Abstract
Free full text

Molecular Analysis of the rfb O Antigen Gene Cluster of Salmonella enterica Serogroup O:6,14 and Development of a Serogroup-Specific PCR Assay
Abstract
The Kauffmann-White scheme for serotyping Salmonella recognizes 46 somatic (O) antigen groups, which together with detection of the flagellar (H) antigens form the basis for serotype identification. Although serotyping has become an invaluable typing method for epidemiological investigations of Salmonella, it does have some practical limitations. We have been characterizing the genes required for O and H antigen biosynthesis with the goal of developing a DNA-based system for the determination of serotype in Salmonella. The majority of the enzymes involved in O antigen biosynthesis are encoded by the rfb gene cluster. We report the sequencing of the rfb region from S. enterica serotype Sundsvall (serogroup O:6,14). The S. enterica serotype Sundsvall rfb region is 8.4 kb in length and comprises six open reading frames. When compared with other previously characterized rfb regions, the serogroup O:6,14 sequence is most related to serogroup C1. On the basis of DNA sequence similarity, we identified two genes from the mannose biosynthetic pathway, two mannosyl transferase genes, the O unit flippase gene and, possibly, the O antigen polymerase. The whole cluster is derived from a low-G+C-content organism. Comparative sequencing of an additional serogroup O:6,14 isolate (S. enterica serotype Carrau) revealed a highly homologous sequence, suggesting that O antigen factors O:24 and O:25 (additional O factors associated with serogroup O:6,14) are encoded outside the rfb gene cluster. We developed a serogroup O:6,14-specific PCR assay based on a region of the putative wzx (O antigen flippase) gene. This provides the basis for a sensitive and specific test for the rapid identification of Salmonella serogroup O:6,14.
An estimated 1.4 million cases of salmonellosis and 556 (31%) estimated food-related deaths are attributed to Salmonella each year in the United States (24). Salmonella isolates are serotyped according to the Kaufmann-White scheme by using somatic (O) and flagellar (H) antigens, with new serotypes registered in annual updates of the scheme as they are detected (29). Since the establishment of this scheme in the early 1930s (33), the public health value of this phenotypic typing method has been well proven (28). State public health laboratories in the United States began reporting the results of Salmonella serotype determination to the Centers for Disease Control and Prevention (CDC) in 1963, and this surveillance system has been critical for improving prevention. It has allowed not only the rapid detection, identification of sources, and control of outbreaks, but also identification of emerging serotypes and new mechanisms of transmission (40).
The 2,523 different serotypes currently described in the Kaufmann-White scheme (29) are derived from the considerable number of permutations of 46 O serogroups, 11 additional O antigens, and 119 H antigens (3). The O antigen is the outermost component of lipopolysaccharide (LPS), an immunogenic glycolipid that is a major component of the outer membrane in gram-negative bacteria (31, 35). A considerable amount of diversity is seen within Salmonella O antigens, which are composed of multiple repeats of an oligosaccharide unit (O unit), and they contribute major antigenic diversity to the cell surface, which is used to serotype Salmonella isolates. The basis of the variation in O antigen structure is represented by differences in sugar composition, arrangement of the sugars in the O unit, the specific linkages between the O units, and the addition of branch sugars and modifying side groups.
Much of the Salmonella O antigen variation is a consequence of the extensive genetic diversity within rfb (O antigen) gene clusters, which encode many of the enzymes involved in O antigen biosynthesis and assembly. Typically, three different types of genes are seen within rfb gene clusters: (i) genes that encode enzymes involved in the synthesis of the sugars that form the O subunit; (ii) genes that encode transferases, which assemble sugar substituents into the O subunit; and (iii) genes that encode proteins involved in processing and assembly steps to build the O antigen from the O subunit, such as the O antigen transporter (wzx) and O antigen polymerase (wzy).
Wzx proteins are involved in the transport of completed O antigen subunits across the cytoplasmic membrane (21). The mechanism of O antigen export in Salmonella enterica has been shown to be Wzx dependent, and all of the Salmonella rfb gene clusters sequenced to date have a wzx gene. The function of the O antigen polymerase is in the polymerization of the O units to form the O antigen, which is encoded by the wzy gene (formerly rfc). While most S. enterica rfb gene clusters contain a wzy homolog, serogroups B and D have a wzy gene elsewhere in the genome.
The rfb gene clusters from 10 Salmonella serogroups have been studied: A (19), B (14, 45), C1 (18), C2 (4), D1 (19), D2 (47), D3 (6), E1 (42), O:54 (16), and O:35 (44). The rfb regions from serogroups A, B, C2, D, and E, which all have a trisaccharide O subunit containing mannose, rhamnose, and galactose, are related. The rfb gene cluster from serogroup C1, whose O subunit is composed of four mannose residues, one N-acetylglucosamine residue, and a glucose side branch, shows little homology to them (18). The S. enterica O:35 rfb gene cluster encodes the same O antigen as Escherichia coli O111, and they have been shown to have closely related gene clusters (44). Synthesis of the O:54 antigen is mediated by a plasmid-encoded gene cluster (16). More recently, the genetic variations in the dDTP-l-rhamnose (rml) (30) and GDP-mannose (man) pathway genes of an additional 11 and 13 serogroups that contain rhamnose and mannose in their respective O antigen structures have been described (13).
Little is known about the rfb gene clusters from other Salmonella O serogroups. As part of a larger project to develop a DNA-based approach for serotyping Salmonella (7), we have sequenced the rfb gene cluster from two serogroup O:6,14 isolates, Salmonella enterica serotype Sundsvall (I 6,14,25:z:e,n,x) and S. enterica serotype Carrau (I 6,14,24:y:1,7), with the aim of identifying serogroup-specific DNA targets. We found that the serogroup O:6,14 rfb gene cluster is most related to serogroup C1. Comparison of the S. enterica serotypes Sundsvall and Carrau rfb sequences, which differ in the expression of O factor 24 or 25, revealed essentially no difference in the rfb region between the two serogroup O:6,14 isolates. We targeted the wzy gene in a PCR detection assay and found the sequence to be specific for serogroup O:6,14.
MATERIALS AND METHODS
Strains.
S. enterica serotype Sundsvall strain #185 and S. enterica serotype Carrau strain 384-90 were obtained from the Salmonella Reference Laboratory collection at the CDC. Serotyping was performed with somatic O and H antisera according to the Kaufmann-White scheme as previously described (2, 3). For the evaluation of the serogroup O:6,14-specific PCR assay, 443 strains were selected from the Salmonella Culture Collection in the National Salmonella Reference Laboratory at CDC. The panel included representatives of all top 100 serotypes and at least one isolate from each of the different subspecies within each of the 46 O antigen serogroups. All isolates were cultivated at 37°C overnight on blood agar base (BAB) medium under aerobic conditions.
DNA manipulation.
General DNA procedures and bacterial transformations were performed as previously described (34). The rfb region was amplified by PCR (Expand Long PCR kit; Roche, Indianapolis, Ind.) with oligonucleotide primers corresponding to the 5′ end of gnd and the middle of the JUMPstart sequence as previously described (43). The 8.4-kb amplicon was doubly digested with EcoRI and ClaI (New England Biolabs), and the restricted fragments were cloned into DH5α (Gibco BRL). Three clones of different sizes were identified.
DNA sequencing.
The ends of each of the fragments were sequenced with universal primers, and the sequences were extended via primer walking. Sequencing was performed on an PE Applied Biosystems 377 automated DNA sequencer with BigDye terminator cycle sequencing ready reaction mix according to the manufacturer's instructions (PE Applied Biosystems, Foster City, Calif.). DNA sequence data were processed by using Lasergene 99 (DNASTAR, Madison, Wis.) software and assembled into a contiguous sequence. The TMpred program (12) was used to predict transmembrane domains. This program uses an algorithm based on the statistical analysis of a database of naturally occurring transmembrane proteins to predict membrane-spanning regions and protein orientation. The National Center for Biotechnology Information (NCBI) BLAST network server was used to search sequence databases for sequences with homology to the open reading frames (ORFs) found in our sequence.
Serogroup O:6,14 PCR assay.
Bacterial genomic DNA extracted with the Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.) was resuspensed in sterile distilled water at a concentration of 50 ng/μl. A total of 46 pools were made, with 1 to 12 samples of DNA per pool. A forward primer (5′-GTCTCCGCTAAGCTATTTCGGTTTGTA-3′) and a reverse complementary sequence primer (5′-TACCGCAATAATTCAATCACAAGGG-3′) were derived from sequence within wzy to generate a 501-bp PCR amplicon. PCR amplification was performed in 25-μl volumes with Ready-to-Go PCR beads (Amersham Biosciences, Piscataway, N.J.). PCR amplification was carried out in a thermal cycler (MJ Research, Waltham, Mass.) with the following cycle parameters: initial denaturation, 96°C for 2 min; followed by 25 cycles of 94°C for 30 s (denaturation), 58°C for 30 s (annealing), and 72°C for 45 s (extension); and a final extension at 72°C for 10 min. Amplification products were analyzed with a bufferless E-Gel 96 high-throughput agarose electrophoresis system (Invitrogen, Carlsbad, Calif.) and a UV transilluminator (GelDoc1000; Bio-Rad). If a positive PCR result was seen for a particular pool, all DNA samples from the pool were individually retested by PCR, and amplification products were then confirmed by conventional agarose gel elecrophoresis with 1% (wt/vol) agarose gels.
Nucleotide sequence accession number.
The sequences of the rfb O antigen gene cluster determined in this study have been deposited in the GenBank database under accession no. AY 334017.
RESULTS
DNA sequence of the rfb region from S. enterica serotypes Sundsvall and Carrau.
Primers located in the middle of the JUMPstart sequence and the 5′ end of the gnd gene, which flank the rfb region, were used to amplify the entire rfb gene cluster from S. enterica serotypes Sundsvall and Carrau, and the nucleotide sequence was determined. Analysis of the DNA sequence revealed 8.4 kb flanked by JUMPstart and gnd and the presence of six ORFs (Fig. (Fig.11 and Table Table1).1). Alignment of the DNA sequences from S. enterica serotypes Sundsvall (O:6,14,25) and Carrau (O:6,14,24) revealed only 3 nucleotide differences between the two sequences. One of these base differences occurred within ORF 3.72, and one occurred within ORF 5.33: they were silent substitutions. The third nucleotide difference was in noncoding sequence.
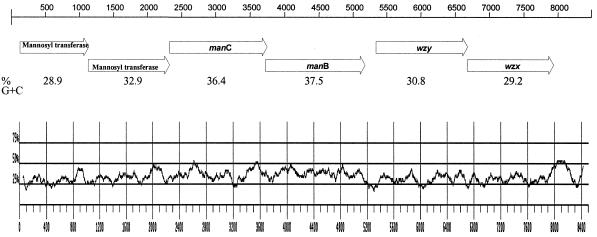
O antigen gene cluster of S. enterica serogroup O:6,14. Putative ORFs are represented by arrows, with the corresponding assignment of gene name. The percentage G+C ratios were calculated and plotted for each 100 bases.
TABLE 1.
Characteristics of each ORF, including gene identity and length, G+C content of each gene, and P values for the corrected average G+C contents for codon positions 1, 2, and 3
| ORF | Gene or gene product | Length (bp) | G+C content (%) | P value for codon position
| ||
|---|---|---|---|---|---|---|
| P1 | P2 | P3 | ||||
| 0.13 | Mannosyl transferase | 1,005 | 28.9 | 38.8 | 28.6 | 19.5 |
| 1.12 | Mannosyl transferase | 1,203 | 32.9 | 43.1 | 31.1 | 24.7 |
| 2.33 | manC | 1,416 | 36.4 | 51.2 | 34.9 | 23.3 |
| 3.72 | manB | 1,428 | 37.5 | 48.0 | 39.6 | 25.0 |
| 5.33 | wzy | 1,344 | 30.8 | 36.1 | 30.7 | 25.7 |
| 6.67 | wzx | 1,254 | 29.2 | 35.6 | 27.7 | 24.4 |
The ORFs of the DNA coding strand all have the same transcriptional direction from JUMPstart to gnd. All ORFs started with an ATG codon. Three ORFs (1.12, 2.33, and 3.72) ended with a TAA codon, two ORFs (5.33 and 6.67) ended with a TGA codon, and one ORF (0.13) ended with a TAG codon. The sites for putative translation starts and potential Shine-Dalgarno ribosomal binding sites are shown in Table Table2.2. Noncoding sequences of 1, 4, and 155 bp were found preceding the start codons of ORFs 2.33, 3.72, and 5.33, respectively. All of the other ORF junctions have overlapping termination and initiation codons. There is a 509-bp region between the end of ORF 6.67, the last gene in the serogroup O:6,14 rfb gene cluster, and the start codon of gnd. The size of the intergenic region between the last rfb gene and gnd in different S. enterica serogroups is shown in Table Table3.3. Putative gene designations were made based on knowledge of O antigen structure homology and to previously characterized genes. The similarities in the sequences of the deduced polypeptides of the encoded phosphomannomutase, guanosine diphosphomannose (GDP-man) pyrophophorylase, and mannosyl transferases are shown in Tables Tables44 and and55.
TABLE 2.
Features of the initiation regions of the ORFs of S. enterica serotype Sundsvall strain #185a
| ORF | Initiation region sequence |
|---|---|
| 0.13 | TTTAAGAGTAAATATG |
| 1.12 | GAATTGAGAAAAATTTATGAGTAGTC |
| 2.33 | ATGGAGAGTGTTAAATAAAATG |
| 3.72 | GTTGGAGAAAAATAAAATTATG |
| 5.33 | TATATTTGTTGATAAAAATG |
| 6.67 | CAGTAGAATCAAAATGATCAA |
TABLE 3.
Comparison of the size of the intergenic region between the last gene in the rfb gene cluster and the start of gnd for a number of Salmonella serogroups
| Salmonella serogroup (serotype) | Last gene in rfb cluster | Distance (bp) from last gene to start of gnd |
|---|---|---|
| B (Typhimurium) | wbaP | 163 |
| E1 (Anatum) | wbaE | 251 |
| C1 (Montevideo) | wzx | 136 |
| O:35 (Adelaide) | wbdM | 180 |
| O:6,14 (Sundsvall) | wzx | 509 |
TABLE 4.
Comparison of GDP-mannose pyrophosphorylases and phosphomannomutases from serogroups B, C1, C2, and O:6,14
| Gene | Protein size (amino acids) | % G+C content | % Amino acid identity |
|---|---|---|---|
| GDP-mannose pyrophosphorylases | |||
    manC (H) manC (H) | 472 | 36.4 | |
    manC (C1) manC (C1) | 472 | 39.0 | 58.5 |
    manC (C2) manC (C2) | 474 | 39.0 | 56.4 |
    manC (B) manC (B) | 480 | 40.3 | 57.0 |
    cpsBa (B) cpsBa (B) | 481 | 60.4 | 59.1 |
| Phosphomannomutases | |||
    manB (O:6,14) manB (O:6,14) | 476 | 37.4 | |
    manB (C1) manB (C1) | 457 | 61.3 | 14.9 |
    manB (C2) manB (C2) | 479 | 37.6 | 58.2 |
    manB (B) manB (B) | 478 | 40.9 | 57.4 |
    cpsGa (B) cpsGa (B) | 457 | 61.7 | 14.9 |
TABLE 5.
Amino acid sequence homologies between Salmonella mannosyl transferases
| Serogroup | BPGN namea | Protein size (amino acids) | % Amino acid identity to serogroup O:6,14
| |
|---|---|---|---|---|
| ORF1 | ORF2 | |||
| O:6,14 (ORF 1) | 334 | 9.6 | ||
| O:6,14 (ORF 2) | 400 | 9.6 | ||
| A/D1 | wbaU | 333 | 7.5 | 10.2 |
| B | wbaU | 353 | 14.4 | 13.6 |
| C1 | wbaB | 333 | 11.4 | 10.8 |
| C1 | wbaC | 335 | 47.3 | 8.1 |
| C1 | wbaD | 399 | 9.0 | 49.1 |
| C2 | wbaW | 33 | 13.5 | 17.0 |
| C2 | wbaZ | 385 | 8.4 | 3.6 |
| D2 | wbaO | 103 | 10.7 | 8.7 |
| E1 | wbaO | 361 | 8.1 | 10.0 |
PCR identification of serogroup O:6,14.
Primers within the coding sequence of wzx were designed. PCR was performed on each of the 46 DNA pools corresponding to the 46 O serogroups. A 501-bp amplicon was generated with only one pool; the one containing serogroup O:6,14 DNA. All other pools, containing DNA from 408 strains representing the other 45 serogroups, were negative in the PCR assay. DNA from all 12 of the strains in the O:6,14 pool were individually tested by PCR, and all generated a PCR product of the expected size (Fig. (Fig.2).2). An additional 21 O:6,14 strains were tested, and all generated the expected 501-bp PCR product. The 33 serogroup O:6,14 strains tested represented all of the five subspecies within this serogroup.
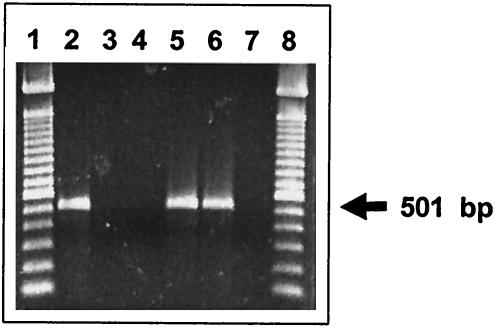
PCR amplification products with serogroup O:6,14-specific primers using template DNA from different S. enterica serogroups. Lanes 1 and 8 contain a 100-bp ladder (Invitrogen, Carlsbad, Calif.). Lanes 2, 5, and 6 represent serogroup H strains; lane 3 represents a serogroup B strain, lane 4 represents a serogroup C2 strain, and lane 7 represents the negative control.
DISCUSSION
We have sequenced the rfb gene cluster from two S. enterica serogroup O:6,14 isolates. Of the previously characterized rfb regions, the serogroup O:6,14 rfb region was most similar to the C1 rfb region. Both of these serogroups have O6 and O14 in common. On the basis of DNA sequence similarity, we have putatively identified all of the six ORFs.
Mannose biosynthetic genes.
manB, encoding phosphomannomutase, and manC, encoding mannose-1-phosphate guanylytransferase, are responsible for the biosynthesis of GDP-mannose from mannose-6-phosphate (13). Mannose is present in the O antigen side chain of serogroup O:6,14 (22), and the rfb gene cluster of this serogroup contained two genes with sufficient similarity to identify them as manB and manC, respectively. At the amino acid level, serogroup O:6,14 manC (ORF 2.33) had 58% identity to manC from serogroup C1 and 57% identity to manC from serogroup B. Serogroup O:6,14 manB (ORF 3.72) had 58 and 57% amino acid identity to the manB genes from serogroups C2 and B, respectively, with much lower homology to manB from serogroup C1 (14.9%). Both genes from the S. enterica serotype Carrau strain had 100% identity to a previously published sequence of manC and manB from S. enterica serotype Carrau (13). Two genes from the cps gene cluster, cpsB and cpsG, are considered isogenes to manC and manB (37). The serogroup O:6,14 manC and manB genes share a similar level of identity to the respective isogenes of serogroup B, as previously reported for LT2 (37).
Mannosyl transferase genes.
ORF 0.13 of serogroup O:6,14 shared 47% identity with wbaC (ORF 6.17) from S. enterica serotype Choleraesuis (serogroup C1). Both ORFs have a low G+C content; 29% for serogroup O:6,14 and 30% for serogroup C1, respectively. ORF 1.12 shared 49% identity with wbaD (ORF 7.17) in serogroup C1 (S. enterica serotype Choleraesuis), and again they have a low G+C content: 33% for serogroup O:6,14 and 31% for serogroup C1. The wbaC and wbaD genes from the rfb gene cluster of serogroup C1 are thought to encode the mannosyl transferases that assemble the serogroup C1 O antigen subunit (18). Until now, however, the sequences of these genes had shown little similarities to the rfb region of other Salmonella. Although mannosyl transferases have been identifed in rfb regions from serogroups A, B, D, C2, and E1 (19, 20), they share little homology to wbaC and wbaD (Table (Table5),5), suggesting that different specific functions are performed by these transferases from the different serogroups.
O antigen transport and polymerase genes.
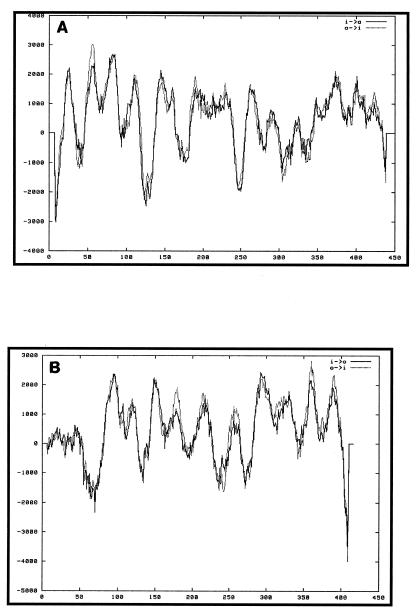
Both ORFs 5.33 and 6.67 had no significant homology to any sequences in GenBank. Proteins Wzx and Wzy from different O antigen gene clusters have little sequence homology even at the amino acid level; however, their predicted structural homology (multiple transmembrane domains) has been used to putatively identify them. Analysis with the TMpred program (12) showed that ORFs 5.33 and 6.67 contained 13 and 12 transmembrane helices, respectively (Fig. 3A and B). As is characteristic of flippases and O antigen polymerases, both of these predicted proteins are hydrophobic.
The serogroup O:6,14 Wzx was most homologous to other wzx gene products from S. enterica serogroups A, B, and C2 and E. coli (09a:k30, O111, K12), Shigella sonnei, and related organisms when a PSI-blast search (1) was performed. The predicted protein also shared some homology to the conserved motif found near the amino-terminal end of wzx gene products (38). We believe that ORF 6.67 is the wzx gene, and as in Salmonella serogroup C1, this gene is at the distal end of the rfb gene cluster.
O antigen polymerases share a characteristic secondary structure of multiple transmembrane regions and a large cytoplasmic loop. Serogroup H wzy encoded a protein predicted to have a similar secondary structure. However, there was no sequence homology between this ORF and any of the other known O antigen polymerases. When the predicted protein of ORF 5.33 was used to then search databases with PROPSEARCH (11), a program designed to find putative protein families for protein sequences with little sequence identity, 87% reliability was found, linking ORF 5.33 to the O antigen polymerases of S. enterica serogroup C2, E. coli K-12, and Shigella flexneri. Other features common to Wzy-like polymerases include similar amino acid compositions and codon usage (25). The predicted protein from ORF 5.33 had a high content of leucine, isoleucine, and phenylalanine and contained a high percentage of rare or modulating codons, 12.9% (10), as seen in other wzy genes (25). Thus, wzy has many features in common with other wzy genes, but definitive identification requires further experimental validation.
Other features of the rfb region.
The intergenic region between the last rfb gene and gnd was much larger in serogroup O:6,14 than in the other Salmonella serogroups (Table (Table3).3). A 103-base sequence starting 3 bp downstream from the end of the wzx gene shared 89 and 82% homology to the IS3-like insertion element IS1230B of E. coli (32) and S. enterica serotype Enteritidis (5), suggesting that it is the remnant of an IS3-like element. Sequences with homology to IS3 have been previously reported in Salmonella, located near the invH gene of S. enterica serotype Choleraesuis (9) and the sef operon of S. enterica serotype Enteritidis (5). The occurrence of an IS5 element located about 500 bp upstream from the gnd promoter in E. coli K-12 has been shown (15) and was thought to play a role in gene expression. Analysis of the promoter region of the gnd revealed that the −10 region of the gnd gene (AGGAG) is identical to the corresponding region of gnd in S. enterica serotype Typhimurium and E. coli K-12, and the 30 bp upstream of the gnd share between a 96 and 100% homology to the corresponding region in S. enterica serotypes Typhimurium and Choleraesuis and E. coli K-12 and O111.
G+C content.
Although bacterial species display large variation in their overall G+C content, the genes within a particular bacterial species' genome are usually relatively similar in base composition (27). However, S. enterica rfb gene clusters that have been sequenced to date all contained segments of different G+C content, which are thought to have evolved in organisms of largely low G+C content that have been independently acquired and incorporated into the same Salmonella locus (31). Consistent with this, Shepard et al. (36) reported interspecies transfer of an entire O antigen gene cluster via a plasmid. All of the genes within the serogroup O:6,14 gene cluster had a low G+C content ranging from 28.9 to 37.4% (Table (Table1),1), which is significantly lower than the 51% GC content seen in the majority of S. enterica coding sequences, suggesting that all of the genes in the serogroup O:6,14 rfb gene cluster were also acquired by transfer from a different species.
Within a bacterial species, codon positions 1, 2, and 3 have a characteristic base composition, with differences due to biases in the mutation rates for the 4 bases (26, 39). Consequently, each species has a characteristic G+C content for each codon position and specific codon preferences. A positive, linear relationship exists between the genomic base composition and the G+C content for each codon position (26), with the effect greatest at position 3, where most changes are synonymous and not under strong selective constraints. Therefore, if base composition and codon usage patterns are primarily caused by mutational biases, horizontally acquired DNA will often have unusual sequence characteristics that distinguish it from ancestral DNA. At the time of introgression, newly acquired DNA will reflect the base composition of the donor genome. However, the introgressed genes will be subject to the same mutational pressures as the recipient genome, and so over time, these sequences will change or “ameliorate” to reflect the base composition of the new genome (17). This is most evident at sites with few functional constraints, such as codon position 3. The P values for the corrected average G+C contents for codon positions 1, 2, and 3 (P1, -2, and -3) (39) are shown in Table Table1.1. These properties are similar to those of the other Salmonella serogroups that have been sequenced, with the P3 value being the lowest. Given that the average G+C content at P3 is 58% for E. coli and S. enterica serotype Typhimurium, the observed P3 value of 23.8% for the serogroup O:6,14 rfb gene cluster suggests that this region is still in the process of amelioration.
Models of amelioration can been used to estimate the time of introgression of foreign genes into a chromosome. The G+C content of the third codon positions can each be back-ameliorated until the sequences conform to the Muto and Osawa relationships (25), providing estimates of both time of introgression and nucleotide composition of the donor genome (15). Unfortunately, however, rfb genes are not good candidates for amelioration because they have seen too many mutational contexts. Moreover, there is likely too little information to use: the amelioration algorithm becomes reliable only when the DNA sequence under scrutiny exceeds 15 to 20 kb; below that, the estimates get very large variances. Wang and Reeves (44) recently applied this algorithm to back-ameliorate the E. coli O111 and S. enterica O35 rfb gene clusters, which they showed to be highly conserved. They found the program did not give meaningful data and suggested the reason was because the genes of interest deviated too much from the Muto and Osawa models or incorrect parameters were used in the amelioration program.
Comparison of S. enterica serotypes Sundsvall and Carrau.
Only minor sequence differences were identified for the rfb region of S. enterica serotypes Sundsvall (O:6,14,25) and Carrau (O:6,14,24), indicating that O antigen factors 24 and 25 are encoded outside the rfb gene cluster, possibly by phage or plasmid. A number of Salmonella O antigens have been reported to be bacteriophage encoded, such as factors O1 (41) and O34 (46). A plasmid-encoded rfb gene cluster has also been reported necessary for biosynthesis of the O54 antigen in S. enterica serotype Borreze (16).
PCR for identification of serogroup O:6,14 isolates.
A PCR based on wzx was developed for the rapid identification of Salmonella serogroup O:6,14. Several serotypes of this serogroup were among the top 20 emerging serotypes in a study investigating the trend in Salmonella serotypes isolated from humans in the United States from 1987 to 1997 (28). Together with the serogroup PCR assays that have been previously described (23), we intend to identify additional rfb targets specific for other Salmonella serogroups (8). The overall goal is to combine this serogroup identification system with DNA targets specific for each H antigen (J. R. McQuiston, M. Ortiz-Rivera, L. Gheesling, F. Brenner, and P. I. Fields, submitted for publication) to establish a comprehensive DNA-based scheme for identification of the major Salmonella serotypes, without the need for serological testing (7). This will allow a rapid and convenient alternative for identification of Salmonella serotypes attainable by nonspecialized laboratories.
REFERENCES
 34. J. Bacteriol. 105:927-936. [Europe PMC free article] [Abstract] [Google Scholar]
34. J. Bacteriol. 105:927-936. [Europe PMC free article] [Abstract] [Google Scholar]Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.69.10.6099-6105.2003
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc201254?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Engineering a Novel Bivalent Oral Vaccine against Enteric Fever.
Int J Mol Sci, 22(6):3287, 23 Mar 2021
Cited by: 3 articles | PMID: 33807097 | PMCID: PMC8005139
Multiplex ligation reaction based on probe melting curve analysis: a pragmatic approach for the identification of 30 common Salmonella serovars.
Ann Clin Microbiol Antimicrob, 18(1):39, 05 Dec 2019
Cited by: 3 articles | PMID: 31805936 | PMCID: PMC6894471
Prevalence and molecular characterization of Salmonella enterica serovar Typhimurium from ice and beverages in Jakarta, Indonesia.
BMC Res Notes, 12(1):45, 21 Jan 2019
Cited by: 2 articles | PMID: 30665448 | PMCID: PMC6341521
Nanoparticle Enhanced Antibody and DNA Biosensors for Sensitive Detection of Salmonella.
Materials (Basel), 11(9):E1541, 27 Aug 2018
Cited by: 15 articles | PMID: 30150524 | PMCID: PMC6163637
In vitro attenuation of a virulent swine isolate of Brachyspira hampsonii.
Pathog Dis, 76(1), 01 Feb 2018
Cited by: 0 articles | PMID: 29069340
Go to all (39) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sequence analysis of the rfb loci, encoding proteins involved in the biosynthesis of the Salmonella enterica O17 and O18 antigens: serogroup-specific identification by PCR.
Appl Environ Microbiol, 72(12):7949-7953, 20 Oct 2006
Cited by: 15 articles | PMID: 17056694 | PMCID: PMC1694211
Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups.
J Clin Microbiol, 45(10):3323-3334, 18 Jul 2007
Cited by: 88 articles | PMID: 17634307 | PMCID: PMC2045348
Escherichia coli O123 O antigen genes and polysaccharide structure are conserved in some Salmonella enterica serogroups.
J Med Microbiol, 58(pt 7):884-894, 05 Jun 2009
Cited by: 6 articles | PMID: 19502376
A complete view of the Escherichia coli O-antigen biosynthesis gene cluster and the development of molecular-based O-serogrouping methods.
Nihon Saikingaku Zasshi, 71(4):209-215, 01 Jan 2016
Cited by: 3 articles | PMID: 27980292
Review