Abstract
Free full text

GeneScan Reverse Transcription-PCR Assay for Detection of Six Common Respiratory Viruses in Young Children Hospitalized with Acute Respiratory Illness
Abstract
A reverse transcription-PCR (RT-PCR) assay based on automated fluorescent capillary electrophoresis and GeneScan software analysis was developed to detect six common respiratory viruses in clinical specimens from young children. Assays for human respiratory syncytial virus (HRSV); human parainfluenza viruses 1, 2, and 3 (HPIV1, -2, and -3, respectively); and influenza A and B viruses were incorporated into a single standard assay format. The optimized assay panel was used to test 470 respiratory specimens obtained from 462 children hospitalized with acute respiratory illness that had been previously tested by viral culture (405 specimens) or direct immunofluorescence staining (DIF) (65 specimens). Of 93 specimens positive for respiratory viruses by culture or DIF, 86 (92%) were positive by RT-PCR, including 66 HRSV, 2 HPIV2, 5 HPIV3, 3 influenza A virus, and 10 influenza B virus specimens. An additional 119 respiratory viruses were identified by RT-PCR in 116 patients for whom results were negative by viral isolation or DIF. We conclude that the GeneScan RT-PCR panel can markedly improve detection of acute respiratory virus infections in young children.
Viruses account for 50 to 90% of lower respiratory tract infections in young children (13), with over 80% of identified infections attributed to six viruses: human respiratory syncytial virus (HRSV); human parainfluenza viruses 1, 2, and 3 (HPIV1, -2, and -3, respectively); and influenza A and B viruses (12, 16). For many years, the diagnosis of respiratory virus infections in young children has been based on virus isolation, antigen detection, and serologic response. Although these methods have usually proven adequate for diagnosis, they have several important limitations. Viral isolation is labor-intensive and often takes several days to complete, making it unhelpful for clinical management of the patient. Antigen detection assays vary widely in sensitivity and specificity. Serologic diagnosis is encumbered by the difficulty and delay of collecting convalescent-phase blood samples and is unreliable in infants, presumably because of their immature immune systems and/or the presence of maternal antibodies that can mask a serologic response (17).
Reverse transcription-PCR (RT-PCR) assays offer the possibility of improved sensitivity and more timely diagnosis of respiratory virus infections. In studies of children (11), adults (31), and the immunocompromised (24, 30), RT-PCR has been shown to provide rapid results with equivalent or greater sensitivity than direct antigen detection or virus isolation for detection of respiratory viruses. Although numerous RT-PCR assays have been developed for individual respiratory viruses, they differ widely in conditions for extraction, amplification, and method of amplicon detection. A more recent trend has been the development of RT-PCR assays that simultaneously detect multiple respiratory pathogens by (i) using primer pairs to different respiratory viruses in separate reactions that share single standard amplification conditions (10, 11, 30, 32), (ii) using broadly reactive primers to highly conserved genes that encompass different virus genera (HRSV and HPIV3) (7), and (iii) combining multiple primer sets into single “multiplexed” amplification reactions (1, 4, 5, 8, 14, 25, 28, 29).
Automated fluorescent capillary electrophoresis and GeneScan software analysis (Applied Biosystems, Foster City, Calif.) has been used extensively in procedures requiring very precise sizing and quantitation of DNA fragments, including single-nucleotide-polymorphism (SNP) validation and screening, microsatellite analysis, amplified fragment length polymorphism analysis, and others. In PCR applications of GeneScan, one dye-labeled primer and one unlabeled primer are used to amplify the specific target, and a small portion of the amplified product is combined with a dye-labeled size standard and electrophoresed on an automated sequencer, where the fluorescent product is sized and quantified. In this study, we combined GeneScan analysis with RT-PCR in a novel application for the sensitive and specific detection of HRSV; HPIV1, -2, and -3; and influenza A and B viruses in clinical specimens and compared this assay with virus isolation and antigen detection for diagnosis of infection with these six common respiratory viruses in young children.
MATERIALS AND METHODS
Specimens.
A total of 470 respiratory specimens collected from 462 children <5 years of age hospitalized with acute respiratory illness were obtained from three hospitals in Nashville, Tenn., two hospitals in Rochester, N.Y., and one hospital in Montevideo, Uruguay. As part of the New Vaccine Surveillance Network population-based surveillance of acute respiratory infections in hospitalized children in Rochester and Nashville, 405 combined nasal and throat swabs from 397 children were collected in viral transport media from August 2000 through March 2001; two specimens each were obtained from eight children who were hospitalized twice in the surveillance period. Specimen collection protocols and procedures were approved by the Institutional Review Boards (IRB) at the Centers for Disease Control and Prevention (CDC), Vanderbilt University, and the University of Rochester. Aliquots of specimens collected at Nashville and Rochester hospitals were immediately placed in NucliSens lysis buffer (bioMérieux, Durham, N.C.) and held at −70°C until being shipped to CDC on dry ice. Nasopharyngeal aspirates collected from 65 children at the Hospital Central de las Fuerzas Armadars in Montevideo, Uruguay, from July to August 2001 were also available for testing. These specimens had been stored at −70°C, and undiluted aliquots were shipped on dry ice to CDC.
The specimens were delinked from personal identifiers and exempted from IRB review.
Virus isolation and antigen detection. Respiratory swab specimens received at Vanderbilt and Rochester university medical centers were inoculated onto tube cultures of primary rhesus monkey kidney, HEp-2, and NCI-H292 cells and monitored for cytopathic effect for 10 to 14 days. All cultures were screened for HRSV; HPIV1, -2, and -3; and influenza A and B viruses by indirect immunofluorescence staining with reagents obtained from Chemicon International, Inc., Temecula, Calif., and Bartels/Trinity Biotech, County Wicklow, Ireland. Direct immunofluorescence staining (DIF) for the same viruses was performed on specimens at the Laboratorio de Analisis Clinicos, Hospital Central de las Fuerzas Armadas, using reagents obtained from Argene Biosoft, Varilhes, France.
Nucleic acid extraction.
Aliquots (100 μl) of each specimen were dispensed into two tubes, each containing 900 μl of prewarmed NucliSens lysis buffer and incubated at room temperature for 10 min. Total nucleic acid was then recovered from the specimens by automated NucliSens extraction following the manufacturer's instructions (bioMérieux). Nucleic acid extracts (40 to 50 μl) were supplemented with 100 μl of nuclease-free water (Ambion, Austin, Tex.) containing RNase inhibitor (Roche) at a final concentration of ~1 U/μl. Extracts were tested by RT-PCR immediately or stored at −70°C.
Primers.
Previously described oligonucleotide primers for RSV (24); HPIV1, -2, and -3 (5); and influenza A and B viruses (3) were reviewed against current GenBank and CDC sequence databases and modified where required to minimize nucleotide sequence mismatches (Table (Table1).1). Primers for human β-actin mRNA (27) were used to test for specimen adequacy, RNA extraction, and the presence of PCR inhibitors. For GeneScan analysis, the positive-strand primer of each primer set was 5′ end labeled with the fluorescent dye 6-carboxyfluorescein (6-FAM).
TABLE 1.
Primers for respiratory virus RT-PCR assay panel
| Virus (reference) | Primer | Length (nt) | Gene | Location | Polarity | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|---|---|---|---|---|
| HRSV (24) | RSVAB-F1-FAM | 23 | F | 1124-1146 | + | AACAGTTTAACATTACCAAGTGA | 380 |
| RSVAB-R1 | 23 | 1481-1503 | − | TCATTGACTTGAGATATTGATGC | |||
| HPIV1 (5) | HPIV1-F1-FAM | 22 | HN | 780-801 | + | CCGGTAATTTCTCATACCTATG | 317 |
| HPIV1-R1 | 21 | 1076-1096 | − | CCTTGGAGCGGAGTTGTTAAG | |||
| HPIV2 (5) | HPIV2-F1-FAM | 22 | HN | 845-866 | + | CCATTTACCTAAGTGATGGAAT | 204 |
| HPIV2-R1 | 22 | 1027-1048 | − | GCCCTGTTGTATTTGGAAGAGA | |||
| HPIV3 (5) | HPIV3-F1-FAM | 22 | HN | 884-905 | + | ACTCCCAAAGTTGATGAAAGAT | 103 |
| HPIV3-R1 | 22 | 965-986 | − | TAAATCTTGTTGTTGAGATTGA | |||
| Influenza A virus (3) | FLUA-F1-FAM | 20 | NS1 | 465-484 | + | CTAAGGGCTTTCACCGAAGA | 192 |
| FLUA-R1 | 20 | 637-656 | − | CCCATTCTCATTACTGCTTC | |||
| Influenza B virus (3) | FLUB-F1-FAM | 21 | NS1 | 746-765 | + | ATGGCCATCGGATCCTCAAC | 241 |
| FLUB-R1 | 20 | 967-986 | − | TGTCAGCTATTATGGAGCTG | |||
| RNA control (27) | BACTIN-F2-FAM | 18 | Human β-actin | −8-26 | + | CCTCGCCTTTGCCGATCC | 626 |
| BACTIN-R2 | 23 | 596-618 | − | GGATCTTCATGAGGTAGTCAGTC |
RT-PCR.
RT-PCR assays were developed with the one-step Access RT-PCR system (Promega Corp., Madison, Wis.). For each specimen, 50-μl reaction mixtures were prepared for each of the six viruses and the β-actin control by adding 5 μl of RNA extract to 45 μl of nuclease-free water containing 1× reaction buffer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 1.5 mM MgSO4, 0.1 U of avian myeloblastosis virus reverse transcriptase per μl, 0.1 U of Tf1 DNA polymerase per μl, and 1.0 μM each primer, and distributed in 0.2-ml volumes to an eight-tube strip. Thermocycling was performed on a MicroAmp 9600 (Applied Biosystems) programmed as follows: 48°C for 45 min for RT and 94°C for 2 min for RT denaturation; 40 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min for cDNA amplification; and 5 min at 72°C for final amplicon extension. Positive and negative RT-PCR controls containing standardized viral RNA extracts and nuclease-free water, respectively, were included in each run.
GeneScan analysis.
Following RT-PCR, 1 μl of the amplification product was transferred to Microamp 96-well tubes (Applied Biosystems) containing 12 μl of deionized formamide (Sigma, St. Louis, Mo.) and 0.5 μl of the internal size standard GenoTYPE TAMARA 50-500 DNA ladder (Invitrogen, Carlsbad, Calif.), denatured at 90°C for 5 min, and cooled on ice. Amplicon analysis was then performed on an ABI Prism 310 genetic analyzer (Applied Biosystems) with GeneScan software (version 3.1.2), using a 47-cm capillary with POP-4 polymer. Electropherograms were examined, and specimens showing discrete amplicon peaks within ≤1 nucleotide (nt) of the respective positive control peak were considered “preliminary” positives. Specimens were designated “true” positives if the same amplification product was identified in the duplicate specimen aliquot after a second independent RT-PCR amplification.
Sensitivity and specificity studies.
The sensitivities of the GeneScan RT-PCR assays were assessed by testing 10-fold serial dilutions of the following: (i) freshly grown cultures of RSV (A2 strain) in HEp-2 cells; HPIV1 (strain C-35), HPIV2 (strain Greer), and HPIV3 (strain C-243) in NCI-H292 cells; and influenza A (strain A/Bangkok/1/79) and influenza B (strain Ann Arbor/1/86) viruses in MDCK cells; and (ii) quantified, cloned PCR amplification products of each virus prepared with the TOPO TA cloning kit (Invitrogen). RT-PCR assays were further evaluated by testing multiple field isolates of the test viruses, other respiratory disease pathogens, including HPIV4 A and B, adenovirus, enterovirus, rhinovirus, measles virus, mumps virus, coronavirus 229E and OC43, Epstein-Barr virus, cytomegalovirus, herpes simplex virus types 1 and 2, Streptococcus pneumoniae, Mycoplasma pneumoniae, and mock-infected human cell cultures.
RESULTS
GeneScan RT-PCR assay design and optimization.
Primers, dNTPs, enzymes, MgCl2 concentrations, and thermocycling conditions were optimized for each assay, and a single standard set of conditions was chosen to accommodate all assays. An electropherogram of typical positive control results for each respective virus and β-actin is shown in Fig. Fig.1.1. GeneScan analysis of dilution series of cloned PCR products was more sensitive than conventional gel electrophoresis and ethidium bromide staining, and product sizing was highly specific, giving reproducible results to within 1 nt (Fig. (Fig.2).2). Standardized RT-PCR assays detected fewer than 50 cloned plasmid copies of each virus and had sensitivity that was identical to (HPIV2 and influenza A and B viruses) or 10-fold-greater (HRSV, HPIV1, and HPIV3) when compared in duplicate viral infectivity assays. Each assay correctly amplified the corresponding field isolates and gave no false amplifications with other respiratory pathogens. Multiplexing primers in various combinations resulted in 10- to 100-fold-lower sensitivity than that of assays using single primer pairs, and for this reason, we chose to use single-primer-pair amplification reactions for each virus.

Electropherogram of GeneScan analysis results showing RT-PCR amplification products for respiratory virus and β-actin controls run under denaturing conditions.
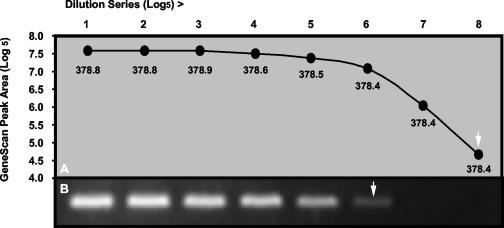
Comparison of GeneScan (A) with ethidium bromide staining (B) for analysis of RT-PCR products obtained from serial fivefold dilutions of a plasmid containing cloned HRSV fragment. The mean product sizes (nucleotides) obtained by GeneScan from duplicate samples for each dilution are indicated under the data points. Arrows indicate detection limits for each method.
GeneScan RT-PCR with clinical specimens.
Of the 470 clinical specimens previously tested by viral isolation or DIF, 93 (19.8%) were positive for one of the six respiratory viruses (Table (Table2).2). Sixty-eight of 405 (16.8%) specimens tested by virus culture were positive: 44 for HRSV, 2 for HPIV2, 8 for HPIV3, 4 for influenza A virus, and 10 for influenza B virus. Twenty-nine other viruses were also isolated from these specimens, including 15 adenovirus, 4 cytomegalovirus, 1 herpes simplex virus, 7 enterovirus, and 2 rhinovirus specimens. Twenty-five of 65 (38.5%) specimens tested by DIF were positive for HRSV only. Overall, the GeneScan RT-PCR assay panel was positive with 86 of 93 (92%) culture- or DIF-positive specimens. Three specimens culture positive for HRSV (4%), three specimens positive for HPIV3 (37.5%), and one specimen positive for influenza A virus (25%) were RT-PCR negative. All specimens that had a detectable amplification product of the expected size from the first sample aliquot had an identical product identified from the duplicate aliquot. All specimens had a detectable β-actin product, confirming successful extraction of cellular RNA. Specimens culture positive for a respiratory virus, but RT-PCR negative, remained negative on repeat testing. With one exception, virus isolates obtained from the culture-positive, RT-PCR-negative specimens were readily amplified by the corresponding RT-PCR assay. One influenza A virus isolate could not be amplified by RT-PCR or confirmed by culture passage. All DIF-positive specimens were also RT-PCR positive. RT-PCR identified an additional 119 respiratory viruses in 116 patients, including 95 HRSV, 4 HPIV1, 2 HPIV2, 5 HPIV3, 9 influenza A virus, and 4 influenza B virus. Dual-virus infections were identified in two patients by RT-PCR that were culture negative: one patient with both influenza A virus and HRSV and one patient with influenza A virus and HPIV1. A dual-virus infection with HRSV and HPIV3 was also identified by RT-PCR in a third patient who was culture positive for only one virus (HRSV).
TABLE 2.
Comparison of GeneScan RT-PCR with culture and DIF
| Culture or DIF result | No. of specimans with RT-PCR assay resulta
| ||||||
|---|---|---|---|---|---|---|---|
| HRSV + | HPIV1 + | HPIV2 + | HPIV3 + | Flu A + | Flu B + | Negative | |
| HRSV + | |||||||
    Culture Culture | 41 | 0 | 0 | 0 | 0 | 0 | 3 |
    DIF DIF | 25 | 0 | 0 | 0 | 0 | 0 | 0 |
| HPIV1 + | |||||||
    Culture Culture | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
    DIF DIF | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HPIV2 + | |||||||
    Culture Culture | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
    DIF DIF | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HPIV3 + | |||||||
    Culture Culture | 0 | 0 | 0 | 5 | 0 | 0 | 3c |
    DIF DIF | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Flu A + | |||||||
    Culture Culture | 0 | 0 | 0 | 0 | 3 | 0 | 1f |
    DIF DIF | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Flu B + | |||||||
    Culture Culture | 0 | 0 | 0 | 0 | 0 | 10 | 0 |
    DIF DIF | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Negative | |||||||
    Culture Culture | 75b | 3c | 2 | 4d | 6be | 1 | 250 |
    DIF DIF | 20 | 1 | 0 | 1 | 3 | 3 | 12 |
DISCUSSION
The objective of this study was to develop an RT-PCR assay panel based on automated fluorescent capillary electrophoresis and GeneScan software analysis for detection of HRSV; HPIV1, -2, and -3; and influenza A and B viruses in clinical specimens. Our results suggest that this assay will achieve this goal and improve the ability of laboratories to detect these viruses in young children. This conclusion is best supported for HRSV, but the limited data obtained for the other respiratory viruses suggest that this will also apply to HPIVs and influenza viruses. Although we expected our assay to be more sensitive than virus isolation or antigen detection, we were surprised at the magnitude of the increased sensitivity. The choice of nasal and throat swabs, rather than nasopharyngeal aspirates or washes, among specimens collected in this study may account for this result. We suspect that swab specimens yield less virus—often below the threshold for detection by virus isolation (2, 22), but still above the threshold for detection by RT-PCR. In older children and adults, who often have low-titer HRSV in respiratory secretions, RT-PCR has also been markedly more sensitive than virus isolation (15, 31). Given the increased sensitivity of our assay, we were concerned that we were unable to detect viral RNA in several culture-positive specimens. Because in most cases we were able to amplify the corresponding virus isolate, these negative results were not likely due to mismatches between our primers and these particular virus strains. We suspect that our inability to detect viral RNA in these specimens may be due to specimen mislabeling or degradation of the RNA during specimen handling or shipping.
The susceptibility of RT-PCR to contamination from previously amplified DNA makes false-positive results an ongoing concern. To reduce the opportunity for amplicon contamination, we separated pre- and postassay work areas, frequently changed gloves, premeasured reagents into aliquots, and used multiple negative specimen controls in each test. In addition, our use of automated NucliSens extraction and GeneScan analysis further reduced the risk of contamination by (i) eliminating manual nucleic acid extraction, which can introduce specimen cross-contamination; and (ii) permitting sensitive amplicon detection without the risks inherent with nested PCR (9). Finally, to rule out contamination, we retested all positive specimens by reextracting RNA from a second previously unopened specimen aliquot.
Most, but not all, studies comparing RT-PCR with virus isolation and antigen detection have shown a substantial increase in sensitivity for detection of respiratory viruses. Ellis et al. (6) achieved a 20% increase in detection of influenza viruses by RT-PCR compared with virus isolation. In contrast, Magnard et al. (23) improved detection of influenza virus with RT-PCR by only 3 and 7% over virus isolation or combined isolation and antigen detection, respectively. RT-PCR assays described by Henkel et al. (15), Freymuth et al. (10), and van Elden et al. (30) showed increased frequency of detection of HRSV from 30% to 112% compared with isolation and antigen detection, whereas little improvement in RSV detection was obtained by Eugene-Ruellan et al. (7), Osiowy (25), or Paton et al. (26). RT-PCR assays described by Osiowy (25), Freymuth et al. (10), Gilbert et al. (11), and Aguilar et al. (1) increased the frequency of detection of HPIVs from 65% to 90%, whereas no improvement in detection of HPIVs was reported by Eugene-Ruellan et al. (7) or Karron et al. (19). The commercially available Hexaplex RT-PCR assay (Prodesse, Inc., Milwaukee, Wis.) has been shown to enhance detection of HRSV, HPIV1 and -3, and influenza A virus (8, 18, 20, 21), but these studies evaluated few or no specimens culture positive for HPIV2 or influenza B virus. Differences in the results obtained by those studies and our study may reflect differences in the patient groups studied, assay performance, or types of specimens tested.
RT-PCR assays permit detection of mixed virus infections that can be missed by virus isolation (7, 10, 11, 20). This capability was illustrated in our study by the detection of a dual-virus infection (HRSV and HPIV3) by RT-PCR in one patient who was culture positive for only one virus (HRSV) and dual infections in two patients (HRSV and influenza A virus and HPIV1 and influenza A virus) who were culture negative for both. However, even though not systematically tested for, other viruses were identified by culture in this study (i.e., adenovirus, cytomegalovirus, herpes simplex virus, enterovirus, and rhinovirus) that may have contributed to the patients' disease, but would not have been detected by our RT-PCR panel.
In conclusion, our GeneScan RT-PCR assay offers a highly sensitive and specific method for detection of common respiratory viruses in young children that will improve infection rate and disease burden estimates for these viruses in this population. Further assessment of our assay with specimens from other high-risk populations, including the elderly and immunocompromised, and expansion of our assay panel to include additional viral pathogens are in progress.
Acknowledgments
We thank Brian Holloway, Karen McCaustland, and Mar Mosquera for assistance in assay development; Graciela Borthagaray, Caroline Hall, Kathy Holland, Diane Kent, Ken Schnabel, Andrea Marino, and Linda Anderson for specimen acquisition, processing, and viral culture; and John O'Connor for reviewing the manuscript.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.41.9.4298-4303.2003
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc193844?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jcm.41.9.4298-4303.2003
Article citations
Genetic analysis of the 2019 coronavirus pandemic with from real-time reverse transcriptase polymerase chain reaction.
Saudi J Biol Sci, 28(1):911-916, 12 Nov 2020
Cited by: 6 articles | PMID: 33199970 | PMCID: PMC7658593
Review Free full text in Europe PMC
Advances in laboratory assays for detecting human metapneumovirus.
Ann Transl Med, 8(9):608, 01 May 2020
Cited by: 6 articles | PMID: 32566634 | PMCID: PMC7290561
Review Free full text in Europe PMC
Viral etiology of pneumonia among severely malnourished under-five children in an urban hospital, Bangladesh.
PLoS One, 15(2):e0228329, 04 Feb 2020
Cited by: 15 articles | PMID: 32017782 | PMCID: PMC6999894
Effectiveness of a Behavior Change Intervention with Hand Sanitizer Use and Respiratory Hygiene in Reducing Laboratory-Confirmed Influenza among Schoolchildren in Bangladesh: A Cluster Randomized Controlled Trial.
Am J Trop Med Hyg, 101(6):1446-1455, 01 Dec 2019
Cited by: 16 articles | PMID: 31701861 | PMCID: PMC6896855
Bacterial load and inflammatory response in sputum of alpha-1 antitrypsin deficiency patients with COPD.
Int J Chron Obstruct Pulmon Dis, 14:1879-1893, 21 Aug 2019
Cited by: 7 articles | PMID: 31686800 | PMCID: PMC6709647
Go to all (82) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Evaluation of the Hexaplex assay for detection of respiratory viruses in children.
J Clin Microbiol, 39(5):1696-1701, 01 May 2001
Cited by: 100 articles | PMID: 11325976 | PMCID: PMC88011
Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses.
J Virol Methods, 126(1-2):53-63, 01 Jun 2005
Cited by: 216 articles | PMID: 15847919 | PMCID: PMC7112904
Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza a and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4.
J Clin Microbiol, 42(4):1564-1569, 01 Apr 2004
Cited by: 292 articles | PMID: 15071005 | PMCID: PMC387552
Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay.
J Clin Microbiol, 36(11):3149-3154, 01 Nov 1998
Cited by: 88 articles | PMID: 9774555 | PMCID: PMC105291





