Abstract
Free full text

Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel–Lindau protein
Abstract
Inactivation of the von Hippel–Lindau (VHL) tumor suppressor gene is linked to the hereditary VHL disease and sporadic clear cell renal cell carcinomas (CCRCC). VHL-associated tumors are highly vascularized, a characteristic associated with overproduction of vascular endothelial growth factor (VEGF). The VHL protein (pVHL) is a component of the ubiquitin ligase E3 complex, targeting substrate proteins for ubiquitylation and subsequent proteasomic degradation. Here, we report that the pVHL can directly bind to the human RNA polymerase II seventh subunit (hsRPB7) through its β-domain, and naturally occurring β-domain mutations can decrease the binding of pVHL to hsRPB7. Introducing wild-type pVHL into human kidney tumor cell lines carrying endogenous mutant non-functional pVHL facilitates ubiquityl ation and proteasomal degradation of hsRPB7, and decreases its nuclear accumulation. pVHL can also suppress hsRPB7-induced VEGF promoter transactivation, mRNA expression and VEGF protein secretion. Together, our results suggest that hsRPB7 is a downstream target of the VHL ubiquitylating complex and pVHL may regulate angiogenesis by targeting hsRPB7 for degradation via the ubiquitylation pathway and preventing VEGF expression.
Introduction
von Hippel–Lindau (VHL) disease is a rare hereditary multiorgan neoplastic disorder, transmitted by the mutant VHL gene (Maher and Kaelin, 1997; Stolle et al., 1998). VHL patients develop tumors in several discrete organ systems, including hemangioblastomas in the central nervous system and retina, clear cell carcinomas of the kidney, pheochromocytomas of the adrenal gland, cysts/adenomas and islet cell tumors of the pancreas, cystadenomas of the epididymis and endolymphatic sac tumors of the inner ear (Maher and Kaelin, 1997; Stolle et al., 1998). More importantly, the VHL gene is inactivated in ~50–80% of the common sporadic form of clear cell renal cell carcinoma (CCRCC) (Foster et al., 1994; Gnarra et al., 1994; Shuin et al., 1994). The VHL protein (pVHL) has been demonstrated to associate with a multiprotein complex with components including elongin C (EC), elongin B (EB), cullin-2 (CUL-2) and Rbx1 to form a complex called VCB–CUL-2 (Pause et al., 1997; Kamura et al., 1999; Skowyra et al., 1999). In light of the structural analogy of VCB–CUL-2 to SCF (Skp1–Cdc53 or Cullin–F-box) ubiquitin ligase, several groups have demonstrated that the VCB–CUL-2 complex functions as a ubiquitin ligase (Iwai et al., 1999; Lisztwan et al., 1999). The ubiquitin–proteasome degradation system has been shown to control the abundance and activity of several oncogene and tumor suppressor gene products, transcription factors and other signaling molecules (Baumeister et al., 1998; Hershko and Ciechanover, 1998). The crucial substrate-recognition step in ubiquitin-dependent proteolysis is mediated by the diverse family of E3 ubiquitin ligases. pVHL contains two domains, an α-domain and a β-domain (Stebbins et al., 1999). While the α-domain serves as the EC-binding site, the β-domain plays a role in substrate recognition (Stebbins et al., 1999). Hypoxia-inducible factors, HIF-1α and HIF-2α, have been demonstrated to be targeted for ubiquitylation and degradation under normoxic conditions by physical interaction of its oxygen-dependent degradation domain with the VCB–CUL-2 ligase (Cockman et al., 2000; Kamura et al., 2000; Ohh et al., 2000; Tanimoto et al., 2000; Ivan et al., 2001; Jaakkola et al., 2001). Without functional pVHL, HIF-1α and HIF-2α accumulate in cells, causing the overexpression of HIF target genes such as vascular endothelial growth factor (VEGF), which helps to explain the highly vascular character of VHL tumors and CCRCC. Besides HIF-1α and HIF-2α, other important downstream targets of VCB-CUL2 E3 ubiquitin ligase may exist. There is considerable interest in identifying new downstream targets or interacting proteins of pVHL, which may provide new insights into the biological functions of pVHL.
In order to explore further the molecular mechanisms of pVHL-related tumorigenesis, especially in sporadic CCRCC, we used yeast two-hybrid screening to identify previously unknown pVHL-interacting proteins from a kidney cDNA library. Two independent clones identified from this screening encoded the full-length human RNA polymerase II seventh subunit (hsRPB7). RNA and protein expression patterns of hsRPB7 have been studied in different tissues and cell lines (Khazak et al., 1995). A particularly high level of expression for hsRPB7 mRNA and protein levels was observed in normal kidney tissue and the kidney-derived cell line 293 (Khazak et al., 1995). These findings, along with recognition that the specific expression pattern of hsRPB7 mRNA in human tissues differs from that of the largest subunit of RNA polymerase II (hsRPB1) (Khazak et al., 1995), suggest a potential regulatory role for hsRPB7 on gene expression, especially in kidney cells. Here we demonstrate that hsRPB7 is a pVHL-interacting protein and that hsRPB7 can be ubiquitylated and rapidly degraded by a pVHL-mediated ubiquitin–proteasomal pathway in kidney cells. Furthermore, we provide evidence that transcription of VEGF may be regulated by hsRPB7. Our data demonstrate that hsRPB7 is a degradation target of pVHL–E3 ubiquitin ligase, which may represent another important mechanism by which pVHL acts as a renal tumor suppressor.
Results
hsRPB7 is a novel pVHL-associated protein
The human pVHL19 (amino acids 54–213), a biologically functional isoform of pVHL, was used as bait to screen pVHL-interacting partners in a human kidney cDNA library. From a screen of 5.5 × 106 clones, two of the positive clones were identified as encoding the full-length cDNA of hsRPB7. These two clones differed in their 3′ untranslated regions, indicating that they were independent. Subsequent yeast two-hybrid assays showed that yeast cells transformed with either the bait pVHL19 (pGBKT7-VHL19) and pGADT7 vector or the full-length hsRPB7 (pGADT7-hsRPB7) plus pGBKT7 vector did not generate any clones on growth-selection plates (data not shown), while yeast two-hybrid assay using independent retransformation of pGBKT7-VHL19 and pGADT7-hsRPB7, followed by growth selection and a β-galactosidase assay, further confirmed the interaction between pVHL and hsRPB7 in the yeast system (Figure 1A). To demonstrate that these two proteins could physically interact with each other, a GST pulldown assay was performed and the results showed that hsRPB7 could bind directly to GST–VHL19 but not GST alone (Figure 1B). Finally, the results from mammalian two-hybrid assay further confirmed that hsRPB7 interacted with pVHL19 in mammalian cells (Figure 1C). Together, these experiments demonstrated that hsRPB7 could interact with pVHL.
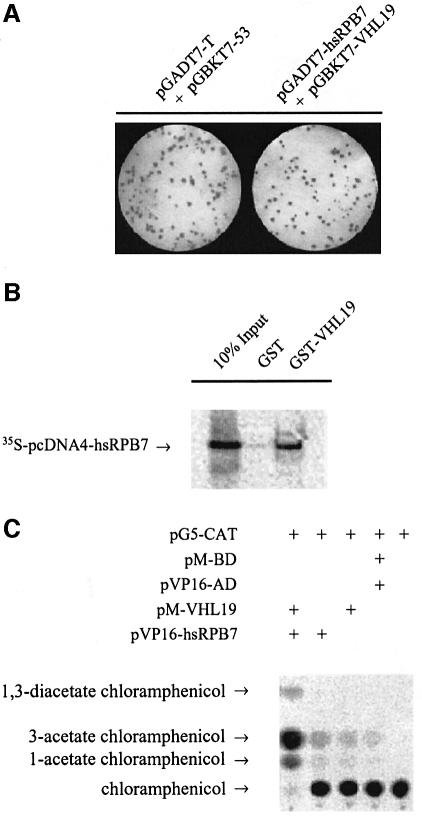
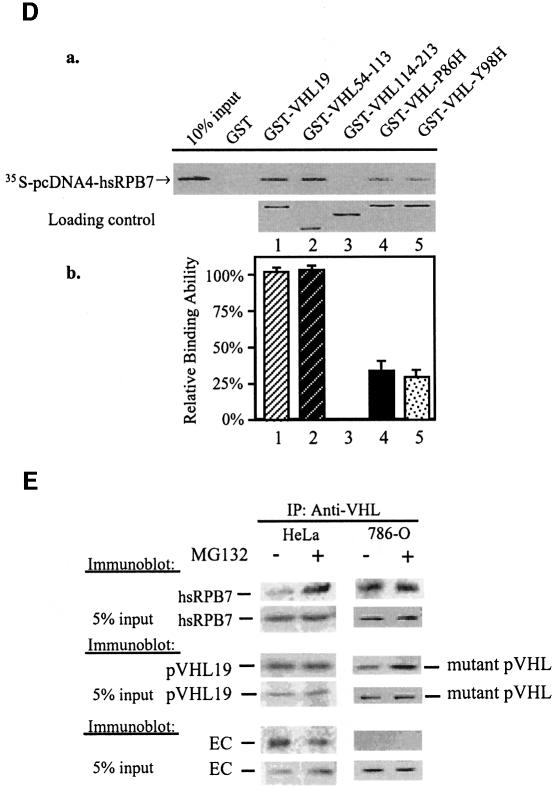
Fig. 1. pVHL interaction with hsRPB7. (A) Yeast two-hybrid assay followed by a colony lift filter β-Gal assay demonstrates the interaction between pVHL and hsRPB7. Yeast strain AH109 was cotransformed with pGBKT7-VHL19 and pGADT7-hsRPB7 or with pGBKT7-p53 and pGADT7-T (served as a positive control) plated on nutrition dropout selection plates and cultured at 30°C for 3 days, followed by colony-lift filter β-Gal assay. Positive colonies turned blue as shown. (B) hsRPB7 interacts with pVHL in vitro. GST protein and GST–VHL19 fusion protein were expressed in bacteria, purified and incubated with [35S]methionine-labeled hsRPB7. Elutes were separated on a SDS–PAGE gel and detected by phosphoimager. (C) hsRPB7 interacts with pVHL in mammalian cells. pM-VHL19 construct was cotransfected with pVP16-hsRPB7 into COS-7 cells, along with a GAL4-dependent CAT reporter pG5-CAT and a pCMV-β-Gal expression vector. The β-Gal vector served as an internal control for normalization of transfection efficiency. Control experiments with pM-BD or pVP16-AD alone were also conducted in parallel. (D) Mapping the hsRPB7-interacting region in pVHL. In (a), 10% of in vitro translated 35S-labeled hsRPB7 was used as a positive control, and in vitro translated 35S-labeled hsRPB7 protein was incubated with beads coated with GST, GST–VHL19, GST–VHL54–113, GST–VHL114–213, GST–VHL-P86H and GST–VHL-Y98H, respectively. The bound proteins were eluted and analyzed on SDS–PAGE gel. Equal amounts of viable GST–VHL fusion proteins were demonstrated. Relative binding abilities of hsRPB7 and pVHL fragments were analyzed in (b). Results are expressed as the mean ± SD of three independent experiments. (E) Extracts of HeLa cells and 786-O cells treated with or without MG132 were used in immunoprecipitation (IP) assay by pVHL antibody. Precipitates were resolved on SDS–PAGE gels and immunoblotted (IB) with hsRPB7, VHL and EC antibodies, respectively.
To determine the hsRPB7-interacting region in pVHL, GST pulldown assays were performed with pVHL fragments covering different regions. GST–VHL19 and GST–VHL54–113, but not GST–VHL114–213, could retain the in vitro translated hsRPB7 protein (Figure 1D). These results suggest that hsRPB7 interacts with pVHL within residues 54–113. This region is a part of the β-domain of pVHL, which is responsible for substrate recognition for ubiquitylation and degradation. Many missense mutations found in VHL disease or renal cell carcinomas occur in this region. We further tested the interactions between hsRPB7 and several common VHL mutants that harbor naturally occurring point mutations in the β-domain. Also demonstrated in Figure 1D, two representative pVHL mutants harboring mutations in the β-domain (P86H and Y98H, respectively) showed a reduced binding capacity between pVHL and hsRPB7 (Figure 1D, a and b).
In vivo co-immunoprecipitation was performed using the HeLa cell line, which contains endogenous wild-type pVHL, and the 786-O cell line, which contains mutant pVHL (truncated pVHL with the intact β-domain but missing the α-domain). pVHL antibody can recognize and precipitate this mutant form of pVHL. To demonstrate that pVHL and hsRPB7 could interact with each other in their natural forms, cultured HeLa cells were treated with or without MG132 (a proteasomal inhibitor) and lysates of these cells were immunoprecipitated with pVHL antibody, and then fractionated on SDS–PAGE gels and immunoblotted with anti-hsRPB7, pVHL and EC antibodies, respectively. As shown in Figure 1E, hsRPB7 can be found in the immunoprecipitates against endogenous pVHL in HeLa cells. In HeLa cells without proteasomal inhibitor MG132 treatment, the amount of hsRPB7 associated with pVHL was significantly less compared with MG132-treated HeLa cells, indicating that rapid turnover of hsRPB7 occurred after a brief interaction with pVHL under normal conditions. As expected, EC was associated with pVHL either with or without MG132 treatment. In contrast, in 786-O cells, endogenous mutant pVHL can be found associated with hsRPB7; MG132 treatment did not make any difference in the interaction. As expected, mutant pVHL without the α-domain failed to precipitate EC. Co-immunoprecipitation was also performed with an anti-RNA polymerase II (anti-Pol II) C-terminal domain (CTD) antibody. hsRPB7 was detected in the precipitates, but not pVHL (data not shown). This indicates that hsRPB7 does not recruit pVHL into the Pol II complex. Taken together, the above in vitro and in vivo data demonstrate that hsRPB7 is a pVHL β-domain-associated protein, and, more importantly, the above results indicate that hsRPB7 may degrade through the proteasome degradation pathway. The following experiments further explore the functional significance of this interaction.
Ubiquitylation of hsRPB7 is pVHL dependent
To test whether hsRPB7 can be ubiquitylated by pVHL–E3 ubiquitin ligase, we performed a series of in vitro and in vivo ubiquitylation assays. HeLa cell cytoplasmic extracts (S-100) were incubated with [35S]methionine-labeled hsRPB7. Incubation of the extracts along with an ATP regeneration system converted the hsRPB7 substrate to multiple slower migrating forms (Figure 2A), indicating that the hsRPB7 could be polyubiquitylated. To determine the role of pVHL19 in hsRPB7 ubiquitylation, we performed similar assays using extracts from 786-O cells. This kidney cancer cell line contains non-functional pVHL. Our results showed that the 786-O cell extracts with transfected wild-type pVHL19 (pcDNA4-VHL19) resulted in the ubiquitylation of hsRPB7, whereas with pcDNA4 vector only, the 786-O cell extracts were unable to lead to polyubiquitylation of hsRPB7. The authenticity of ubiquitylation was confirmed by incorporating UbK48R and methylated ubiquitin into the assays (Figure 2A and B). This pVHL-dependent ubiquitylation was also specific to hsRPB7, because the ubiquitylation of p53, which was tested under the same conditions, was not affected by the addition of pVHL19 (data not shown). Furthermore, this pVHL-dependent ubiquitylation process was diminished when using naturally occurring VHL missense mutations in its β-domain that disrupt the interaction between hsRPB7 and pVHL (Figure 2C). In vitro ubiquitylation of hsRPB7 using recombinant E1 and E2 (UbcH5a) was also tested (Figure 2D). VBC complex was immunoprecipitated from HeLa cells (Iwai et al., 1999). hsRPB7 could only be ubiquitylated in the presence of E1, E2 and VBC complex, and the intensity of ubiquitylation is directly dependent on the amount of VBC complex (Figure 2E). Finally, in vivo ubiquitylation was performed using kidney 786-O cells. Transfection of wild-type pVHL19 led hsRPB7 to become a ubiquitylated form, which further confirmed the previous in vitro results (Figure 2F) and demonstrated that ubiquitylation of hsRPB7 is pVHL dependent.
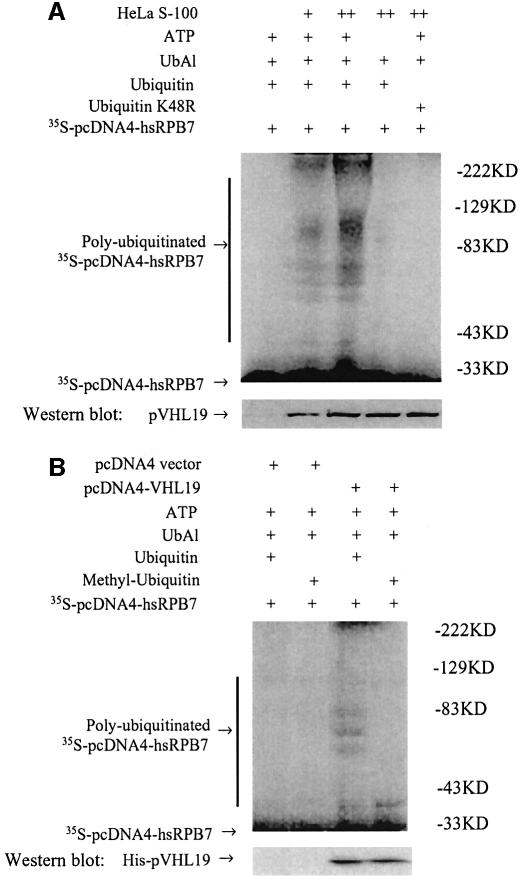

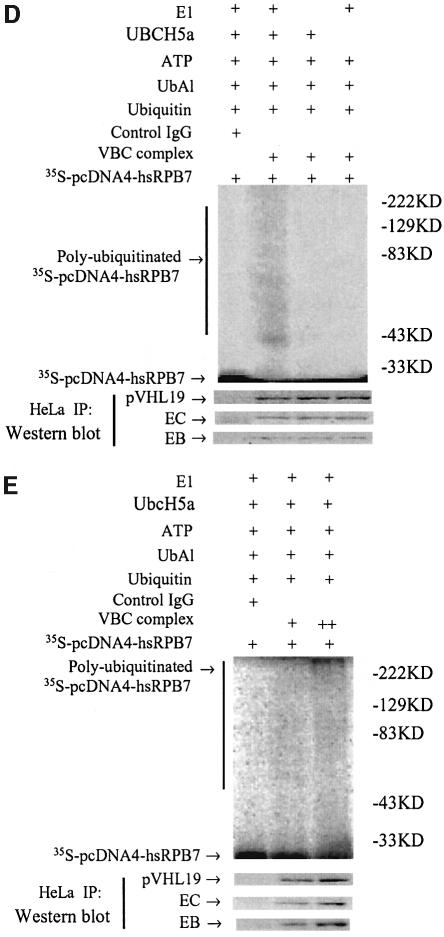
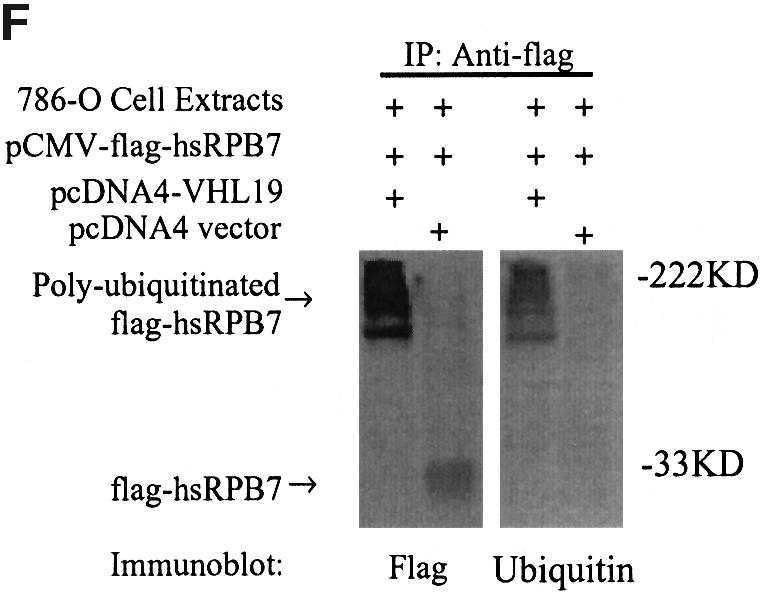
Fig. 2. pVHL-dependent ubiquitylation of hsRPB7. (A) In vitro ubiquitylation of hsRPB7 by HeLa cell cytoplasmic extracts (S-100). 35S-labeled hsRPB7 was used as a substrate in reactions of different compositions for 2 h. The positions of the non-ubiquitylated and ubiquitylated hsRPB7 are indicated. The amounts of endogenous pVHL19 are depicted by western blot. (B) pVHL-dependent hsRPB7 ubiquitylation. 35S-labeled hsRPB7 was incubated at 37°C for 2 h in reactions consisting of 786-O cell extracts transfected with pcDNA4 vector or pcDNA4-VHL19, ATP-regenerating system, ubiquitin or methylated ubiquitin and ubiquitin-aldehyde (UbAl). Ubiquitylation of hsRPB7 was only found in the presence of pVHL19. The expression of His–pVHL19 was detected by anti-His immunoblot. (C) The effect of several pVHL mutations on the hsRPB7 ubiquitylation. 786-O cells were tranfected with wild-type pVHL or pVHL mutant constructs. No ubiquitylation was detected in the presence of pVHL mutants (pcDNA4-VHLP86H and pcDNA4-VHLY98H). The expressions of variable His–pVHL19 mutants were detected by anti-His immunoblot. (D) pVHL-dependent ubiquitylation of hsRPB7 in vitro by recombinant E1 and E2. 35S-labeled hsRPB7 was incubated at 37°C for 2 h in reactions consisting of E1, E2 or VBC complex or control IgG immunoprecipitates. Ubiquitylation of hsRPB7 was only found in the presence of E1, E2 and VBC complex. Endogenous VBC complex (VHL, EB, EC) was detected by western blot. (E) Ubiquitylation of hsRPB7 in vitro is in a VBC complex-dependent fashion. The amount of hsRPB7 ubiquitylation is VBC complex dose dependent. (F) pVHL-dependent ubiquitylation of hsRPB7 in vivo. 786-O cells were cotransfected with pCMV-flag-hsRPB7 and pcDNA4-VHL19 or pCMV-flag-hsRPB7 and pcDNA4 vector. Cell extracts were immunoprecipitated by flag antibody, resolved on SDS–PAGE gel and detected by anti-flag and anti-ubiquitin antibodies independently.
pVHL mediates the proteasomic degradation of hsRPB7
To test whether this ubiquitylation mediated by pVHL will result in a rapid turnover of hsRPB7, we examined the degradation of hsRPB7 in kidney tumor cell lines A498 and 786-O. A498 cells were transiently transfected with an empty vector (pcDNA4), wild-type pVHL19 (pcDNA4-pVHL19) and pVHL114–213 (without the hsRPB7 interaction region), respectively. MG132 treatment was also used in a separate panel in combination with wild-type pVHL19 (pcDNA4-VHL19). Equal amounts of total protein were loaded into each lane. As shown in Figure 3A, overexpression of pVHL19 decreased the amount of endogenous hsRPB7 in A498 cells and, importantly, this effect was reversible by a combined treatment with MG132. MG132 alone did not significantly increase the level of hsRPB7 protein (data not shown). pVHL114–213 without the hsRPB7-binding domain did not exhibit the ability to decrease hsRPB7 levels. As a control, a northern blot was performed and no change in hsRPB7 mRNA levels was observed following transfection with wild-type pVHL19, making it very unlikely that pVHL19 influences hsRPB7 at the mRNA level (data not shown). To evaluate further the effect of pVHL on the degradation of hsRPB7, pulse–chase assays were performed in 786-O cells. As shown in Figure 3B (a), after cycloheximide treatment, hsRPB7 was quite stable in the group transfected with pcDNA4 vector only. In contrast, the hsRPB7 level decreased dramatically when transfected with wild-type pVHL. The cycloheximide treatment led to a significant decrease in hsRPB7 protein level and MG132 treatment restored the hsRPB7 protein level (Figure 3B, b). Taken together, these data demonstrated that the hsRPB7 protein was undergoing proteasome-dependent degradation in a pVHL-dependent manner, indicating that hsRPB7 is a downstream target of pVHL–E3 ligase for ubiquitylation and rapid degradation by proteasome.
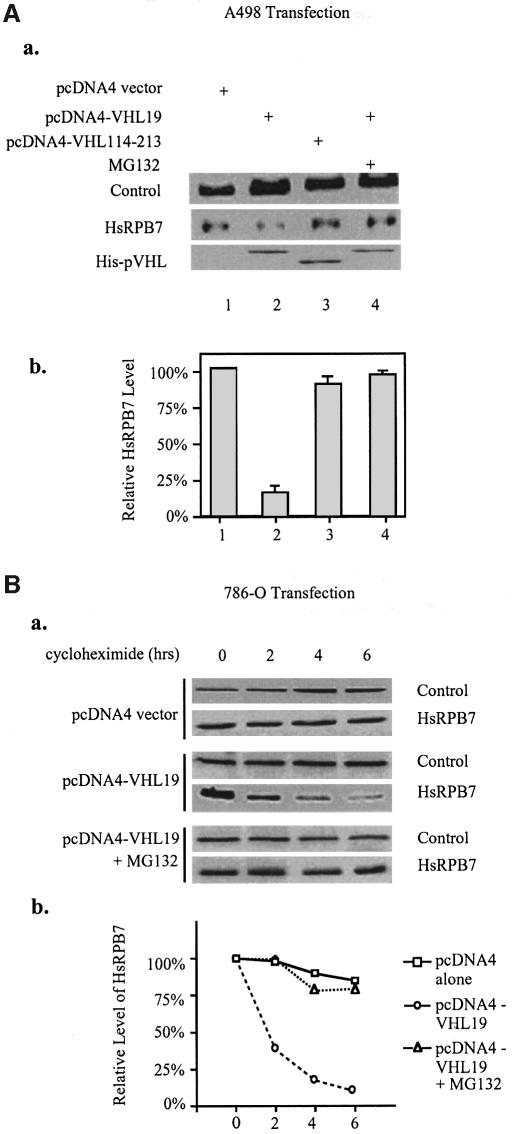
Fig. 3. pVHL mediates proteasomal degradation of hsRPB7. (A) pVHL-dependent proteasomal degradation of hsRPB7 in A498 cells. In (a), A498 cells were transfected with empty pcDNA4 vectors, pcDNA4-VHL19 or pcDNA4-VHL114–213 with or without MG132 treatment as indicated. Fifty micrograms of whole-cell lysates were loaded in each lane. The expressions of pVHL fragments were detected by anti-His western blot. Immunoblots (IB) were performed against hsRPB7 and p62 (the p62 subunit of TFIIH was used as a loading control). Relative hsRPB7 amounts were quantitated and are shown in (b). Results are expressed as the mean ± SD of three independent experiments. (B) pVHL-dependent proteasomal degradation of hsRPB7 in 786-O cells. In (a), 786-O cells were transfected with pcDNA4 vector or pcDNA4-VHL19. Cells were treated with 5 µM MG132 or DMSO. Six hours later, cells were incubated with cycloheximide (CHX) for 0, 2, 4 and 6 h, as indicated above. Fifty micrograms of whole-cell lysates were loaded in each lane. Immunoblotting (IB) was performed by using hsRPB7 antibody. P62 was detected to demonstrate equal loading of each sample and relative hsRPB7 amounts are quantitated in (b).
pVHL decreases nuclear accumulation of endogenous hsRPB7
To test whether pVHL has any influence on the subcellular localization of hsRPB7, dual immunofluorescence staining with pVHL antibody (for endogenous pVHL protein when transfected with empty vector only) or anti-His6 antibody (for His6 wild-type or mutant pVHL fusion proteins) and hsRPB7 antibody was performed using 786-O tumor cells, which express an endogenous mutant non-functional pVHL. FITC green fluorescence staining of endogenous hsRPB7 is shown in Figure 4, column 1 (A1–E1) and rhodamine red staining of endogenous pVHL (A2) or transfected His6-tagged wild-type or mutant pVHL (B2–F2) is shown in Figure 4, column 2. Merged images of column 1 and column 2 are shown in column 3. DAPI staining in column 4 demonstrates the nuclear positions of corresponding cells. Cells in Figure 4 (rows A–E) were transfected with pcDNA4 vector (row A), pcDNA4-VHL19 (row B), pcDNA4-VHL114-213 (row C), pcDNA4-VHL-P86H (row D) and pcDNA4-VHL-Y98H (row E), respectively. As shown in Figure 4 (A1–A4), with transfection of empty pcDNA4 vectors alone, endogenous hsRPB7 was localized in both the nucleus and cytoplasm with strong staining in the nucleus (A1 and A3). In contrast, after introducing wild-type pVHL19 (pcDNA4-VHL19), which was mainly localized in the cytoplasm (B2), into the cells, hsRPB7 nuclear accumulation was decreased dramatically (B1 and B3). This effect cannot be found by using mutant pVHL, including cells transfected with pcDNA4-VHL114–213 (C1 and C3), pcDNA4-VHL-P86H (D1 and D3) or pcDNA4-VHL-Y98H (E1 and E3), which either lost or had a reduced binding capacity to hsRPB7, as demonstrated in our earlier results (Figure 1D). To test whether the effect of pVHL on the distribution of hsRPB7 is specific, another RNA Pol II subunit, hsRPB4, was tested. As shown in Figure 4 (F1–F4), transfection of wild-type pVHL did not decrease the nuclear accumulation of this protein, confirming the specificity of this process. These phenomena suggest that wild-type pVHL19 facilitates degradation of hsRPB7 and intact interaction region of the β-domain is required for this process.
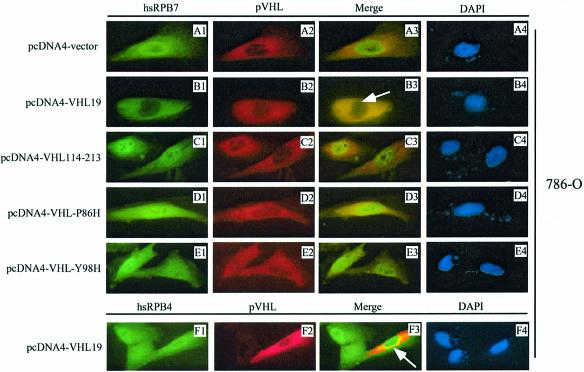
Fig. 4. Decreased nuclear accumulation of endogenous hsRPB7 by pVHL. 786-O cells were transfected with pcDNA4 vector, pcDNA4-VHL19, pcDNA4-VHL114–213, pcDNA4-VHL-P86H and pcDNA4-VHL-Y98H, respectively, as indicated. Endogenous hsRPB7, hsRPB4 and pVHL or His6-tagged wild-type or mutant pVHL fusion proteins were detected by double-immunostaining with hsRPB7 or hsRPB4 rabbit polyclonal antibody and pVHL mouse monoclonal antibody or anti-His6 mouse monoclonal antibody (for His6–pVHL fusion proteins), followed by FITC green conjugated secondary antibody for visualizing hsRPB7 (column 1, A1–E1) and hsRPB4 (column 1, F1), and rhodamine red-conjugated secondary antibody for endogenous pVHL (column 2, A2) or His6-tagged pVHL (column 2, B2–F2). Merged images of columns 1 and 2 are shown in column 3. DAPI staining in column 4 demonstrates the nucleus positions of corresponding cells. Note that the nuclear intensity of hsRPB7, but not hsRPB4, dramatically decreases only in the presence of wild-type pVHL19 (indicated by arrow).
hsRPB7 enhances VEGF transactivation, VEGF mRNA expression and VEGF protein secretion in kidney cancer cells
To explore the biological consequence of pVHL-mediated hsRPB7 degradation, we examined whether hsRPB7 could influence VEGF expression by testing the effects of hsRPB7 on VEGF promoter transactivation, endogenous VEGF mRNA expression and protein secretion. A498 and 786-O cell lines were used in our study. Transfection of hsRPB7 plasmid at a 3:1 ratio relative to the VEGF promoter plasmid caused a 1.5- to 2-fold increase in the VEGF promoter activity in A498 and 786-O cells (Figure 5A). Introducing a pVHL expression plasmid at a 3:1 ratio to the VEGF promoter plasmid decreased VEGF transactivation by 30–50%. Cotransfection of pVHL with hsRPB7 expression plasmids at a 3:3:1 ratio to the VEGF promoter plasmid abolished the hsRPB7-mediated transactivation of the VEGF promoter in A498 and 786-O cells (Figure 5A). Our results demonstrated that hsRPB7 could enhance VEGF transactivation and this effect could be reversed by wild-type pVHL. The specificity of the transactivation effect of hsRPB7 on the VEGF promoter was further tested by using the androgen response element promoter and the fibrinogen promoter. No transactivation effects were observed (data not shown).
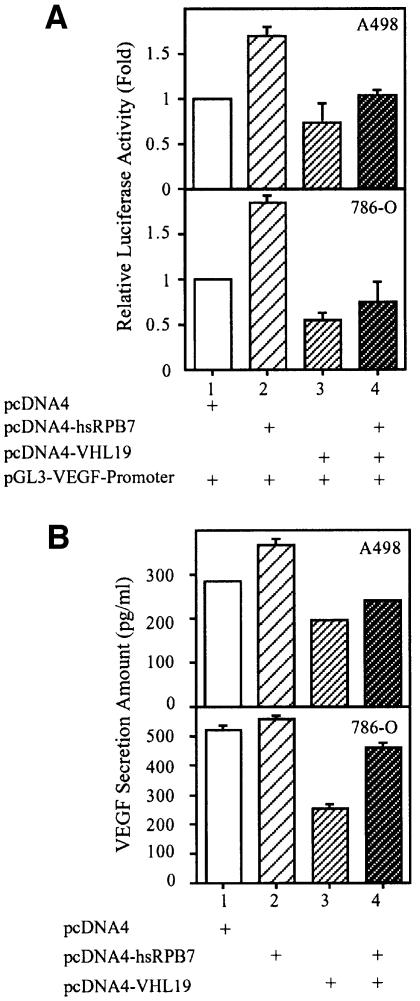


Fig. 5. The effects of hsRPB7 on transactivation of the VEGF promoter, VEGF mRNA expression and VEGF secretion. (A) hsRPB7 enhances VEGF transactivation. A498 and 786-O cell lines were cotransfected with combinations of pGL3-VEGF promoter construct, pRL-SV40, pcDNA4-hsRPB7 and pcDNA4-VHL19 as indicated. The firefly luciferase activity from the VEGF promoter was normalized by Renilla luciferase activity. The relative luciferase activity of cells transfected with vector only was set as basal level. Results are expressed as the mean ± SD of three independent experiments. (B) hsRPB7 promotes VEGF secretion. A498 and 786-O cell lines were transfected with pcDNA4-hsRPB7 and/or pcDNA4-VHL19 as indicated. VEGF secretion into the medium was quantitated using ELISA assays and normalized by an equal amount of total proteins from each sample. Results are expressed as the mean ± SD of three independent experiments. (C) hsRPB7 enhances VEGF mRNA expression. A498, 786-O and HeLa cells were transfected with pcDNA4-hsRPB7 and/or pcDNA4-VHL19 as indicated. VEGF mRNA isoforms expression were analyzed by RT–PCR with total RNA and normalized by GAPDH as control (a). Results are expressed as the mean ± SD of three independent experiments (b). (D) Silencer RNA of hsRPB7 (sihsRPB7) downregulates VEGF mRNA expression. 786-O and HeLa cells were transfected with increasing amounts of hsRPB7 silencer RNA as indicated. Expression of hsRPB7, HIF-2α and β-actin was studied by western blotting (a); VEGF mRNA isoform expression was analyzed by RT–PCR with total RNA and normalized by GAPDH. Results are expressed as the mean ± SD of three independent experiments (b).
The VEGF protein secretion level was further analyzed in these kidney cancer cell lines. Transient transfection of hsRPB7 could promote the VEGF secretion into the medium in these two cell lines. Transfection with pVHL decreased VEGF secretion by 40–60% in all cell lines. Cotransfection of pVHL with hsRPB7 expression plasmid at a 1:1 ratio abolished VEGF oversecretion enhanced by hsRPB7 (Figure 5B). These data demonstrated that hsRPB7 could regulate VEGF expression and this effect could be reversed by pVHL.
To rule out the possible artifacts associated with reporter gene assays, we analyzed the effect of hsRPB7 and wild-type pVHL19 on endogenous VEGF mRNA expression and VEGF secretion in the kidney cancer cell lines A498 and 786-O. HeLa cells with endogenous wild-type pVHL were also incorporated in our study. As shown in Figure 5C, naturally occurring isoforms of VEGF mRNA were elevated by overexpression of hsRPB7. Introducing wild-type pVHL into these tumor cell lines resulted in decreased levels of VEGF mRNA expression. Cotransfection of pVHL and hsRPB7 resulted in a significant decrease of hsRPB7-dependent overexpression of VEGF mRNA.
To specify that the downregulation of VEGF expression by pVHL is through target degradation of hsRPB7, we used silencer RNA of hsRPB7 (sihsRPB7) to specifically diminish endogenous hsRPB7 expression. As shown in Figure 5D for 786-O cells, when increasing amounts of sihsRPB7 are introduced, the endogenous expression of hsRPB7 protein (Figure 5D, a) and VEGF mRNA (Figure 5D, b) can be decreased in a dose-dependent manner. To rule out the possibility of the influence on VEGF expression by HIF-1α and HIF-2α, we also studied the expression of these proteins under the effect of sihsRPB7. HIF-1α expression is not detectable in this cell line (Maxwell et al., 1999), and it has no effects on HIF-2α expression (Figure 5D, a). This effect is also specific to VEGF but not to GAPDH. HeLa cells with endogenous wild-type pVHL demonstrated similar patterns. Taken together, our results indicated a novel functional association between pVHL and hsRPB7.
Discussion
Studies of the yeast Saccharomyces cerevisiae Pol II have provided a remarkable view of the detailed structures and functional aspects of eukaryotic Pol II (Cramer et al., 2000, 2001). In yeast, the Pol II complex is comprised of 12 subunits (RPB1–RPB12), of which RPB4 and RPB7 form a dissociable subcomplex and are present in substoichiometric amounts during exponential growth (Cramer et al., 2000, 2001). This subcomplex has been linked to the stress response and is required for promoter recognition in transcription in vitro, which is similar to the prokaryotic sigma factor (Edwards et al., 1991; Orlicky et al., 2001). The facts that some of the stress-sensitive phenotypes linked to an rpb4– mutation are suppressed by overexpression of RPB7 (Sheffer et al., 1999) and that deletion of RPB7 is lethal (Young, 1991; McKune et al., 1993), while deletion of RPB4 is not (Woychik and Young, 1989; Choder, 1993; Rosenheck and Choder, 1998), suggest that RPB7 is the key factor of the RPB4–RPB7 subcomplex function(s). Recent studies also demonstrate that RPB7 is capable of interacting with Pol II independently without RPB4 (Sheffer et al., 1999). The fact that RPB7 may be found in some, but not all, Pol II complexes suggests that RPB7-dependent and -independent regulatory mechanisms may exist (Khazak et al., 1995). Three-dimensional structure studies of yeast Pol II have revealed that the RPB4–RPB7 subcomplex is located between the two major domains that form the DNA-binding cleft of the Pol II complex and suggested that RPB7 might be involved in inducing tighter closure of the DNA-binding cleft and thus stabilizing the Pol II complex (Jensen et al., 1998). This may help explain why RPB7 is important in the initiating transcription for a subset of genes during the stress response. How RPB7 is regulated during the stress response could be crucial for understanding how Pol II functions in the regulation of gene expression. Recently, it has been found that hsRPB7 is able to interact with two transcriptional factors, the Ews-Fli oncogenic fusion protein and the retinoic acid receptor, and modulate their transcriptional activity (Petermann et al., 1998; Shen et al., 1999). These studies further support the current idea that hsRPB4-hsRPB7 may function as a site of interaction between enhancer-binding transcription factors and the central Pol II complex (Petermann et al., 1998; Shen et al., 1999).
Using yeast two-hybrid screening, we have identified hsRPB7 as a novel pVHL-interacting partner from a kidney cDNA library. We have confirmed this interaction with a series of in vitro and in vivo experiments and demonstrated that hsRPB7 interacts with the β-domain of pVHL and naturally occurring pVHL β-domain mutants impair their ability to bind hsRPB7. Our subsequent experiments demonstrated that hsRPB7 does indeed undergo pVHL-dependent ubiquitylation and rapid degradation via the proteasomal pathway. Our double immunostaining revealed that in kidney 786-O tumor cells carrying a non-functional pVHL, the endogenous hsRPB7 was accumulated in the nucleus. In contrast, after introducing wild-type pVHL into the cells, endogenous nuclear accumulation of hsRPB7 was dramatically reduced; this could be due to rapid degradation targeted by wild-type pVHL19. Since overexpression of VEGF mRNA has been recognized as one of the signatures of pVHL loss-of-function mutation, we tested whether hsRPB7 could selectively transactivate the VEGF promoter. Using the fibrinogen promoter and the androgen response element as controls, we found that hsRPB7 could transactivate the VEGF promoter but not two control promoters in kidney A498 and 786-O cell lines. This effect could be reversed by cotransfection of wild-type pVHL19. Parallel experiments demonstrating that endogenous mRNA expression and VEGF secretions in these cell lines could be influenced by hsRPB7 and pVHL also supported the above data. Our results suggest that the VEGF promoter may be one gene component that hsRPB7 can selectively regulate. Using sihsRPB7, we also demonstrated that the regulation of VEGF by hsRPB7 and by HIF proteins are separate processes. Recent studies using authentic renal tissues demonstrated that the overaccumulation of HIFs cannot be observed in all CCRCC with VHL gene mutations and high VEGF expression, indicating the existence of other potential regulators of VEGF (Wiesener et al., 2001; Turner et al., 2002).
Overall, our studies provide important evidence on the regulation of hsRPB7 at the protein level by pVHL and its potential regulatory role in renal carcinogenesis. A recent study reported that pVHL could target the large subunit (Rpb1) of RNA Pol II for ubiquitylation and degradation (Kuznetsova et al., 2003). Together with our study, it indicates that pVHL could regulate gene expression through RNA Pol II subunits. It would be important to identify other genes that could be specifically regulated by hsRPB7 other than VEGF, as well as determining their roles in tumorigenesis and tumor development. It would also be crucial to explore the mechanism of hsRPB7 regulation on those specific genes, whether through direct binding to conserved promoter sequence or bridging specific transcription factors into the central RNA Pol II complex. HIF-1α is an important transcription factor targeted by pVHL–E3 ligase for rapid turnover (Ohh et al., 2000). HIF-1α induces expression of hypoxia-inducible genes, such as VEGF. Based on amino acid sequence analysis, hsRPB7 does not contain the oxygen-dependent degradation (ODD) domains that are present in HIF-1α, indicating that a novel mechanism might exist in the interaction between hsRPB7 and pVHL. Whether prolyl hydroxylation of hsRPB7 is required for pVHL-mediated ubiquitylation and degradation remains to be explored. It would be interesting to explore whether HIF-1α and hsRPB7 can regulate a subset of stress-response genes synergistically, and whether there are temporal and spatial differences in the regulation of stress-response-related transcription.
In summary, our current data suggest that hsRPB7 is a natural degradation target of the pVHL–E3 ligase complex and hsRPB7 may participate in the regulation of VEGF expression. It is conceivable that, under normal physiological conditions, hsRPB7 is regulated by pVHL and maintained at an optimal level. Once pVHL becomes mutated, hsRPB7 overaccumulation occurs, resulting in overexpression of hsRPB7-targeted genes, such as VEGF, which may be relevant to the VHL inactivation-related phenotype.
Materials and methods
Yeast two-hybrid screening
Using the Yeast Two-Hybrid System 3 (Clontech Laboratories, Palo Alto, CA), a pretransformed human kidney cDNA library in the Y187 yeast strain (Clontech Laboratories) was screened via yeast mating with AH109, a yeast strain transformed with a VHL bait construct (pGBKT7-VHL19) for high-stringency selection according to the manufacturer’s protocol. Positive clones were sequenced and the corresponding cDNAs were subcloned into appropriate vectors for subsequent experiments. A yeast two-hybrid assay using independent retransformation of the bait construct and the candidate clones together followed by a β-Gal assay was used to confirm the interaction between pVHL and newly identified proteins.
Plasmid construction
pGBKT7-VHL19, pcDNA4-VHL19, pGEX6p1-VHL19 and pM-VHL19 were constructed by inserting human pVHL19 cDNA (amino acid 54–213) into pGBKT7, pcDNA4, pGEX6p1 and pM vectors, respectively. pGEX6p1-VHL54–113 and pGEX6p1-VHL114–213, derived from pGEX6p1-VHL19, were constructed to express pVHL fragments containing residues 54–113 and 114–213, respectively. Missense mutation constructs (P86H and Y98H, respectively) were derived from pGEX6p1-VHL19 by PCR-based site-directed mutagenesis. These mutant VHL constructs were subsequently transferred into pcDNA4 vectors to facilitate expression of His6 tag fusion protein in mammalian cells. pGADT7-hsRPB7, pVP16-hsRPB7, pcDNA4-hsRPB7 and pCMV-hsRPB7 were constructed by transferring the full-length insert from the pACT2-hsRPB7 clone identified by yeast two-hybrid screening. pSG5-p53 was constructed by inserting a p53 coding sequence into the pSG5 vector. All inserts of constructs involved in PCR amplification were confirmed by complete sequencing.
GST pulldown assay
Overnight cultures of bacteria strain BL21 transformed with wild-type or mutant VHL GST fusion proteins were diluted 1:100 in a Luria–Bertani medium plus ampicillin and cultured at 30°C until A600 reached 0.4. The fusion proteins were then induced with 1 mM ispropyl-β-d-thiogalactopyranoside (IPTG) and purified. Two-hundred micrograms of GST or GST fusion protein were added to glutathione–Sepharose 4B MicroSpin Columns (Amersham Pharmacia, Cleveland, OH). Total volume was adjusted to 300 µl with H400 buffer [20 mM HEPES pH 7.8, 0.5 mM EDTA, 20% glycerol, 400 mM KCl, 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and proteinase inhibitor cocktail] and incubated for 3 h at 4°C. After washing three times in H400 buffer, 10 µl of in vitro translated [35S]methionine-labeled hsRPB7 were added to the reaction and incubated at 4°C for an additional 6 h. After washing five times with H400 buffer, the proteins were eluted in 20 µl of 2× SDS buffer and resolved on a 10% SDS–PAGE gel followed by autoradiography.
Mammalian two-hybrid assay
pM-VHL19, pVP16-hsRPB7 and pG5-CAT were cotransfected into COS-7 cells with a constant amount of a second reporter, pCMV-β-gal, for normalization of the transfection efficiency. Parallel transfections with negative and positive controls were performed. After 36 h, cells were harvested and lysed. The lysates were used for radioactive CAT assay to demonstrate the interaction between the two proteins.
In vivo co-immunoprecipitation
HeLa cells were grown to 90% confluence in T-150 flasks with or without MG132 treatment. Cells were lysed in RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS). Total protein concentration was measured (Bio-Rad Laboratories, Hercules, CA). Five hundred micrograms of total protein were incubated with 50 µl of protein A/G–Sepharose beads (Santa Cruz Laboratories, Santa Cruz, CA), together with 50 µl of human VHL antibody (PharMingen, San Diego, CA) at 4°C. Total volume was adjusted to 300 µl by H200 buffer (20mM HEPES pH 7.8, 0.5 mM EDTA, 20% glycerol, 200 mM KCl, 0.5 mM PMSF and proteinase inhibitor cocktail). After incubation for 6 h, the beads were washed with H200 buffer three times, eluted in 20 µl of 2× SDS buffer twice and resolved on a 10% SDS–PAGE gel followed by immunoblotting (IB) using VHL, hsRPB7 and EC (Santa Cruz Laboratories) antibodies, respectively. Twenty-five micrograms of total protein (5% input) were loaded on the gel for comparison.
In vitro ubiquitylation assay
The method for in vitro ubiquitylation assay has been described previously (Ohh et al., 2000). Ubiquitylation assays were carried out at 37°C for 2 h in a total volume of 15 µl. 35S-labelled hsRPB7 (1 µl) was incubated in the presence of S-100 extracts (45 µg) supplemented with 8 µg/µl ubiquitin (BostonBiochem, Cambridge, MA), 100 ng/µl ubiquitin aldehyde (BostonBiochem, Cambridge, MA) and an energy-regenerating system (20 mM Tris pH 7.4, 2 mM ATP, 5 mM MgCl2, 40 mM creatine phosphate and 0.5 µg/µl creatine kinase). In vitro ubiquitylation using recombinant E1 and E2 was described previously (Iwai et al., 1999). VBC complex was immunoprecipitated from HeLa cells (Iwai et al., 1999). E1 and E2 were purchased from A.G. Scientific Inc. (San Diego, CA).
In vivo ubiquitylation assay
768-O cells were grown to near 50% confluence in 35 mm culture dishes and cotransfected with pCMV-flag-hsRPB7 and pcDNA4-VHL19 or pCMV-flag-hsRPB7 and pcDNA4 vector. Cells were lysed in RIPA buffer 36 h after transfection and centrifuged at 14 000 r.p.m. for 20 min at 4°C. The supernatants were incubated with antiflag antibodies (Santa Cruz Laboratories) for 6 h at 4°C as described above (in vivo co-immunoprecipitation). Washed immunoprecipitated complex and crude lysates were separated by SDS–PAGE and detected by anti-flag or anti-ubiquitin antibodies, respectively.
Fluorescence immunostaining
786-O cells were seeded on two-well slides (Nalge, Rochester, NY) overnight. Transient transfection was performed using pcDNA4, pcDNA4-VHL19, pcDNA4-VHL-P86H, pcDNA4-VHL-Y98H or pcDNA4-VHL114–213. After incubation for 36 h, cells were fixed and double-immunostained using anti-hsRPB7 rabbit polyclonal antibody (Khazak et al., 1998) and anti-VHL mouse monoclonal or anti-His mouse monoclonal antibody (Invitrogen, Carlsbad, CA) as the primary antibodies, and followed by FITC green-conjugated goat anti-rabbit and rhodamine red-conjugated goat anti-mouse antibodies. Blue DAPI staining was used to show the location of nuclei.
Degradation and pulse–chase assay
A498 cells and 786-O cells were transiently transfected with pcDNA4 vector (a negative control) or pcDNA4-VHL19 in 35 mm culture dishes with a total of 2 µg of plasmid DNA per transfection. After 18 h, cells were treated with MG132 (5 µM) (Calbiochem, La Jolla, CA) or DMSO for 6 h, followed by incubation with cycloheximide (100 µg/ml) (Sigma, St Louis, MO), and then lysed at 0, 2, 4 and 6 h. Proteins were collected from each time point and an equal amount of total protein was resolved by SDS–PAGE. Western blots were performed using an anti-hsRPB7 and an anti-p62 antibody (the p62 subunit of TFIIH used as a loading control), respectively.
VEGF promoter transactivation assay
The VEGF promoter construct covering the –194 to +153 region (Pal et al., 1998) was constructed into the pGL3-luciferase vector (Promega Corporation, Madison, WI) to make pGL3-VEGF-Promoter. Cells were seeded on six-well plates overnight and then transfected with 0.3 µg plasmid of pGL3-VEGF-Promoter, 0.9 µg plasmid of pcDNA4-VHL19 and/or pcDNA4-hsRPB7. A Renilla luciferase expression plasmid, pRL-SV40 (Promega Corporation, Madison, WI), was used as an internal control for transfection efficiency. Total amounts of DNA were adjusted to 2 µg by pcDNA4 vector. Cells were lysed for dual-luciferase assay after 36 h of transfection. At least three independent experiments were performed to ensure consistency.
VEGF mRNA expression
A498, 786-O and HeLa cells were seeded on 100 mm plates overnight and then transfected with 5 µg plasmid of pcDNA4-VHL19 and/or pcDNA4-hsRPB7. Total amounts of DNA were adjusted to 10 µg by adding pcDNA4 vector. Total RNA was extracted 36 h after transfection, and RT–PCR was performed using 2 µg of total RNA from each sample to semiquantitate endogenous VEGF mRNA expression. GAPDH was used as an internal control. VEGF primers, 5′-CGAAGTGGTGAAGTT CATGGATG-3′ (sense) and 5′-TTCTGTATCAGTCTTTCCTGGTGA-3′ (antisense), and GAPDH primers, 5′-CCAAAACTGGCAATTCCAT GGCA-3′ (sense) and 5′-TCTAGACGGCAGGTCAGGTCCACC-3′ (antisense), were used in our experiments.
VEGF secretion assay
A498 and 786-O cells were seeded on six-well plates overnight and then transfected with 1 µg plasmid of pcDNA4-VHL19 and/or pcDNA4-hsRPB7. Total amounts of DNA were adjusted to 2 µg by adding pcDNA4 vector. The media were collected 36 h after transfection and the protein concentration was measured. An ELISA assay for VEGF was performed using equal amounts of total protein according to the manufacturer’s protocol (R&D Systems, Minneapolis, MN). At least three independent experiments were performed to ensure consistency.
hsRPB7 silencer RNA construction and transfection
sihsRPB7 was constructed according to the standard protocol from Ambion (Ambion Inc., Austin, TX). sihsRPB7 primer, 5′-AATTGCTG TCACCACCATTGA-3′, was used in our experiments. 786-O and HeLa cells were seeded on 100 mm plates overnight and then transfected with concentrations of 25, 50 and 100 nM sihsRPB7 with oligofectamine (Invitrogen Life Technologies, Carlsbad, CA). Total RNA and protein were extracted 36 h after transfection. Western blot analysis of hsRPB7 and HIF-2α expression and RT–PCR analysis of VEGF were performed as described above.
Acknowledgements
This work was partially supported by NIH core grant CA-06927 to Fox Chase Cancer Center.
References
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [Abstract] [Google Scholar]
- Choder M. (1993) A growth rate-limiting process in the last growth phase of the yeast life cycle involves RPB4, a subunit of RNA polymerase II. J. Bacteriol., 175, 6358–6363. [Europe PMC free article] [Abstract] [Google Scholar]
- Cockman M.E. et al. (2000) Hypoxia inducible factor-α binding and ubiquitylation by the von Hippel–Lindau tumor suppressor protein. J. Biol. Chem., 275, 25733–25741. [Abstract] [Google Scholar]
- Cramer P. et al. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science, 288, 640–649. [Abstract] [Google Scholar]
- Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science, 292, 1863–1876. [Abstract] [Google Scholar]
- Edwards A.M., Kane,C.M., Young,R.A. and Kornberg,R.D. (1991) Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem., 266, 71–75. [Abstract] [Google Scholar]
- Foster K. et al. (1994) Somatic mutations of the von Hippel–Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum. Mol. Genet., 3, 2169–2173. [Abstract] [Google Scholar]
- Gnarra J.R. et al. (1994) Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet., 7, 85–90. [Abstract] [Google Scholar]
- Hershko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [Abstract] [Google Scholar]
- Ivan M. et al. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science, 292, 464–468. [Abstract] [Google Scholar]
- Iwai K., Yamanaka,K., Kamura,T., Minato,N., Conaway,R.C., Conaway,J.W., Klausner,R.D. and Pause,A. (1999) Identification of the von Hippel–Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA, 96, 12436–41. [Europe PMC free article] [Abstract] [Google Scholar]
- Jaakkola P. et al. (2001) Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science, 292, 468–472. [Abstract] [Google Scholar]
- Jensen G.J., Meredith,G., Bushnell,D.A. and Kornberg,R.D. (1998) Structure of wild-type yeast RNA polymerase II and location of Rpb4 and Rpb7. EMBO J., 17, 2353–2358. [Europe PMC free article] [Abstract] [Google Scholar]
- Kamura T. et al. (1999) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science, 284, 657–661. [Abstract] [Google Scholar]
- Kamura T., Sato,S., Iwai,K., Czyzyk-Krzeska,M., Conaway,R.C. and Conaway,J.W. (2000) Activation of HIF1α ubiquitination by a reconstituted von Hippel- Lindau (VHL) tumor suppressor complex. Proc. Natl Acad. Sci. USA, 97, 10430–10435. [Europe PMC free article] [Abstract] [Google Scholar]
- Khazak V., Sadhale,P.P., Woychik,N.A., Brent,R. and Golemis,E.A. (1995) Human RNA polymerase II subunit hsRPB7 functions in yeast and influences stress survival and cell morphology. Mol. Biol. Cell, 6, 759–775. [Europe PMC free article] [Abstract] [Google Scholar]
- Khazak V., Estojak,J., Cho,H., Majors,J., Sonoda,G., Testa,J.R. and Golemis,E.A. (1998) Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II. Mol. Cell. Biol., 18, 1935–1945. [Europe PMC free article] [Abstract] [Google Scholar]
- Kuznetsova A.V., Meller,J., Schnell,P.O., Nash,J.A., Ignacak,M.L., Sanchez,Y., Conaway,J.W., Conaway,R.C. and Czyzyk-Krzeska,M.F. (2003) von Hippel–Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination. Proc. Natl Acad. Sci. USA, 100, 2706–2711. [Europe PMC free article] [Abstract] [Google Scholar]
- Lisztwan J., Imbert,G., Wirbelauer,C., Gstaiger,M. and Krek,W. (1999) The von Hippel–Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev., 13, 1822–1833. [Europe PMC free article] [Abstract] [Google Scholar]
- Maher E.R. and Kaelin,W.G.,Jr (1997) von Hippel–Lindau disease. Medicine (Baltimore), 76, 381–391. [Abstract] [Google Scholar]
- Maxwell P.H. et al. (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature, 399, 271–275. [Abstract] [Google Scholar]
- McKune K., Richards,K.L., Edwards,A.M., Young,R.A. and Woychik,N.A. (1993) RPB7, one of two dissociable subunits of yeast RNA polymerase II, is essential for cell viability. Yeast, 9, 295–299. [Abstract] [Google Scholar]
- Ohh M., Park,C.W., Ivan,M., Hoffman,M.A., Kim,T.Y., Huang,L.E., Pavletich,N., Chau,V. and Kaelin,W.G. (2000) Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel–Lindau protein. Nat. Cell Biol., 2, 423–427. [Abstract] [Google Scholar]
- Orlicky S.M., Tran,P.T., Sayre,M.H. and Edwards,A.M. (2001) Dissociable Rpb4-Rpb7 subassembly of RNA polymerase II binds to single- strand nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem., 276, 10097–10102. [Abstract] [Google Scholar]
- Pal S., Claffey,K.P., Cohen,H.T. and Mukhopadhyay,D. (1998) Activation of Sp1-mediated vascular permeability factor/vascular endothelial growth factor transcription requires specific interaction with protein kinase Cζ. J. Biol. Chem., 273, 26277–26280. [Abstract] [Google Scholar]
- Pause A., Lee,S., Worrell,R.A., Chen,D.Y., Burgess,W.H., Linehan,W.M. and Klausner,R.D. (1997) The von Hippel–Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc. Natl Acad. Sci. USA, 94, 2156–2161. [Europe PMC free article] [Abstract] [Google Scholar]
- Petermann R., Mossier,B.M., Aryee,D.N., Khazak,V., Golemis,E.A. and Kovar,H. (1998) Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene, 17, 603–610. [Abstract] [Google Scholar]
- Rosenheck S. and Choder,M. (1998) Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J. Bacteriol., 180, 6187–6192. [Europe PMC free article] [Abstract] [Google Scholar]
- Sheffer A., Varon,M. and Choder,M. (1999) Rpb7 can interact with RNA polymerase II and support transcription during some stresses independently of Rpb4. Mol. Cell. Biol., 19, 2672–2680. [Europe PMC free article] [Abstract] [Google Scholar]
- Shen X.Q., Bubulya,A., Zhou,X.F., Khazak,V., Golemis,E.A. and Shemshedini,L. (1999) Ligand-free RAR can interact with the RNA polymerase II subunit hsRPB7 and repress transcription. Endocrine, 10, 281–289. [Abstract] [Google Scholar]
- Shuin T. et al. (1994) Frequent somatic mutations and loss of heterozygosity of the von Hippel–Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res., 54, 2852–2855. [Abstract] [Google Scholar]
- Skowyra D., Koepp,D.M., Kamura,T., Conrad,M.N., Conaway,R.C., Conaway,J.W., Elledge,S.J. and Harper,J.W. (1999) Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science, 284, 662–665. [Abstract] [Google Scholar]
- Stebbins C.E., Kaelin,W.G.,Jr and Pavletich,N.P. (1999) Structure of the VHL–elongin C–elongin B complex: implications for VHL tumor suppressor function. Science, 284, 455–461. [Abstract] [Google Scholar]
- Stolle C. et al. (1998) Improved detection of germline mutations in the von Hippel–Lindau disease tumor suppressor gene. Hum. Mutat., 12, 417–423. [Abstract] [Google Scholar]
- Tanimoto K., Makino,Y., Pereira,T. and Poellinger,L. (2000) Mechanism of regulation of the hypoxia-inducible factor-1α by the von Hippel–Lindau tumor suppressor protein. EMBO J., 19, 4298–4309. [Europe PMC free article] [Abstract] [Google Scholar]
- Turner K.J. et al (2002) Expression of hypoxia-inducible factors in human renal cancer: relationship to angiogenesis and to the von Hippel–Lindau gene mutation. Cancer Res., 62, 2957–2961. [Abstract] [Google Scholar]
- Wiesener M.S. et al. (2001) Constitutive activation of hypoxia-inducible genes related to overexpression of hypoxia-inducible factor-1α in clear cell renal carcinomas. Cancer Res., 61, 5215–5222. [Abstract] [Google Scholar]
- Woychik N.A. and Young,R.A. (1989) RNA polymerase II subunit RPB4 is essential for high- and low- temperature yeast cell growth. Mol. Cell. Biol., 9, 2854–2859. [Europe PMC free article] [Abstract] [Google Scholar]
- Young R.A. (1991) RNA polymerase II. Annu. Rev. Biochem., 60, 689–715. [Abstract] [Google Scholar]
Articles from The EMBO Journal are provided here courtesy of Nature Publishing Group
Full text links
Read article at publisher's site: https://doi.org/10.1093/emboj/cdg410
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc175799?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/emboj/cdg410
Article citations
ARMC5 controls the degradation of most Pol II subunits, and ARMC5 mutation increases neural tube defect risks in mice and humans.
Genome Biol, 25(1):19, 15 Jan 2024
Cited by: 0 articles | PMID: 38225631 | PMCID: PMC10789052
A multi-model based on radiogenomics and deep learning techniques associated with histological grade and survival in clear cell renal cell carcinoma.
Insights Imaging, 14(1):207, 27 Nov 2023
Cited by: 1 article | PMID: 38010567 | PMCID: PMC10682311
Cleavage fragments of the C-terminal tail of polycystin-1 are regulated by oxidative stress and induce mitochondrial dysfunction.
J Biol Chem, 299(9):105158, 12 Aug 2023
Cited by: 4 articles | PMID: 37579949 | PMCID: PMC10502374
Function, mechanism and drug discovery of ubiquitin and ubiquitin-like modification with multiomics profiling for cancer therapy.
Acta Pharm Sin B, 13(11):4341-4372, 22 Jul 2023
Cited by: 1 article | PMID: 37969742 | PMCID: PMC10638515
Review Free full text in Europe PMC
Heterogeneous nuclear ribonucleoprotein U-actin complex derived from extracellular vesicles facilitates proliferation and migration of human coronary artery endothelial cells by promoting RNA polymerase II transcription.
Bioengineered, 13(5):11469-11486, 01 May 2022
Cited by: 2 articles | PMID: 35535400 | PMCID: PMC9276035
Go to all (68) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease.
Cancer Res, 62(13):3803-3811, 01 Jul 2002
Cited by: 90 articles | PMID: 12097293
The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity.
Mol Cell Biol, 17(9):5629-5639, 01 Sep 1997
Cited by: 182 articles | PMID: 9271438 | PMCID: PMC232411
Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein.
J Biol Chem, 275(33):25733-25741, 01 Aug 2000
Cited by: 625 articles | PMID: 10823831
The von Hippel-Lindau tumor suppressor gene and kidney cancer.
Clin Cancer Res, 10(18 pt 2):6290S-5S, 01 Sep 2004
Cited by: 173 articles | PMID: 15448019
Review
Funding
Funders who supported this work.
NCI NIH HHS (2)
Grant ID: CA-06927
Grant ID: P30 CA006927




