Abstract
Free full text

Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli.
Abstract
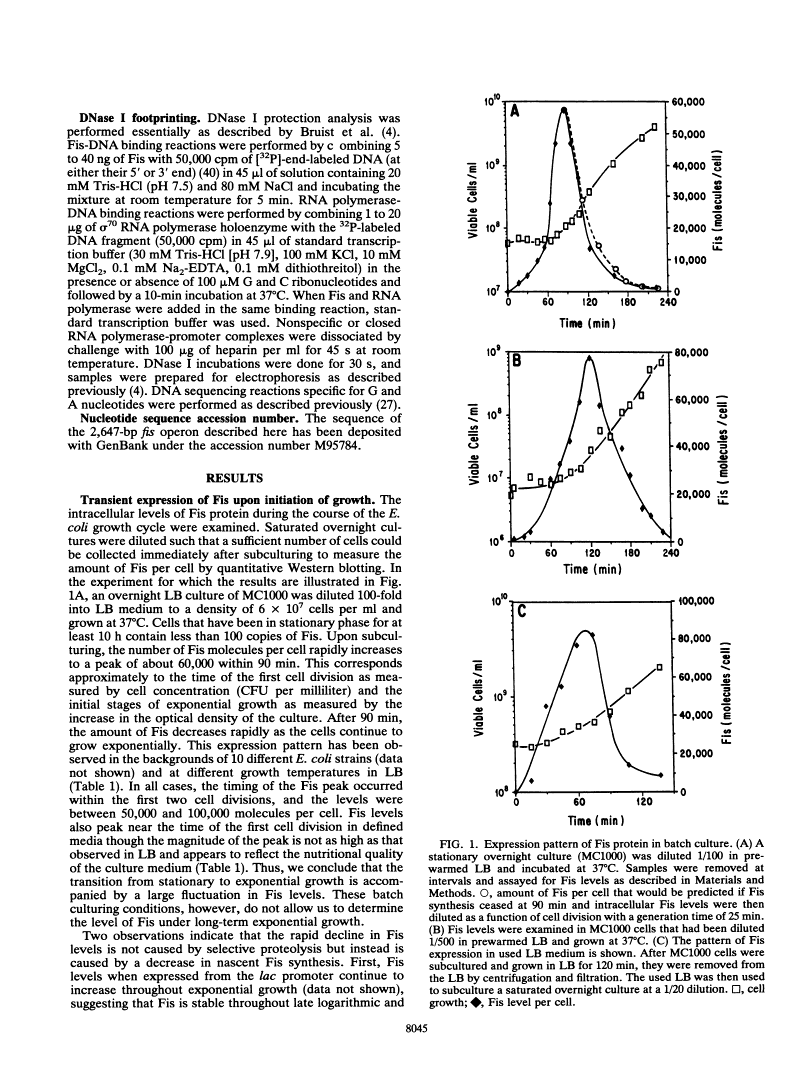
Fis is a small basic DNA-binding protein from Escherichia coli that was identified because of its role in site-specific DNA recombination reactions. Recent evidence indicates that Fis also participates in essential cell processes such as rRNA and tRNA transcription and chromosomal DNA replication. In this report, we show that Fis levels vary dramatically during the course of cell growth and in response to changing environmental conditions. When stationary-phase cells are subcultured into a rich medium, Fis levels increase from less than 100 to over 50,000 copies per cell prior to the first cell division. As cells enter exponential growth, nascent synthesis is largely shut off, and intracellular Fis levels decrease as a function of cell division. Fis synthesis also transiently increases when exponentially growing cells are shifted to a richer medium. The magnitude of the peak of Fis synthesis appears to reflect the extent of the nutritional upshift. fis mRNA levels closely resemble the protein expression pattern, suggesting that regulation occurs largely at the transcriptional level. Two RNA polymerase-binding sites and at least six high-affinity Fis-binding sites are present in the fis promoter region. We show that expression of the fis operon is negatively regulated by Fis in vivo and that purified Fis can prevent stable complex formation by RNA polymerase at the fis promoter in vitro. However, autoregulation only partially accounts for the expression pattern of Fis. We suggest that the fluctuations in Fis levels may serve as an early signal of a nutritional upshift and may be important in the physiological roles Fis plays in the cell.
Full text
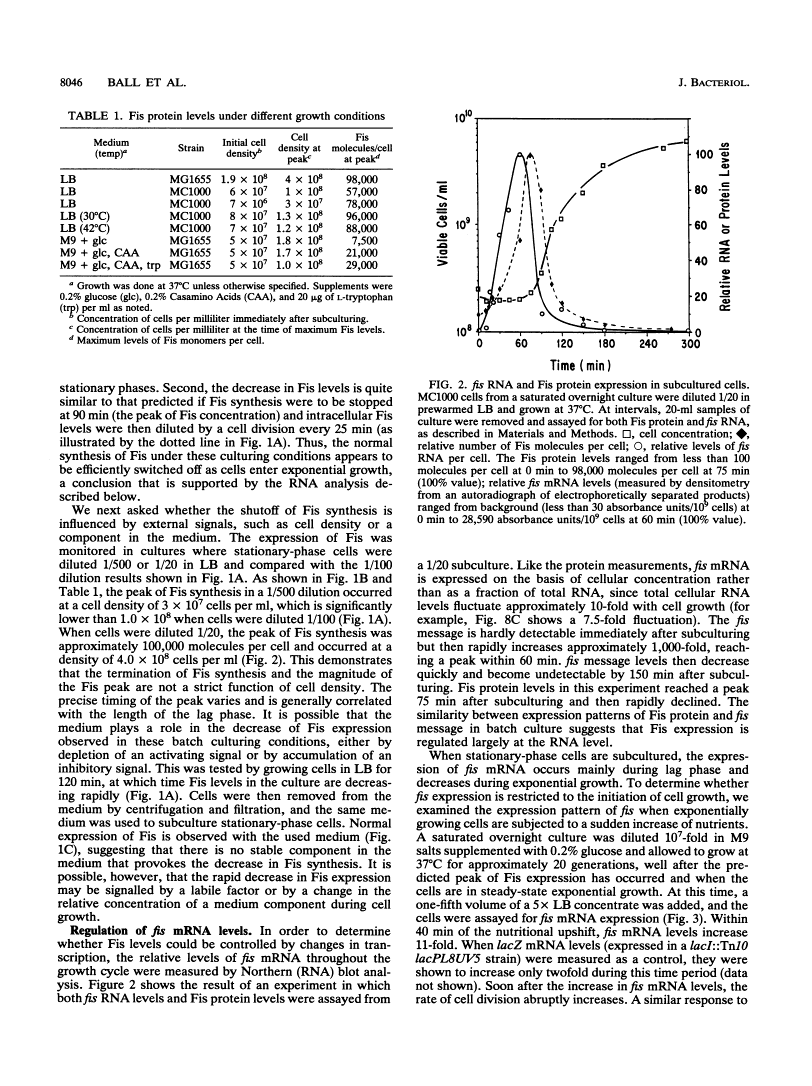
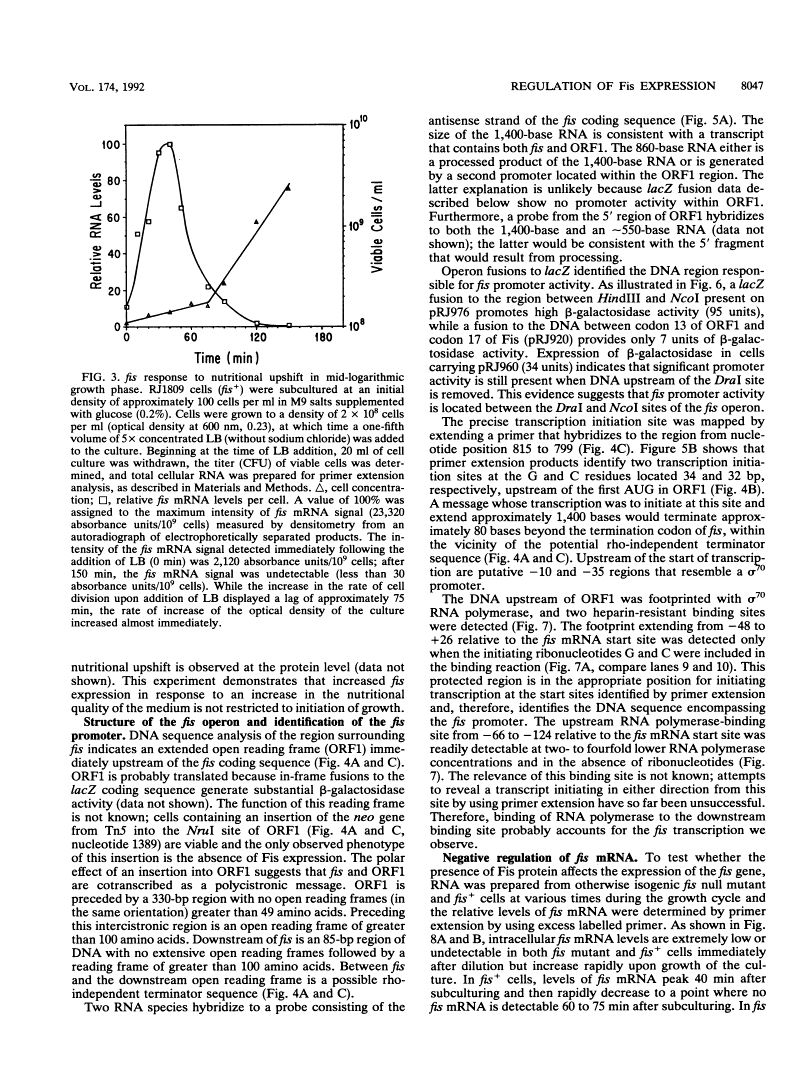
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (3.2M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
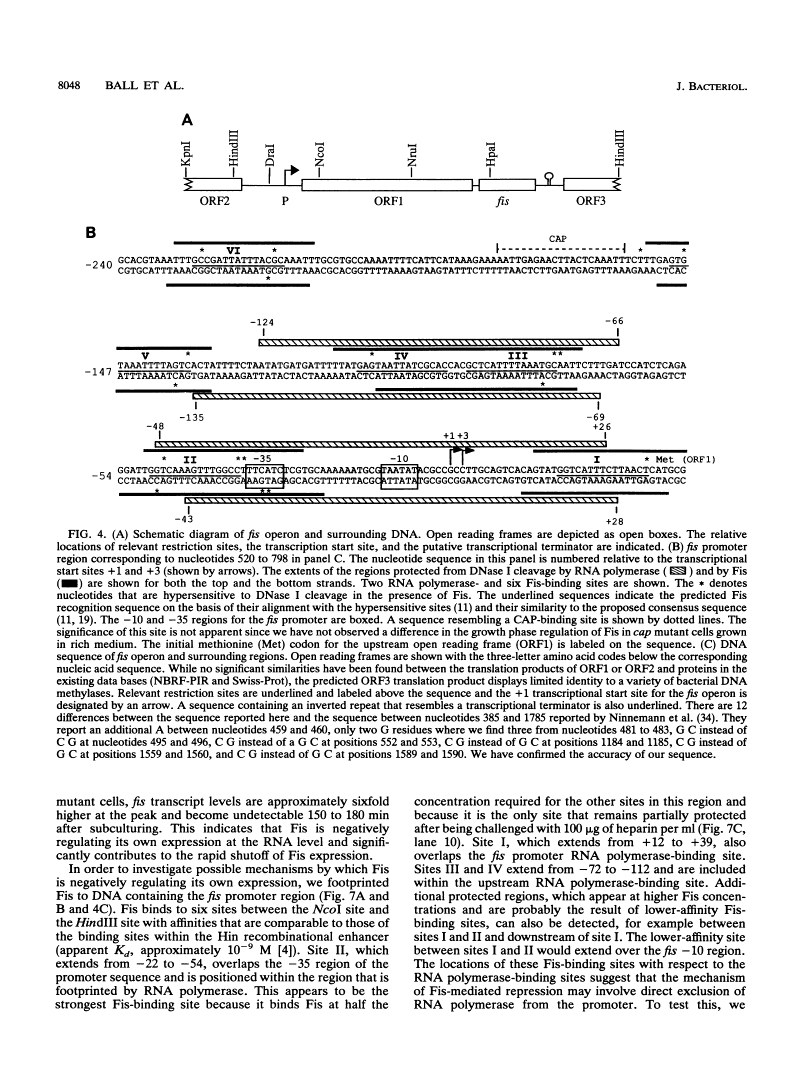
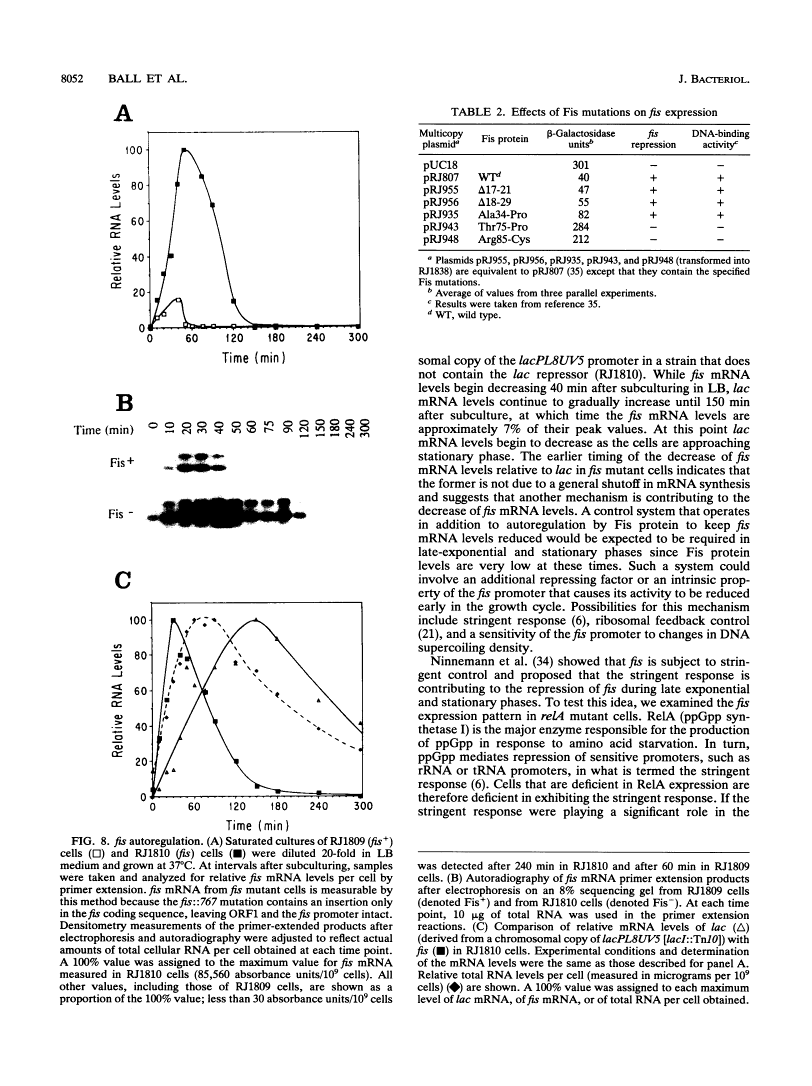
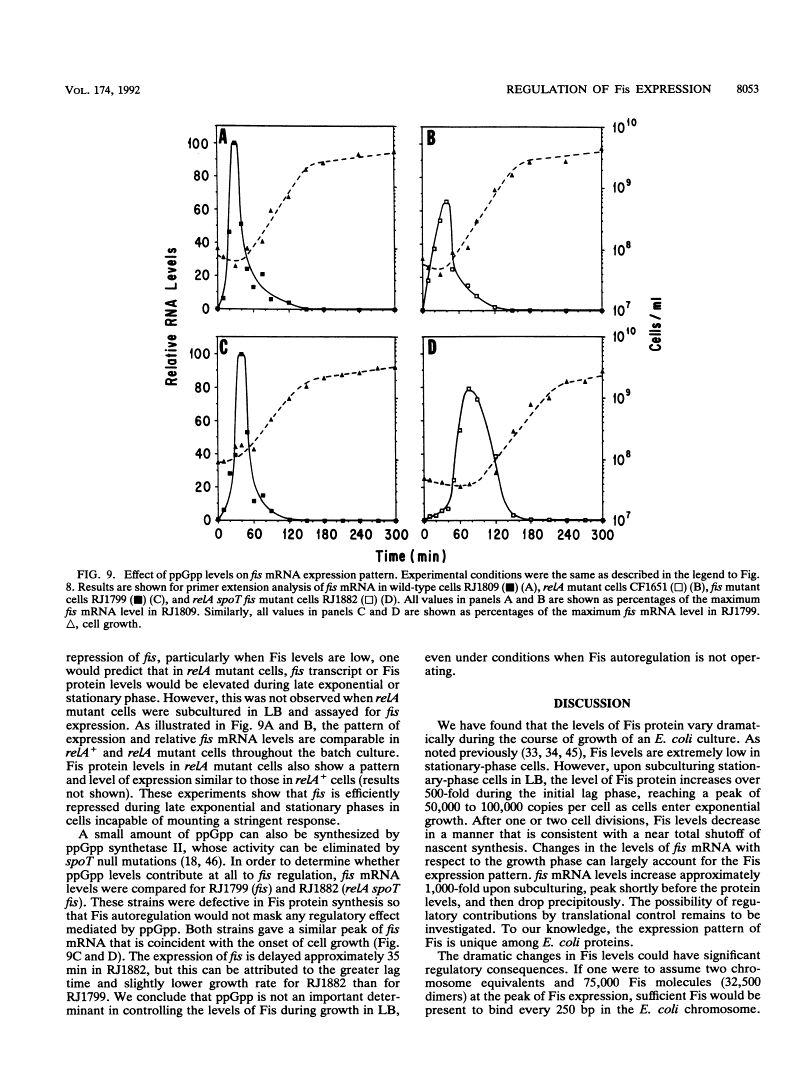
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball CA, Johnson RC. Efficient excision of phage lambda from the Escherichia coli chromosome requires the Fis protein. J Bacteriol. 1991 Jul;173(13):4027–4031. [Europe PMC free article] [Abstract] [Google Scholar]
- Ball CA, Johnson RC. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J Bacteriol. 1991 Jul;173(13):4032–4038. [Europe PMC free article] [Abstract] [Google Scholar]
- Bruist MF, Glasgow AC, Johnson RC, Simon MI. Fis binding to the recombinational enhancer of the Hin DNA inversion system. Genes Dev. 1987 Oct;1(8):762–772. [Abstract] [Google Scholar]
- Case CC, Roels SM, González JE, Simons EL, Simons RW. Analysis of the promoters and transcripts involved in IS10 anti-sense RNA control. Gene. 1988 Dec 10;72(1-2):219–236. [Abstract] [Google Scholar]
- Cole JR, Olsson CL, Hershey JW, Grunberg-Manago M, Nomura M. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J Mol Biol. 1987 Dec 5;198(3):383–392. [Abstract] [Google Scholar]
- Drlica K, Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. [Europe PMC free article] [Abstract] [Google Scholar]
- Filutowicz M, Ross W, Wild J, Gourse RL. Involvement of Fis protein in replication of the Escherichia coli chromosome. J Bacteriol. 1992 Jan;174(2):398–407. [Europe PMC free article] [Abstract] [Google Scholar]
- Finkel SE, Johnson RC. The Fis protein: it's not just for DNA inversion anymore. Mol Microbiol. 1992 Nov;6(22):3257–3265. [Abstract] [Google Scholar]
- Gille H, Egan JB, Roth A, Messer W. The FIS protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 1991 Aug 11;19(15):4167–4172. [Europe PMC free article] [Abstract] [Google Scholar]
- Haffter P, Bickle TA. Purification and DNA-binding properties of FIS and Cin, two proteins required for the bacteriophage P1 site-specific recombination system, cin. J Mol Biol. 1987 Dec 20;198(4):579–587. [Abstract] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. [Abstract] [Google Scholar]
- Hernandez VJ, Bremer H. Escherichia coli ppGpp synthetase II activity requires spoT. J Biol Chem. 1991 Mar 25;266(9):5991–5999. [Abstract] [Google Scholar]
- Hübner P, Arber W. Mutational analysis of a prokaryotic recombinational enhancer element with two functions. EMBO J. 1989 Feb;8(2):577–585. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson RC, Ball CA, Pfeffer D, Simon MI. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci U S A. 1988 May;85(10):3484–3488. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson RC, Bruist MF, Simon MI. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986 Aug 15;46(4):531–539. [Abstract] [Google Scholar]
- Koch C, Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986 Nov 25;261(33):15673–15678. [Abstract] [Google Scholar]
- Koch C, Ninnemann O, Fuss H, Kahmann R. The N-terminal part of the E.coli DNA binding protein FIS is essential for stimulating site-specific DNA inversion but is not required for specific DNA binding. Nucleic Acids Res. 1991 Nov 11;19(21):5915–5922. [Europe PMC free article] [Abstract] [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. [Europe PMC free article] [Abstract] [Google Scholar]
- Metzger S, Schreiber G, Aizenman E, Cashel M, Glaser G. Characterization of the relA1 mutation and a comparison of relA1 with new relA null alleles in Escherichia coli. J Biol Chem. 1989 Dec 15;264(35):21146–21152. [Abstract] [Google Scholar]
- Mulligan ME, Hawley DK, Entriken R, McClure WR. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. [Europe PMC free article] [Abstract] [Google Scholar]
- Nilsson L, Vanet A, Vijgenboom E, Bosch L. The role of FIS in trans activation of stable RNA operons of E. coli. EMBO J. 1990 Mar;9(3):727–734. [Europe PMC free article] [Abstract] [Google Scholar]
- Nilsson L, Verbeek H, Vijgenboom E, van Drunen C, Vanet A, Bosch L. FIS-dependent trans activation of stable RNA operons of Escherichia coli under various growth conditions. J Bacteriol. 1992 Feb;174(3):921–929. [Europe PMC free article] [Abstract] [Google Scholar]
- Ninnemann O, Koch C, Kahmann R. The E.coli fis promoter is subject to stringent control and autoregulation. EMBO J. 1992 Mar;11(3):1075–1083. [Europe PMC free article] [Abstract] [Google Scholar]
- Osuna R, Finkel SE, Johnson RC. Identification of two functional regions in Fis: the N-terminus is required to promote Hin-mediated DNA inversion but not lambda excision. EMBO J. 1991 Jun;10(6):1593–1603. [Europe PMC free article] [Abstract] [Google Scholar]
- Prentki P, Krisch HM. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. [Abstract] [Google Scholar]
- Ross W, Thompson JF, Newlands JT, Gourse RL. E.coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990 Nov;9(11):3733–3742. [Europe PMC free article] [Abstract] [Google Scholar]
- Schmid MB. More than just "histone-like" proteins. Cell. 1990 Nov 2;63(3):451–453. [Abstract] [Google Scholar]
- Siegele DA, Kolter R. Life after log. J Bacteriol. 1992 Jan;174(2):345–348. [Europe PMC free article] [Abstract] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. [Abstract] [Google Scholar]
- Thompson JF, Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. [Europe PMC free article] [Abstract] [Google Scholar]
- Thompson JF, Moitoso de Vargas L, Koch C, Kahmann R, Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. [Abstract] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991 Mar 25;266(9):5980–5990. [Abstract] [Google Scholar]
- Yuan HS, Finkel SE, Feng JA, Kaczor-Grzeskowiak M, Johnson RC, Dickerson RE. The molecular structure of wild-type and a mutant Fis protein: relationship between mutational changes and recombinational enhancer function or DNA binding. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9558–9562. [Europe PMC free article] [Abstract] [Google Scholar]
Associated Data
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.174.24.8043-8056.1992
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/jb/174/24/8043.full.pdf
Free to read at jb.asm.org
http://jb.asm.org/cgi/content/abstract/174/24/8043
Free after 4 months at jb.asm.org
http://jb.asm.org/cgi/reprint/174/24/8043
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Bacteriophage lambda site-specific recombination.
Mol Microbiol, 121(5):895-911, 19 Feb 2024
Cited by: 0 articles | PMID: 38372210
Review
IHF and Fis as Escherichia coli Cell Cycle Regulators: Activation of the Replication Origin oriC and the Regulatory Cycle of the DnaA Initiator.
Int J Mol Sci, 24(14):11572, 18 Jul 2023
Cited by: 3 articles | PMID: 37511331 | PMCID: PMC10380432
Review Free full text in Europe PMC
The morphology and metabolic changes of Actinobacillus pleuropneumoniae during its growth as a biofilm.
Vet Res, 54(1):42, 26 May 2023
Cited by: 3 articles | PMID: 37237397 | PMCID: PMC10224306
Inactivation of ackA and pta Genes Reduces GlpT Expression and Susceptibility to Fosfomycin in Escherichia coli.
Microbiol Spectr, 11(3):e0506922, 18 May 2023
Cited by: 2 articles | PMID: 37199605 | PMCID: PMC10269713
The DNA relaxation-dependent OFF-to-ON biasing of the type 1 fimbrial genetic switch requires the Fis nucleoid-associated protein.
Microbiology (Reading), 169(1), 01 Jan 2023
Cited by: 2 articles | PMID: 36748578 | PMCID: PMC9993118
Go to all (247) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Deletion analysis of the fis promoter region in Escherichia coli: antagonistic effects of integration host factor and Fis.
J Bacteriol, 179(20):6367-6377, 01 Oct 1997
Cited by: 34 articles | PMID: 9335285 | PMCID: PMC179552
Sequence, regulation, and functions of fis in Salmonella typhimurium.
J Bacteriol, 177(8):2021-2032, 01 Apr 1995
Cited by: 69 articles | PMID: 7536730 | PMCID: PMC176845
Fis activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli.
J Bacteriol, 177(18):5222-5231, 01 Sep 1995
Cited by: 59 articles | PMID: 7545153 | PMCID: PMC177312
The Fis protein: it's not just for DNA inversion anymore.
Mol Microbiol, 6(22):3257-3265, 01 Nov 1992
Cited by: 206 articles | PMID: 1484481
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (3)
Grant ID: GM-07104
Grant ID: GM13657
Grant ID: GM38509