the environment. Particularly important are Pseudomonas aeruginosa, which are commonly encountered aerobic microbes that have evolved the remarkable capacity to inhabit diverse natural environments and infect higher organisms such as insects, plants, and animals (1). In humans, P. aeruginosa can colonize virtually any mucosal surface and invade tissues and blood (2). Its ability to thrive in a broad range of environments is in part caused by the fact that it possesses a large and diverse genome (3). Furthermore, P. aeruginosa possesses the largest proportion of regulatory genes (≈1 in 10) of all of the sequenced bacterial genomes (3). This striking feature likely provides a means for coordinating the expression of its many genes in response to a wide range of environmental demands.
Comparative genomics based on the analysis of complete genomes of different strains of the same species can provide valuable insights into the acquisition or loss of genes through horizontal gene transfer and evolution of genes through changes at the nucleotide sequence level. With the availability of DNA microarrays, a larger number of genomes can be compared (4). When representative subsets of strains are examined, useful evolutionary questions can be answered by genome content analysis. Particularly informative have been those studies that used genomic microarray hybridization technology to investigate the epidemiology of disease and correlate the loss of genes with changes in virulence. Specifically, microarray analysis has been used to demonstrate a correlation between certain genes in Vibrio cholerae and specific pandemics (5). In addition, a microarray hybridization-based study of Staphylococcus aureus variants associated with toxic shock syndrome and methicillin resistance revealed that these strains evolved in parallel with changing conditions in the host environment and antibiotic treatment (6). The same approach was used to demonstrate the genetic basis for attenuation of virulence in strains of Mycobacterium tuberculosis including those used for vaccination (7). Analysis of Helicobacter pylori genomes from a wide range of strains has resulted in the identification of a limited number of variable chromosomal segments and their association with specific virulence factors (8). Further studies of this organism have provided evidence for the evolution of the genome of individual clones (including deletions of genes) in a highly restricted niche (9).
To correlate the genome content of P. aeruginosa strains with the various niches that it can inhabit and the different diseases that it can cause, we assembled a collection of strains and assessed their genomic repertoire by using a whole-genome DNA microarray. We have shown that the genome of this organism is highly conserved and that there exists a core set of genes, including nearly all known virulence factors, which are present in all strains regardless of disease source. Interestingly, the same genes are conserved among environmental isolates. We further developed a general strategy, including a specialized cloning vector, for the interrogation of chromosomal regions that show a high degree of polymorphism and often serve as sites of large deletions and insertions. The approach of combining microarray hybridization analysis and targeted capture of identified variable chromosomal segments should facilitate the analysis of many variants of a single species without the need for large-scale sequencing of entire genomes.
Materials and Methods
Genome Content Analysis. GeneChip P. aeruginosa Genome Arrays (Affymetrix) were used to assess genome content. Chromosomal DNA hybridizations were performed as described (10). Hybridization intensity data were extracted from the scanned array images, and intrachip normalizations were performed by using Affymetrix microarray suite 5.0 software. The average signal intensity of the probe sets was scaled to 500 for interchip comparisons. A presence/absence determination was made by comparing hybridization signal between strains. Values derived from the PAO1 genomic DNA hybridization were used as baseline. Probe sets with a ratio of >0.25 were considered present (blue). Ratios between 0.1 and 0.25 were considered indeterminate (gray), and ratios <0.1 were designated absent (yellow).
Design and Construction of a Multifunctional Yeast Capture Vector. A detailed description of the construction of the yeast capture vector p0975–0989capture is provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Recombinational Cloning. The variable genomic segments of P. aeruginosa strains PAO1, PAK, CF127, 6077, and JJ692, flanked by conserved genes PA0975 and PA0989, were cloned by cotransforming 200 μl of competent Saccharomyces cerevisiae strain CRY1–2 spheroplasts with 10 μg of unsheared chromosomal DNA and 1 μg of MluI-linearized plasmid p0975–0989capture. Competent spheroplasts were prepared as described with the exception of using 10 units of Zymolyase 20T (ICN) instead of lyticase and 10 mM DTT instead of 2-mercaptoethanol (11). Transformation mixtures were plated on uracil-deficient media containing cycloheximide (2.5 μg/ml).
Selection of Recombinant Clones. S. cerevisiae colonies were screened by PCR for the presence of captured P. aeruginosa sequences. A portion of each colony was suspended in 15 μl of zymolyase buffer (10 mM sodium phosphate buffer, pH 7.5/1 M sorbitol/2.5 mg/ml zymolyase 20T) and incubated with shaking for 1 h at 37°C. Two microliters of suspension was used as template for each PCR. Primers used to detect an insert were 5′-GGCTCGACCTCAATGGCATGGGCG and 5′-TCAGAAATATGGCGTCGGGTCGGA, which amplified a 500-bp portion of PA0976 that is not included in the original vector but is predicted to be present in the captured sequences. Plasmid DNA from yeast colonies that produced a PCR product of the correct size was purified and electrotransformed into Escherichia coli Genehogs (Invitrogen) for subsequent analysis.
Results and Discussion
Strain Variability and Genome Conservation. We have examined the genome composition of a collection of 18 P. aeruginosa strains by using a whole-genome DNA microarray consisting of probes for 5,549 nonredundant genes that constitute the genome of the sequenced strain PAO1 (3). The collection included four environmental isolates, a laboratory strain, and strains isolated from the most common human infections, including four respiratory isolates from children (<24 months) with cystic fibrosis, five strains associated with urinary tract infections, and two isolates each from ocular and blood infections (Table 1, which is published as supporting information on the PNAS web site). Fig. 1 indicates the predicted presence or absence of each PAO1 gene in the interrogated strains, organized relative to their chromosomal location in the reference PAO1 genome. The presence or absence of these genes was predicted based on the relative hybridization of chromosomal DNA to the microarray (see Materials and Methods). Detailed results for each gene are provided in Table 2, which is published as supporting information on the PNAS web site.

Analysis of genome content indicates a high level of gene conservation. The diagram indicates the presence and absence of genes found in the sequenced genome of strain PAO1 in clinical and environmental isolates of P. aeruginosa as detected by microarray hybridization. Strains are indicated at the top. The source of each strain is as follows: PAO1, reference strain; PAK, laboratory strain; environmental isolates (62, E2, MSH3, MSH10); respiratory isolates from children (<24 months) with cystic fibrosis (CF127, CF18, CF27, CF5); isolates from urinary tract infections (X24509, UDL, S54485, JJ692, U2504); strains isolated from ocular infections (19660, 6077), and blood isolates (S35004, X13273). Horizontal lines represent genes. Blue indicates that a gene is present, yellow represents absence, and gray indicates that presence was indeterminate. Red lines represent the location of tRNA genes in strain PAO1. The scale represents 5,613 genes (including 5,549 ORFs and 64 tRNA-encoding genes) organized according to the PAO1 chromosome. Green circles indicate variable segments in the P. aeruginosa genome as discussed in the text. Red triangles indicate known sites of insertion of horizontally acquired genetic material. References are indicated in parentheses.
The fraction of PAO1 genes detected in the various strains ranged from 96.1% to 97.7% (Fig. 4, which is published as supporting information on the PNAS web site), with 5,183 (93.4%) of the 5,549 PA01 genes present in all strains tested. These 5,183 genes represent the core set of genes, presumably encoding proteins that function in the diverse range of environments that this organism can inhabit. This finding represents a conservative estimate because our analysis does not compensate for sequence polymorphisms or strain-specific genes (see below) that are unrelated at the nucleotide sequence level but encode proteins with conserved function.
The distribution of core genes based on functional classification relative to the PAO1 genome (annotation tables available at www.Pseudomonas.com) indicates a broad conservation of functional diversity (Fig. 5, which is published as supporting information on the PNAS web site). The only functional class of genes that showed significant underrepresentation in all strains included genes associated with horizontal gene transfer (classified as related to phage, transposon, or plasmid). Only 8.6% of these genes were conserved in all strains examined. Of the 53 variable genes in this category 42 constitute two cryptic prophages identified in strain PAO1 but not found in most of the examined strains (Fig. 1, segments 3 and 4) (3).
Of particular interest are those genes associated with P. aeruginosa virulence. These genes are almost exclusively found in the category called secreted factors. Additional genes related to the expression of virulence determinants are found in the categories motility and attachment, protein secretion/export apparatus, and transcriptional regulators. Examination of the genes that play a direct role or are predicted to play a direct role in virulence (267 genes; Table 3, which is published as supporting information on the PNAS web site) indicates a high level of conservation (≈97%). The extensive conservation of virulence genes in the genomes of strains regardless of their clinical source suggests that the disease-causing ability of this opportunistic pathogen during human infections relies, in general, on a set of highly conserved pathogenic mechanisms. However, specific features of each disease may be influenced by phenotypic characteristics provided by individual or a small number of strain-specific virulence genes (for example, exoU and exoS, see below). The conservation of virulence determinants also extends to environmental isolates, suggesting that selection for the maintenance of such traits exists in the environmental reservoir. Based on this conservation it is likely that the environmental strains studied possess the ability to cause human infections despite the low probability of encountering a human host.
The 368 genes that are absent from the genome of at least one strain can be considered strain specific (Table 4, which is published as supporting information on the PNAS web site). This set of genes was subjected to a phylogenic analysis, which shows that strains from similar environmental or clinical sources are in general no more related than those from different sources (Fig. 2A). Not surprisingly, the most closely related strains are the only two isolates from the same locale, the two environmental strains isolated from Spirit Lake, Washington (MSH3 and MSH10). These strains were selected from a larger collection based on unique colony appearance and differential sensitivity to the pilus-specific phage PO4. The second most highly related strains are a pair of clinical isolates from a cystic fibrosis patient and a blood infection (strains CF127 and S35004, respectively) collected at hospitals in different cities (Table 1). Perhaps the most interesting relationship is demonstrated by a group of five strains, three of which were isolated from urinary tract infections (S54485, JJ692, and U2504), one from an ocular infection (6077), and the last from a blood infection (X13273). These strains share several epidemiological markers including the presence of genes for the O11-serotype capsular polysaccharide (data not shown). In addition, these strains are characterized by the presence of the exoU gene and its cognate chaperone spcU in their genomes (Fig. 2B). ExoU is a cytotoxin secreted by the type III secretion system of P. aeruginosa (13). Its expression is associated with increased virulence in model infection systems and poor clinical prognosis in patients (14, 15). A relationship between ExoU-expressing strains has been previously recognized among ocular isolates based on the coincidence of exoU with several phenotypic and genetic traits (16). It is apparent from our studies that this clonal relationship, based on gene content, includes isolates from a wider range of disease states. Another ocular isolate (19660) also carries the exoU and spcU genes; however, it is phylogenetically unrelated to the rest of the strains in this survey. It was previously reported that carriage of exoU is accompanied by the absence of exoS, which encodes another type III secreted protein (17, 18). We also found that the six strains, in this study, with exoU in their genome all lacked exoS (Fig. 2B). The relative conservation of the genomes of these strains, including the exoU carriage and deletion of exoS (see below), implies that conserved selective pressures contributed to the evolution of these genomes in different environmental niches. What selective advantage arises from mutual exclusion of one of two genes encoding virulence factors with distinct activities is unclear. Because the host cell contact-dependent type III secretion system secretes both ExoU and ExoS, the advantage afforded by expression of either one of these genes almost certainly involves interaction with a target eukaryotic organism.

Phylogenetic analysis of P. aeruginosa isolates based on genome content indicates a limited clonal relationship but no correlation with disease. (A) Dendrogram showing the relationship between strains based on the presence and absence of genes designated as strain specific. The source of each strain is indicated. Analysis was performed by using a previously described algorithm for the hierarchical clustering of microarray hybridization data (12). (B) Correlation between a group of clonally related strains and the presence of certain strain-specific virulence genes. The presence (blue) and absence (yellow) of genes encoding ExoU, SpcU, and ExoS are indicated. Strains are arranged according to their position in the accompanying dendrogram.
Distribution and Pattern of Strain-Specific Genes Relative to the Reference Genome. Distinct patterns of variability were evident when the genomes of multiple strains were compared (Fig. 1). Strain-specific genes were localized to 90 discrete regions relative to the PAO1 genome (Table 2). Many of these regions are composed of small gene blocks (one to four genes) that showed variability in one or more strains. These variable blocks likely contain genes that are highly polymorphic at the nucleotide sequence level or are gained or lost through local recombination events (see below).
A second pattern, which is more readily apparent, is characterized by large clusters of tandem genes that show varying levels of polymorphism between strains. Twenty-four of these regions (termed variable segments) were identified (Fig. 1, Table 2). These variable segments are scattered throughout the genome; however, a significant number are immediately adjacent to tRNA or tmRNA genes (Fig. 1, segments 2, 4–6, 16–19, and 24). The use of these genes as targeting sites for the integration of mobile genetic elements is well documented (19). In fact, four different P. aeruginosa tRNA genes have been previously shown to act as integration sites for bacteriophage, plasmids, and genomic islands (19–23) (Fig. 1). A number of additional genomic islands identified in P. aeruginosa are not directly associated with tRNA genes but do localize to regions of the chromosomal that show a high degree of variability (24–26) (Fig. 1). Based on the association of known genomic islands and other horizontally transferred genetic elements with regions of variability, we predict that the variable segments identified here may represent additional sites for integration of novel genetic material.
Deviation in the percentage (G+C) content of genes from that of the genome average is another marker of horizontal gene transfer. Annotation of the sequenced PAO1 genome revealed 10 such regions (3). With the exception of a cluster of low percentage (G+C) content genes encoding ribosomal proteins, all were associated with regions of P. aeruginosa genomes that contain strain-specific genes (Table 2). In fact, strain-specific genes as a group (Table 4) have an average percentage (G+C) of 61.8%, which is significantly lower than the average for core genes (67.1%). This finding suggests that many of these genes are associated with recently acquired genetic material.
Interrogation of Strain-Specific Genes. An “absent” call based on the microarray hybridization results suggests that the particular locus in the test strain, relative to PAO1, either lacks the corresponding gene altogether (deletion) or it contains a gene or genes that significantly differ from that in the reference strain. To investigate several such loci we designed oligonucleotides that corresponded to conserved flanking genes and used them to prime PCRs such that the resulting amplification products would span the polymorphic regions. Sequence and size analysis of the products allowed us to delineate whether an undetected gene had been deleted or replaced by a small insertion. Two such examples involving genes encoding proteins secreted by the type III secretion system are presented. The example in Fig. 3A shows that the exoY gene (PA2191), encoding a secreted adenylate cyclase (27), is flanked in PAO1 by two ORFs (PA2190 and PA2192) that encode conserved hypothetical proteins. These two flanking genes are absent from the chromosome of four isolates (PAK, CF27, S54485, and X13273). Six strains (CF127, CF5, JJ692, U2504, 6077, and S35004) lack only PA2190. Finally, one of the strains (19660) lacks all three genes (PA2190, exoY, and PA2192). Sequence analysis allowed us to identify the precise site of deletion or insertion in each case relative to the PAO1 genome (Fig. 3A).
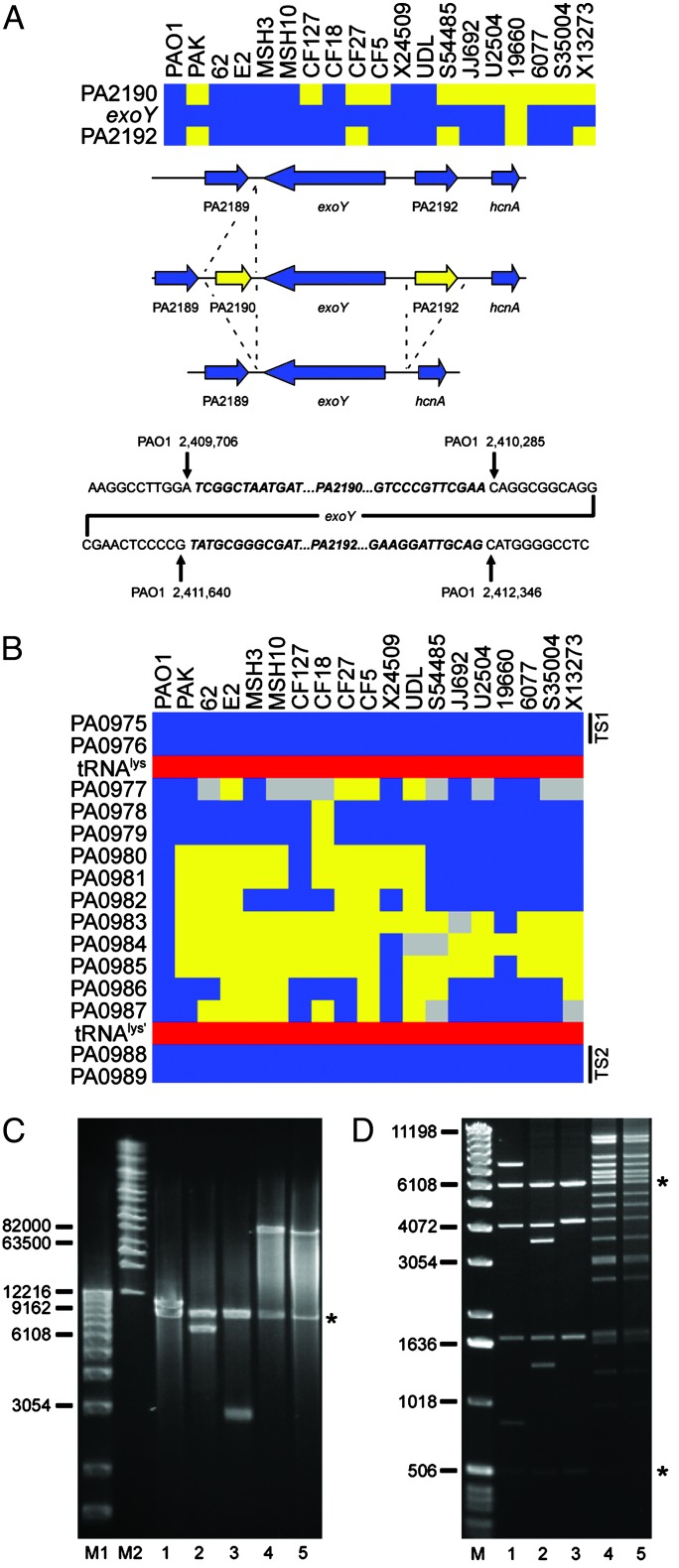
Analysis of polymorphic genes and variable segments associated with type III secreted effector-encoding genes. (A) Loss of genes in the flanking region of exoY. (Top) Diagram showing the presence (blue) and absence (yellow) of exoY and flanking genes in the indicated strains. (Middle) Summary of the specific deletion/insertion events detected by PCR analysis. Not shown is the deletion of all three genes in strain 19660. (Bottom) Sequence analysis of the exoY flanking regions indicates the specific deletion/insertion junctions for strains PAK, CF127, CF27, CF5, S54485, JJ692, U2504, 6077, S35004, and X13273. Sequences in bold/italics are absent in the corresponding deletion strains indicated as yellow (Top). Coordinates of the junction sites relative to the PAO1 chromosome are given. (B) Diagram showing a variable segment of P. aeruginosa genomes associated with the acquisition of exoU. Blue boxes indicate the presence of PAO1 genes in the interrogated strains. Yellow indicates undetected genes, and gray represents genes for which no definitive determination could be made. Red boxes show the location of an intact tRNAlys gene and a partial duplication of the same gene in the PAO1 genome. Black bars indicate the location of conserved targeting sequences (TS1 and TS2) used in the construction of the capture vector. (C) Size determination of captured sequences between conserved genes PA0975 and PA0989 in five P. aeruginosa strains. DNA fragments were analyzed on a 1% agarose gel by pulsed-field gel electrophoresis using the Bio-Rad Chef Mapper System. Lane M1, 1-kb DNA ladder (Invitrogen); lane M2, Midrange I PFG Marker (New England Biolabs). Subsequent lanes contain plasmids with captured sequences from the indicated strains. Inserts were separated from the vector (indicated by *) by digestion with I-SceI. Lane 1, PAO1; lane 2, PAK; lane 3, CF127; lane 4, 6077; lane 5, JJ692. (D) Restriction fragment fingerprinting of captured sequences. Lane M, 1-kb DNA ladder. The following lanes contain NcoI digested plasmids with captured sequences from the indicated strains: 1, PAO1; 2, PAK; 3, CF127; 4, 6077; and 5, JJ692. Asterisks denote the location of vector fragments.
A similar analysis of the exoS region confirmed that a number of strains lack this particular gene (Fig. 2B and Fig. 6, which is published as supporting information on the PNAS web site). In the PAO1 genome exoS is flanked by a near perfect (9 of 10 bp) direct repeat (Fig. 6). Sequence analysis indicates that a recombination event between these repeats could account for the loss of exoS in strains S54485, JJ692, U2504, 19660, 6077, and X13273. In all cases the adjacent gene (orf1) is unaffected (Fig. 6). Interestingly, orf1 encodes a specific chaperone essential for ExoS secretion from P. aeruginosa by the type III secretion system (28). It is therefore likely that the absence of exoS is the result of a specific deletion event. Given that nearly all strains that lack exoS contain exoU (Fig. 2B) it is conceivable that acquisition of exoU leads to the deletion of exoS by an unknown mechanism.
Interrogation of Variable Segments in the Genomes of P. aeruginosa by Recombinational Cloning. The inability to span a region identified as absent by PCR may indicate the presence of a large insertion. Clusters of unique genes at a particular location in the genome called “genomic islands” are not uncommon. As described above, microarray analysis can be used to identify preferential sites of integration of such islands. We constructed a capture vector for yeast recombinational cloning of genomic islands. The vector is a modified version of the plasmid used for cloning P. aeruginosa LPS islands (26). It includes several features that will greatly facilitate the analysis of bacterial genomes and simultaneously provide a tool for further downstream functional studies of the genes within the genomic islands. The salient features of the vector are outlined in Fig. 7, which is published as supporting information on the PNAS web site. PCR amplification of regions flanking a variable chromosomal segment results in two targeting sequences that are cloned into the capture vector to provide specificity. After cotransformation of competent S. cerevisiae with P. aeruginosa chromosomal DNA and the capture vector, recombination mediated by endogenous yeast proteins facilitates cloning of the P. aeruginosa chromosomal DNA flanked by the homologous targeting sequences. The captured chromosomal DNA is bordered by a short sequence specifying the recognition site for the I-SceI restriction endonuclease. Because this sequence is absent from the P. aeruginosa genome (and from the sequenced genomes of most bacteria) it can be used in preliminary estimation of the size of the insert.
A single capture vector can be constructed and used to interrogate the same region in a large number of strains. The captured sequences can be mobilized into any recipient P. aeruginosa strain by conjugation. Transfer of the captured island into a recipient strain lacking the equivalent sequences will allow for functional assessment. After targeted disruption of a particular gene or transposon mutagenesis of the entire segment, the inactivated gene(s) can be readily introduced onto the P. aeruginosa chromosome by allelic exchange using the method of SCE jumping (29).
We used this vector to investigate whether acquisition of exoU and its linked chaperone spcU involves integration of a genomic island into a common locus in the P. aeruginosa chromosome. The published sequence of exoU from strain PA103 (GenBank accession no. AF027291) (30) contained a segment 3′ of the spcU-coding sequence that was nearly identical to the position 1,069,669–1,069,934 bp of the P. aeruginosa strain PAO1 chromosome. This segment is located in a highly polymorphic region of the chromosome (Fig. 1, segment 6). Closer examination reveals that this variable segment (encompassing genes PA0977–PA0987) is flanked by highly conserved genes (Fig. 3B). To interrogate this region of the chromosome in different P. aeruginosa strains we designed a capture vector that included targeting sequences derived from the conserved flanking genes (Fig. 3B). As a control we cotransformed competent yeast with the capture vector and chromosomal DNA from strain PAO1. The resulting clone was analyzed by digestion with I-SceI, which released a 12-kb fragment from the capture vector, corresponding to the predicted size of the PAO1 sequence (Fig. 3C, lane 1). Restriction fragment fingerprinting of the cloned insert with the NcoI restriction endonuclease generated fragments that agreed with the predicted location of NcoI recognition sites in this region of the PAO1 chromosome (Fig. 3D, lane 1). Chromosomal DNA from four additional strains was captured and the recovered plasmids were analyzed as above (Fig. 3 C and D). Two exoU-carrying strains (6077 and JJ692) possessed a large insert of >80 kb (Fig. 3C, lanes 4 and 5). The NcoI digestion patterns confirmed that these inserts are very similar or perhaps even identical (Fig. 3D, lanes 4 and 5). PCR analysis confirmed that they indeed carried the exoU and spcU genes (data not shown). Analysis of sequences captured from strains lacking exoU (PAK and CF127) indicated that these strains each carried a different-sized chromosomal segment (≈7 and 3 kb, respectively; Fig. 3C, lanes 2 and 3). The NcoI restriction pattern for these segments suggests that they may have arisen through the deletion of sequences from the larger PAO1 sequence. Because a tRNA gene flanks this region, integration of the acquired DNA in the exoU-carrying strains may involve a site-specific recombination system (19). Interestingly, this locus has been previously described as one of two sites for semistable incorporation of a large plasmid present in many genomes of European isolates of P. aeruginosa (22). Collectively, these data indicate that this region of the P. aeruginosa chromosome may be a “hotspot” for integration of horizontally acquired DNA.
Conclusion
The work presented here shows that the genomes of P. aeruginosa strains, representing distinct clinical or environmental sources, are highly conserved. These results support a basic model whereby conservation of a large genome allows this organism to inhabit the widest possible range of environments and confers the ability to cause human infections after relatively infrequent encounters. Although we cannot predict the total gene content of individual strains or whether detected genes are functional, the remarkable conservation of genes encoding proteins associated with virulence indicates that most strains, regardless of source, possess the basic pathogenic mechanisms necessary to cause a wide variety of human infections. The fact that this conservation extends to environmental strains indicates the existence of a natural environmental eukaryotic host and further suggests that environmental strains of P. aeruginosa can serve as the source for human infections.
Analysis of the microarray hybridization patterns revealed a number of variable segments in the P. aeruginosa genome that may serve as sites of integration of horizontally transferred genetic material. Given the limitations of the microarray hybridization technique we cannot determine which of these sites are occupied or the nature of the acquired sequences. To investigate these sites we developed a multifunctional capture vector and used it to identify an 80-kb genomic island carrying a known virulence gene (exoU). This approach will be useful for identifying additional virulence genes that may enhance the ability of P. aeruginosa to survive in specific host niches; however, we expect this number to be small, as extensive work over the past 25 years using a variety of clinical isolates has led to the discovery of very few virulence genes that are not part of the core set present in nearly every P. aeruginosa strain analyzed.
Our study demonstrates the power of whole genome-based microarrays to analyze genome evolution, based on the gene content relative to a reference strain. We further developed a useful tool for the isolation and interrogation of strain-specific islands in bacterial genomes. When these technologies are combined, they should provide essential tools for large-scale analysis of bacterial populations and lead to a better understanding of the selective pressures that control genome maintenance and the spread of novel genetic material.
Acknowledgments
This work was supported by a grant from the Cystic Fibrosis Foundation and Grant DK53369 from the National Institutes of Diabetes and Digestive and Kidney Diseases. GeneChip P. aeruginosa Genome Arrays were subsidized by Cystic Fibrosis Foundation Therapeutics. M.C.W. was supported by a Cystic Fibrosis Foundation postdoctoral research fellowship.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0832438100
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc166255?pdf=render
Citations & impact
Impact metrics
Article citations
Modeling Cystic Fibrosis Chronic Infection Using Engineered Mucus-like Hydrogels.
ACS Biomater Sci Eng, 10(10):6558-6568, 19 Sep 2024
Cited by: 0 articles | PMID: 39297972
'Wild Type'.
Microbiology (Reading), 170(8), 01 Aug 2024
Cited by: 0 articles | PMID: 39212644 | PMCID: PMC11364142
Review Free full text in Europe PMC
Genomic Differences Associated with Resistance and Virulence in Pseudomonas aeruginosa Isolates from Clinical and Environmental Sites.
Microorganisms, 12(6):1116, 30 May 2024
Cited by: 0 articles | PMID: 38930498
Pseudomonas aeruginosa Isolates from Water Samples of the Gulf of Mexico Show Similar Virulence Properties but Different Antibiotic Susceptibility Profiles than Clinical Isolates.
Int J Microbiol, 2024:6959403, 16 May 2024
Cited by: 0 articles | PMID: 38784405 | PMCID: PMC11115996
Cytotoxic rhamnolipid micelles drive acute virulence in Pseudomonas aeruginosa.
Infect Immun, 92(3):e0040723, 23 Feb 2024
Cited by: 2 articles | PMID: 38391248 | PMCID: PMC10929412
Go to all (247) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (2 citations) ENA - X13273
- (1 citation) ENA - AF027291
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment.
J Clin Microbiol, 42(12):5783-5792, 01 Dec 2004
Cited by: 105 articles | PMID: 15583313 | PMCID: PMC535267
Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread.
Infect Immun, 68(7):3998-4004, 01 Jul 2000
Cited by: 120 articles | PMID: 10858214 | PMCID: PMC101680
Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa.
J Bacteriol, 183(3):843-853, 01 Feb 2001
Cited by: 77 articles | PMID: 11208781 | PMCID: PMC94950
Comparative genomic analysis of Pseudomonas aeruginosa virulence.
Trends Microbiol, 12(10):437-439, 01 Oct 2004
Cited by: 9 articles | PMID: 15381190
Review
Funding
Funders who supported this work.
NIDDK NIH HHS (2)
Grant ID: DK53369
Grant ID: P01 DK053369





