Abstract
Free full text

Twist2, a novel ADD1/SREBP1c interacting protein, represses the transcriptional activity of ADD1/SREBP1c
Abstract
Adipocyte determination and differentiation dependent factor 1 (ADD1)/sterol regulatory element binding protein isoform (SREBP1c) is a key transcription factor in fatty acid metabolism and insulin- dependent gene expression. Although its transcriptional and post-translational regulation has been extensively studied, its regulation by interacting proteins is not well understood. To identify cellular proteins that associate with ADD1/SREBP1c, we employed the yeast two-hybrid system with an adipocyte cDNA library. Using the N-terminal domain of ADD1/SREBP1c as bait, we identified Twist2 (also known as Dermo-1), a basic helix–loop–helix (bHLH) protein, as a novel ADD1/SREBP1c interacting protein. Over-expression of Twist2 strongly repressed the transcriptional activity of ADD1/SREBP1c, primarily by reducing its binding to target sequences. Inhibition of histone deacetylase (HDAC) activity with HDAC inhibitors relieved this repression. Our data suggest that physical interaction between Twist2 and ADD1/SREBP1c attenuates transcriptional activation by ADD1/SREBP1c by inhibiting its binding to DNA, and that this inhibition is at least partly dependent on chromatin modification by HDACs.
INTRODUCTION
Sterol regulatory element binding proteins (SREBPs) play a critical role in lipid homeostasis by regulating genes involved in cholesterol and fatty acid metabolism. Three isoforms of SREBPs have been identified, SREBP1a, SREBP1c (also known as adipocyte determination and differentiation dependent factor 1 [ADD1]) and SREBP2, constituting a family of basic helix–loop–helix (bHLH) transcription factors (1–3). SREBP1a and ADD1/SREBP1c are generated by alternative promoter usage and alternative splicing from a single gene, while SREBP2 is encoded by a distinct gene with 47% identity with SREBP1 (3). Unlike other transcription factors, SREBPs are inserted into the endoplasmic reticulum (ER) membrane in a transcriptionally inactive form (4,5). When cellular cholesterol concentrations are low, SREBP cleavage-activating protein, together with Insig-1 or Insig-2, escorts SREBPs from the ER to the Golgi, where they are cleaved in sequence by Site 1 and Site 2 proteases. The mature forms of SREBPs, with N-terminal fragments, are then released from the Golgi and translocate to the nucleus where they bind to the promoters or enhancers of SREBP target genes (3,5–9).
Of the three SREBPs, it is ADD1/SREBP1c that mainly activates genes responsible for fatty acid synthesis, including fatty acid synthase (FAS), lipoprotein lipase (LPL), acetyl Co A carboxylase (ACC) and steroyl Co A desaturase (SCD) in fat and liver (3). ADD1/SREBP1c is predominantly expressed in white adipose tissue, brown adipose tissue and liver, and its mRNA is induced at an early stage of adipocyte differentiation (2,10). Ectopic expression of ADD1/SREBP1c in preadipocyte cell lines promotes adipocyte differentiation (10) and stimulates the activity of peroxisome proliferator activated receptor (PPAR) γ, a master transcription factor for adipogenesis, by producing activators (or endogenous ligands) of PPARγ (11). In addition, ADD1/SREBP1c expression is modulated by nutritional status and regulated in an insulin-sensitive manner in which it regulates transcription of insulin-dependent lipogenic genes (12). It is therefore likely that ADD1/SREBP1c is a major transcription factor coordinating fatty acid and glucose metabolism in insulin-sensitive organs. On the contrary, several studies have suggested that ADD1/SREBP1c is dispensable for the development of adipose tissue in vivo (13–15). For example, aP2-ADD1/SREBP1c transgenic animals show lipodystrophy rather than increased fat mass (13). However, ADD1/SREBP1c-specific knockout mice have decreased fat mass and severely decreased hepatic and plasma triglycerides, suggesting that ADD1/SREBP1c is critical for lipogenesis (14,15).
Compared with other SREBPs, ADD1/SREBP1c is a relatively weak transcription activator. This can be explained by the fact that it has few acidic amino acids in its N-terminal region compared with SREBP1a (16). SREBP1a and SREBP2 associate through their N-terminal domains with various transcriptional co-activators including p300, CBP and ARC/DRIP, but no interaction between ADD1/SREBP1c and co-activators has been demonstrated (17,18). In addition, it has been suggested that the transcriptional activity of SREBP1 isoforms is stimulated by interaction with other co-regulatory factors such as Sp1 and NF-Y (19–21).
Although the transcriptional and post-translational regulation of ADD1/SREBP1c has been investigated intensively, little is known of proteins that interact with it and regulate its transcriptional activity. To address this issue, we used the yeast two-hybrid system. Here we demonstrate that Twist2, a bHLH protein, is a novel ADD1/SREBP1c interacting protein. Twist2 (also called Dermo-1) is expressed in fat and liver where ADD1/SREBP1c is also highly expressed. Over-expression of Twist2 specifically repressed the transcriptional activity of ADD1/SREBP1c by inhibiting its binding to target promoters. Furthermore, histone deacetylase (HDAC)- inhibiting drugs such as sodium butyrate abolished Twist2-dependent inhibition, implying that Twist2 interferes with ADD1/SREBP1c in cooperation with HDACs. Together, our observations suggest that Twist2 antagonizes the transcriptional activity of ADD1/SREBP1c, and that together Twist2 and ADD1/SREBP1c may regulate energy homeostasis.
MATERIALS AND METHODS
Yeast two-hybrid screening
As bait, the N-terminal region of ADD1/SREBP1c containing the acidic domain (amino acids 1–153) was cloned in-frame with the Gal4 DNA-binding domain into vector pAS2 (Clonetech). The yeast strain, PJ69-4A, was simultaneously transformed with the bait plasmid pAS2-Gal4-A153 and a murine HIB-IB cDNA library, cloned into vector pACT2 according to the manufacturer’s instructions (Clonetech). Positive interactions were tested for lacZ activity and survival on selective media, and DNA was extracted from positive clones and sequenced with pACT2-specific oligonucleotides. DNA and protein databases were analyzed with the BLAST package of search algorithms (http://ncbi.nlm.nih.gov/cgi.bin/BLAST).
GST pull-down
35S-labeled active forms of ADD1/SREBP-1c and Twist2 were generated with the TNT®-coupled reticulocyte lysate system (Promega) under conditions suggested by the manufacturer. Each of the in vitro translated proteins (5 µl) was mixed with 1 µg of recombinant GST–ADD1/SREBP1c (amino acids 1–403) fusion protein and glutathione–sepharose beads. Reaction volumes were made up to 150 µl with binding buffer [10 mM Tris–HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.1% Nonidet P-40 (NP-40) and 1% aprotinin] and incubated for 2 h at 4°C. As negative control, GST protein was also added to a mixture with in vitro translated 35S-labeled ADD1/SREBP-1c or 35S-labeled Twist2. The beads were pelleted, washed with binding buffer and mixed with SDS-gel sample buffer (125 mM Tris–HCl pH 6.8, 50% glycerol, 2% SDS, 5% β-mercaptoethanol and 0.01% Bromophenol Blue). Boiled samples were subjected to SDS–PAGE and autoradiography.
Plasmid constructs
The FAS-luciferase plasmid containing a –220 to +25 bp fragment of the FAS promoter in front of a luciferase reporter gene was described previously (12). The ADD1/SREBP1c-luciferase plasmid contains a –2.7 kb to +1 bp fragment of the ADD1/SREBP1c promoter in front of a luciferase reporter gene. Plasmid pRSV-β-gal was used as an internal control. The active form of the ADD1/SREBP1c expression vector encoding amino acids 1–403 of rat ADD1, was cloned in-frame into the pcDNA3.1-Myc/HisA plasmid and pGEX4T-1 vector (Pharmacia). A Twist2 full-length cDNA was cloned into pRc-CMV vector (Invitrogen). Expression vectors for HDACs were gifts from Dr Kyu W. Kim. Mature forms of SREBP1a and SREBP2 cDNAs were cloned into pCMV plasmid (22).
Cell culture
Murine 3T3-L1 preadipocytes were grown to confluence in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. At confluence (day 0), differentiation of 3T3-L1 cells was induced with DMEM containing 10% FBS, methyl-isobutylxanthine (500 µM), dexamethasone (1 µM) and insulin (5 µg/ml) for 48 h. The culture medium was changed every other day with DMEM containing 10% FBS and 1 µg/ml insulin. The 3T3-F442A cell line was grown in DMEM and 10% fetal calf serum. Differentiation of the 3T3-F442A cells to adipocytes was induced by addition of the same medium with 5 µg/ml insulin, and the medium was changed every other day.
Transient transfections and luciferase assays
H293 cells were transfected 1 day before confluence by the calcium phosphate method described previously (12,23). After incubation for 24 h, cell extracts were prepared with lysis buffer (25 mM Tris–phosphate pH 7.8, 2 mM dithiothreitol, 2 mM 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100), and activities of β-galactosidase and luciferase were determined according to the manufacturer’s instructions (Promega). Luciferase activities were normalized to the corresponding β-galactosidase activities and/or protein concentration of each lysate. All the transfection experiments were performed in duplicate and repeated independently at least three times.
Northern blot analysis
Total RNA was isolated with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Twenty micrograms of each RNA was denatured in formamide and formaldehyde, and separated by electrophoresis on formaldehyde-containing agarose gels. It was transferred to nylon membranes (Schleicher and Schuell, Germany), which were cross-linked, hybridized and washed as described by the manufacturer. DNA probes were labeled by random priming using the Klenow fragment of DNA polymerase I (Promega) and [α-32P]dCTP (Amersham-Pharmacia). The cDNAs used as probes were: ADD1/SREBP1c, Twist2, PPARγ, β-actin and aP2.
RT–PCR
The cDNA was synthesized using Moloney leukemia virus reverse transcriptase with dNTPs and oligo(dT) primers (Stratagene). The cDNAs provided templates for PCRs using specific primers at annealing temperatures ranging between 54 and 58°C in the presence of dNTPs and Taq DNA polymerase. PCR synthesis for each primer pair was quantified at 20, 25 and 30 cycles in a test reaction to ensure that the quantitative PCR amplification was in the linear range. The primer sequences used for PCR are available upon request.
Electrophoretic mobility shift assay (EMSA)
One microgram of plasmid DNA expressing active ADD1/SREBP1c or Twist2 was used as template for in vitro translation. In order to measure translation efficiency from the different plasmid templates, l-[35S]methionine was included in separate reactions and radio-labeled proteins were analyzed by SDS–PAGE. The DNA sequences of the double-stranded oligonucleotides used as probes were as follows (only one strand is shown): ABS (ADD1/SREBP1c binding site), 5′-GATCCTGATCACGTGATCGAGGAG-3′; SRE-1, 5′-GATCCTGATCACCCCACTGAGGAG-3′; ARE7, 5′-GATCTGGAACTCTGATCCAGTAAG-3′. Oligonucle otides were end-labeled with [γ-32P]ATP. T4 polynucleotide kinase and 0.1 pmol [γ-32P]ATP (~30 000 c.p.m.) were used in 20 µl binding reactions containing reaction buffer [10 mM Tris pH 7.5, 50 mM KCl, 2.5 mM MgCl2, 0.05 mM EDTA, 0.1% (v/v) Triton X-100, 8.5% (v/v) glycerol, 1 µg of poly (dI–dC), 1 mM dithiothreitol and 0.1% (w/v) non-fat dry milk]. Samples were loaded onto a non-denaturing 4% polyacrylamide gel. The gels were dried and autoradiographed.
Chromatin immunoprecipitation assay
Subconfluent H293 cells were transiently transfected with plasmids ADD1/SREBP1c-luciferase, pcDNA-myc/His-ADD1 (amino acids 1–403) and/or pRC-CMV-Flag-Twist2, and incubated at 37°C for 48 h. The transfected cells were cross-linked in 1% formaldehyde at 37°C for 10 min and resuspended in 200 µl of NP-40-containing buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40). Crude nuclei were precipitated and lysed in 200 µl of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8.1). Nuclear lysates were sonicated and diluted 10-fold with IP buffer (16.7 mM Tris–HCl pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100). They were then incubated with protein A–Sepharose CL-4B (Amersham-Pharmacia) and anti-Myc antibodies for 2 h at 4°C. The immunoprecipitates were washed for 5 min each with 1 ml of TSE 150 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 150 mM NaCl), 1 ml of TSE 500 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 500 mM NaCl), buffer III (0.25 M LiCl, 1% NP40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl pH 8.1), and 1 ml of TE (10 mM Tris–HCl at pH 8.0, 1 mM EDTA). The immune complexes were eluted with 2 vol of 250 µl elution buffer (1% SDS, 0.1 M NaHCO3), and 20 µl of 5 M NaCl was added to reverse formaldehyde cross-linking. DNA was extracted with phenol–chloroform and precipitated with isopropyl alcohol and 80 µg of glycogen. The precipitated DNA was amplified by PCR. Conditions for PCR reactions were: 0.25 µM each primer, 0.1 mM each dNTP, 1× PCR buffer, 1 U Ex Taq polymerase (TaKaRa, Japan), 0.06 mCi/ml [α-32P]dCTP in 20 µl reaction volume. The PCR products were resolved on 8% polyacrylamide/0.5× TBE (Tris–borate–EDTA) gels.
RESULTS
Interaction between Twist2 and ADD1/SREBP1c
To isolate and identify proteins interacting with ADD1/SREBP1c, the N-terminal region of ADD1/SREBP1c (amino acids 1–153) was used as bait in the yeast two-hybrid system together with an adipocyte cDNA library. Twist2 (also known as Dermo-1), one of several apparently positive clones, was isolated and shown to bind to ADD1/SREBP1c.
To determine the domain responsible for the interaction, we generated deletions of ADD1/SREBP1c in-frame with the DNA-binding domain of Gal4 (Fig. (Fig.1A),1A), and examined interaction between these derivatives and Twist2 by measuring LacZ activities and testing survival on selective medium. As shown in Figure Figure1B,1B, when the A60 or A153 bait was present with Twist2, yeast cells were β-gal positive and survived on selective medium. In contrast, the proline rich domain, residues 60–210, or the bHLH domain of ADD1/SREBP1c, did not have β-gal activity with Twist2. Thus, the N-terminal region (amino acids 1–60) of ADD1/SREBP1c appears to be responsible for the interaction with Twist2.
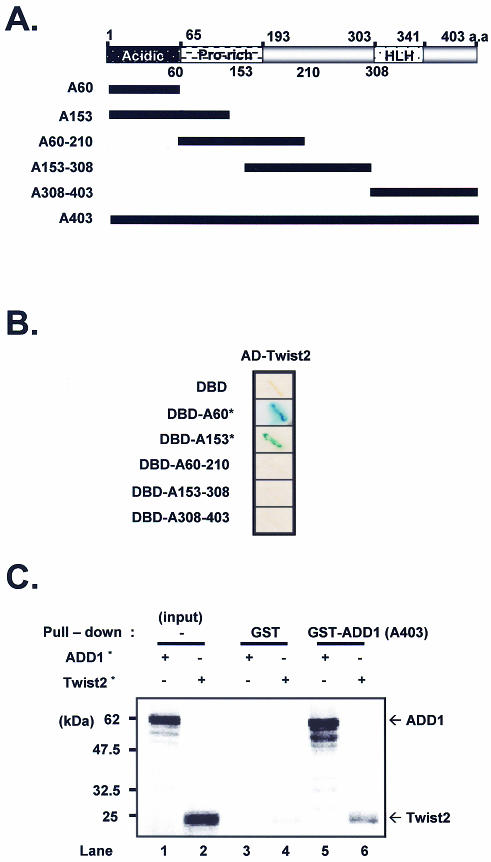
Interaction between Twist2 and ADD1/SREBP1c. (A) Linear representation of each of the ADD1/SREBP1c constructs with the regions indicated that were used in the deletion constructs. (B) The Twist2 interacting domain in ADD1/SREBP1c. Each deletion construct was tested for interaction with full-length Twist2 in the yeast two-hybrid system. cDNAs encoding amino acids 1–60 (A60, containing the N-terminus acidic domain which extends from amino acids 1 to 65), amino acids 1–153 (A153), amino acids 60–210 (A60–210, containing the proline-rich domain from amino acids 65 to 193.), amino acids 153–308 (A153–308), amino acids 308–403 (A308–403, containing the HLH domain from amino acids 303 to 341) and amino acids 1–403 (A403) of ADD1/SREBP1c, were fused to the GAL4 DNA binding domain (pAS2), while Twist2 was fused to the transcriptional activation domain (pACT2). Transformants were checked for interaction by β-gal assay and survival test. Asterisk denotes lacZ activity, >50% blue colonies in <2 h. A403 bait alone showed strong β-gal activity (self transcriptional activity, data not shown). (C) GST pull-down assay. Input of labeled ADD1/SREBP1c (A403) and Twist2 is shown in lanes 1 and 2, respectively. The fraction of labeled proteins that were pulled down by GST alone, and GST–ADD1/SREBP1c are shown in the later lanes. Asterisks denote radiolabeled proteins.
To confirm the interaction between Twist2 and ADD1/SREBP1c, we performed a GST pull-down assay. 35S-labeled Twist2 was incubated with either GST alone or GST–ADD1/SREBP1c in the presence of glutathione–Sepharose beads. Since ADD1/SREBP1c forms homodimers, 35S-labeled ADD1/SREBP1c protein was used as positive control for the GST pull-down analysis (Fig. (Fig.1C,1C, lane 5). As shown in Figure Figure1C,1C, 21% of the input full length Twist2 bound to immobilized GST–ADD1/SREBP1c (lane 6), indicating that Twist2 forms a stable complex with the N-terminus of ADD1/SREBP1c in vitro. Twist2 was not detected when GST was used on its own (Fig. (Fig.1C,1C, lane 4).
Expression of Twist2 in liver and adipose tissue
As a key transcription factor for fatty acid metabolism, ADD1/SREBP1c is highly expressed in fat and liver. To investigate whether Twist2 is also expressed in tissues where ADD1/SREBP1c is abundant, we performed northern blot analyses and semi-quantitative RT–PCR. As shown in Figure Figure2A,2A, expression of Twist2 mRNA was detected in both fat and liver tissue and was also induced during adipocyte differentiation (Fig. (Fig.2B2B and C). Western blot analysis also showed that Twist2 is expressed in 3T3-L1 adipocytes, fat tissue and liver (data not known). These results imply that Twist2 may modulate the activity of ADD1/SREBP1c in fat and liver tissue.
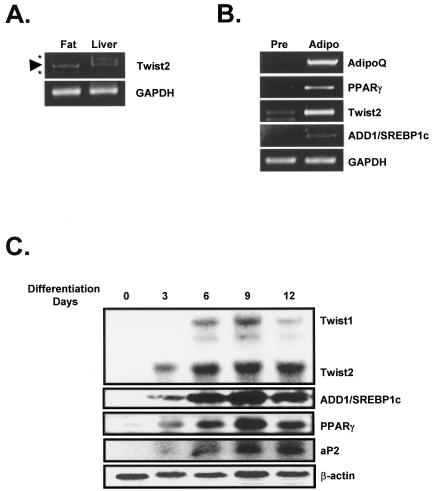
Expression of Twist2 in liver, fat and adipocyte. (A) RT–PCR analysis of Twist2 mRNA expression in fat and liver tissue. (B) Semi-quantitative RT–PCR analysis of Twist2 mRNA in 3T3-L1 preadipocytes (pre) and adipocytes (adipo). (C) Northern blot analysis of Twist2 mRNA during adipocyte differentiation in 3T3-F442A. At the indicated time points, cells were harvested and total RNA isolated for northern blot analysis (see Materials and Methods for details). The β-actin expression level was used to correct for differences in RNA loading. Asterisks denote non-specific PCR products. Arrowhead indicates the Twist2 PCR product. Cycles for the PCR amplifications are 32 for Twist2, 23 for AdipoQ, 25 for PPARγ, 22 for GAPDH and 28 for ADD1/SREBP1c.
Suppression of transcriptional activity of ADD1/SREBP1c by Twist2
In order to investigate the consequences of the interaction between ADD1/SREBP1c and Twist2, we examined the transcriptional activity of ADD1/SREBP1c in the presence and absence of Twist2. Previously, we and others have demonstrated that the promoters of the FAS and ADD1/SREBP1c genes contain binding sites for ADD1/SREBP1c and that they are transactivated by ectopic expression of ADD1/SREBP1c (3,12,24). As shown in Figure Figure3A3A and B, expression of ADD1/SREBP1c activated the promoters of FAS and ADD1/SREBP1c and co-expression of Twist2 suppressed this activation, while Twist on its own had no effect. This observation is consistent with recent reports that Twist2 is a transcriptional repressor of p65 (an NF-κB subunit) and MEF2 (25,26). The proximal promoter region of ADD1/SREBP1c contains binding sites for LXRα/RXRα as well as ADD1/SREBP1c and both ADD1/SREBP1c and LXRα/RXRα independently transactivate ADD1/SREBP1c (24,27,28). As shown in Figure Figure3C,3C, co-expression of Twist2 with LXRα/RXRα did not affect the transcriptional activity of LXRα/RXRα at the ADD1/SREBP1c promoter, indicating that Twist2 specifically represses the transcriptional activity of ADD1/SREBP1c. Because SREBP1a and SREBP2 also can transactivate the FAS promoter, we tested the effect of Twist2 on the transcriptional activity of SREBP1a and SREBP2. As shown in Figure Figure3D,3D, while co-expression of Twist2 did not affect significantly on the transcriptional activity of SREBP2 at the FAS promoter, Twist2 reduced the transcriptional activity of SREBP1a, whose N-terminal acidic domain includes almost all amino acid sequences in the acidic region of ADD1/SREBP1c but for five residues (29).
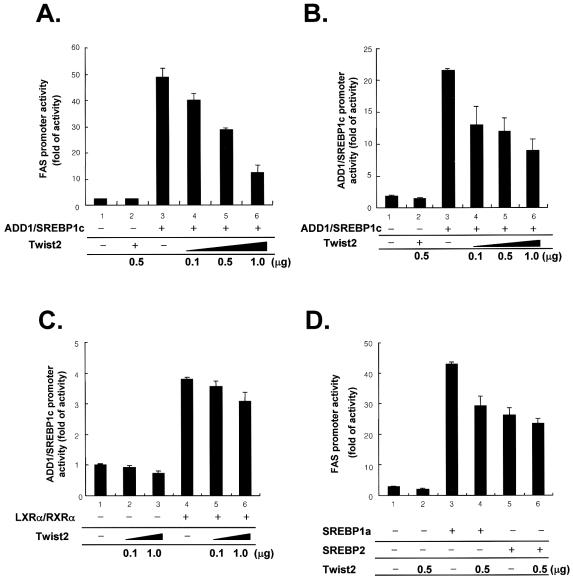
Suppression of the transcriptional activity of ADD1/SREBP1c by Twist2. (A–D) H293 cells were transfected with the FAS-promoter reporter gene (A and D) or ADD1/SREBP1c-promoter reporter gene (B and C) along with pRSV-β-gal, ADD1/SREBP1c, Twist2 and/or other expression vectors, as indicated. Empty expression plasmids were added to give identical amounts of DNA in the transfection assays. (C) LXRα-dependent activation of ADD1/SREBP1c promoter is not influenced by co-expression with Twist2. (D) Twist2 effect on the transcriptional activity of other SREBPs. Amount of DNAs used in each well for the luciferase reporter assay is as follows: ADD1/SREBP1c (200 ng), Twist2 (used as indicated in the figure), LXRα/RXRα (500 ng for each), SREBP1 (200 ng), SREBP2 (200 ng).
Twist2 reduces target DNA binding of ADD1/SREBP1c
We tested whether Twist2 interferes with binding of ADD1/SREBP1c to its target DNA. Unlike other bHLH proteins, ADD1/SREBP1c has dual DNA-binding specificity to both E-boxes (CANNTG) and SRE boxes (ATCACCCCAC), because it contains an atypical tyrosine residue in place of the arginine in the basic domain (23). EMSA demonstrated that Twist2 decreased the DNA-binding ability of ADD1/SREBP1c at both E-box (Fig. (Fig.4A,4A, lanes 3, 8 and 9) and SRE motifs (Fig. (Fig.4B,4B, lanes 3, 7, 8 and 9). Although Twist2 is a bHLH protein, it did not bind to the SRE or E-box motifs, but instead interfered with binding by ADD1/SREBP1c by complexing with it. When Twist2 was reacted with antibody before mixing with ADD1/SREBP1c, its repressive effect was relieved (Fig. (Fig.4A,4A, lanes 9 versus 10 and B, lanes 9 versus 12).

Inhibition of ADD1/SREBP1c DNA-binding by Twist2 in vitro. EMSAs were preformed with 32P-labeled ABS (E-Box) (A) and SRE (B) oligonucleotide probes and in vitro translated ADD1/SREBP1c-Myc/His and/or Flag-Twist2 proteins. The specific probe–ADD1/SREBP1c complex was abolished by addition of a 100-fold molar excess of unlabeled ABS or SRE-1 oligonucleotides (lane 4 of each panel), but not ARE7 (lane 5 of each panel). (A) The specificity of the probe–ADD1/SREBP1c complex confirmed by supershift (lane 6). The amount of the ABS probe–ADD1/SREBP1c complex was decreased by the addition of Twist2 (lanes 8 and 9), but not by mock protein (lanes 11 and 12). α-Flag antibody neutralized repression by Twist2 (lane10). (B) Addition of Twist2 reduced the amount of the SRE probe–ADD1/SREBP1c complex (lane 7–9). Asterisks indicate non-specific binding to each probe by unprogrammed lysate (mock protein). S.C, specific competition with cold SRE or E-box oligonucleotide; N.C, non-specific competition with ARE7 oligonucleotide probe.
To establish that Twist2 inhibits DNA binding by ADD1/SREBP1c in vivo, we performed a chromatin immunoprecipitation (ChIP) assay. It has been shown previously that ADD1/SREBP1c transactivates its own promoter by binding to the SRE motif (24). When ADD1/SREBP1c protein was immunoprecipitated in the ChIP assay, DNA fragments containing the SRE element of the ADD1/SREBP1c promoter were amplified from the immunoprecipitates (Fig. (Fig.5A,5A, lane 3). Expression of Twist2 with ADD1/SREBP1c inhibited the binding of ADD1/SREBP1c to its target (Fig. (Fig.5A,5A, lane 4). Western blot analysis showed that the expression levels of ADD1/SREBP1c and Twist2 were similar (Fig. (Fig.55B).
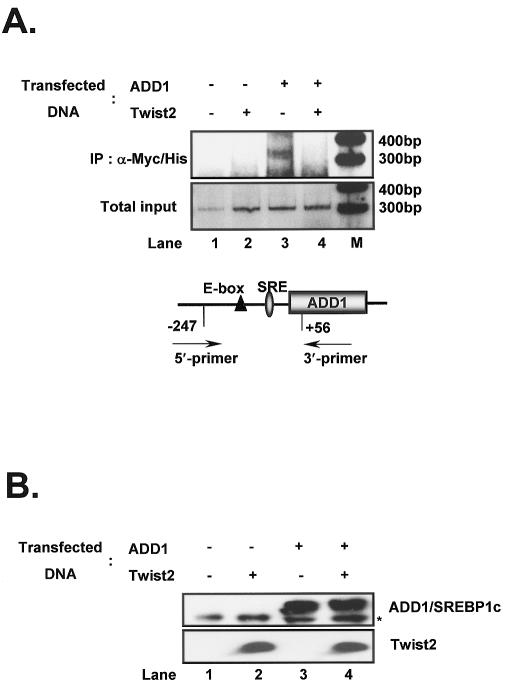
Inhibition of ADD1/SREBP1c DNA binding by Twist2 in vivo. (A) ChIP analysis of Twist2-mediated repression of the DNA-binding ability of ADD1/SREBP1c. H293 cells were transfected in duplicate (for use in western blot analysis) with the DNAs indicated. After cross-linking with formaldehyde, cells were lysed, pelleted and sonicated, and ADD1/SREBP1c was immunoprecipitated with anti-Myc antibody. Complexes were eluted, cross-linking was reversed and purified DNA was used as a template for PCR, using primers for the promoter region of ADD1/SREBP1c containing a SRE site. [α-32P]dCTP-labeled PCR products were resolved by non-denaturing PAGE, and analyzed using a phosphoimager. (B) Western blot analysis of transfected proteins. Transfected cells were lysed, harvested and total protein was resolved by SDS–PAGE and evaluated using α-Myc and α-Flag antibodies to detect ADD1/SREBP1c and Twist2, respectively. Asterisk denotes a non-specific protein band.
Histone deacetylation is involved in the repression of the transcriptional activity of ADD1/SREBP1c by Twist2
It has been reported recently, that repression of the transcriptional activity of MEF2 by Twist2 is dependent on HDAC and that Twist2 forms a complex with E12. We therefore examined the effects of HDAC inhibitor, sodium butyrate, on the transcriptional regulation of ADD1/SREBP1c by Twist2, and found that it prevented Twist2-mediated repression of the transcriptional activity of ADD1/SREBP1c (Fig. (Fig.6A).6A). In addition, Twist2-dependent repression of the transcriptional activity of ADD1/SREBP1c was enhanced by over-expression of HDACs (Fig. (Fig.6B).6B). These results imply that Twist2 contributes to the transcriptional suppression of ADD1/SREBP1c not only by decreasing the DNA-binding ability of ADD1/SREBP1c but also by associating with HDACs.
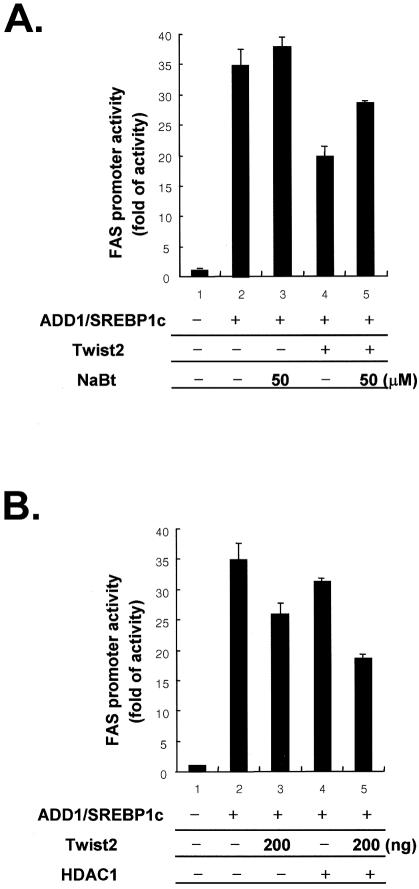
HDAC-dependent repression of the transcriptional activity of ADD1/SREBP1c by Twist2. (A) and (B) H293 cells were transfected with the indicated expression vectors in the presence of a FAS-luciferase reporter gene. (A) Transfected cells were incubated overnight in DMEM medium supplemented with 10% bovine calf serum and/or 5 and 50 mM of sodium butyrate (NaBt) before harvesting for luciferase assay. (B) Co-transfection of HDAC1 enhances the repressive effect of Twist2 on the transcriptional activity of ADD1/SREBP1c. Amount of DNAs used in each well for the luciferase reporter assay is as follows: ADD1/SREBP1c (200 ng), Twist2 (500 ng or as indicated in the figure), HDAC1 (200 ng).
DISCUSSION
Twist2 is a bHLH protein that was cloned as an E12-binding partner by yeast two-hybrid screening (30). When heterodimerized with E12, Twist2 binds to E-boxes and Twist-boxes without displaying any substantial transcriptional activity (30). Recent studies have revealed that Twist2 represses many transcription factors. For example, it inhibits MyoD, a myogenic bHLH transcription factor that is also inhibited by the Ids family of HLH proteins. Because Id family proteins have only the HLH domain and not a basic region, they function as dominant-negative partners of MyoD to inhibit muscle differentiation (31). Unlike Id proteins, Twist proteins are able to block DNA binding of MyoD by squelching E12 proteins, which are necessary as partners for efficient DNA binding of MyoD, and bind to MyoD indirectly to form a transcriptionally inactive complex (25,32,33). Interestingly, it has been suggested that HDACs are involved in the repression of MyoD by Twist2 (25). Twist2 is also reported to repress osteoblast differentiation at the transcriptional level (34–35).
It appears that Twist2 also functions as an oncoprotein, antagonizing p53-dependent apoptosis (36). Consistent with this, Twist2 knockout mice show severe growth retardation and apparently atrophic dermis, thymus, liver and fat tissue, with increased numbers of apoptotic bodies (26). Loss of Twist2 results in elevated expression of pro-inflammatory cytokines and perinatal death from cachexia (26). Twist2 mRNA expression is induced by TNFα via the p65 subunit of NF-κB, and Twist2 appears to repress cytokine gene expression by a negative feedback loop (26).
We have shown above that Twist2 also regulates ADD1/SREBP1c by physical interaction. Twist2 was expressed in both fat and liver tissues where ADD1/SREBP1c is highly expressed (Fig. (Fig.2),2), and ectopic expression of Twist2 repressed transcription by ADD1/SREBP1c at both FAS and ADD1/SREBP1c promoters (Fig. (Fig.3).3). It is likely that repression by Twist2 is due to inhibition of DNA binding, because over-expression of Twist2 attenuated the DNA-binding ability of ADD1/SREBP1c in vivo and in vitro (Figs (Figs44 and and5).5). Furthermore, HDAC inhibitors such as sodium butyrate abolished Twist2-dependent repression, implying that the transcriptional activity of ADD1/SREBP1c may be regulated by Twist2 via HDACs.
These observations suggest that bHLH transcription factors, which regulate cell type determination and differentiation by Ids and/or Twists, act by a common mechanism. It has been reported previously that the transcription by ADD1/SREBP1c, like MyoD, is down-regulated by Ids (37). However, it is likely that the mechanism by which Twist2 inhibits MyoD- and ADD1/SREBP1c-dependent transcription are not identical. Twist2 inhibits MyoD indirectly by squelching E12 protein while Twist2 forms a protein complex with ADD1/SREBP1c and interferes with the domains of ADD1/SREBP1c necessary for transcriptional activation.
As described above, the N-terminal acidic domains of SREBPs are important for their transcriptional activity by associating with co-activators. The acidic domain (amino acids 1–50) of SREBP1a interacts with the ARC (activator-recruited cofactor) complex for transcriptional activation (17). As shown here by yeast two-hybrid analysis, it is likely that ADD1/SREBP1c interacts with Twist2 through its short acidic domain (amino acids 1–60), but not its other domains that include the bHLH domain (amino acids 308–403). In addition, Twist2 expression did affect the transcriptional activity of SREBP1a, as well as ADD1/SREBP1c, but not SREBP2 (Fig. (Fig.3).3). The acidic region of SREBP1a includes almost all amino acid sequences, which that of ADD1/SREBP1c have. Thus, this result also supports that Twist2-mediated transcriptional repression might be mediated by the interaction through the N-terminal acidic region of ADD1/SREBP1c. Therefore, we can exclude the possibility that the interaction between Twist2 and ADD1/SREBP1c is via the HLH domain. Instead, it appears that Twist2 binding to the N-terminal region of ADD1/SREBP1c down-regulates transcription by ADD1/SREBP1c either by squelching co-activator binding to ADD1/SREBP1c, recruiting co-repressors to the acidic domain, or changing the conformation of ADD1/SREBP1c into an inactive form that cannot bind to DNA. On the basis of our observations, the latter two models seem to be plausible explanations of how Twist2 regulates the transcriptional activity of ADD1/SREBP1c. As assessed by in vivo ChIP assay and in vitro EMSA, the interaction between ADD1/SREBP1c and Twist2 clearly reduced the DNA-binding ability of ADD1/SREBP1c (Figs (Figs44 and and5).5). A DNA–ADD1/SREBP1c–Twist2 triple complex was not observed with E-box or SRE probes in EMSA (Fig. (Fig.4).4). Instead, addition of Twist2 reduced the amount of DNA-bound ADD1/SREBP1c just like Ids. These results suggest that Twist2 binding to ADD1/SREBP1c itself reduces the DNA-binding affinity of ADD1/SREBP1c, possibly by changing the conformation of ADD1/SREBP1c into a transcriptionally inactive form.
Furthermore, as proposed by Gong et al. in the repression of transcriptional activity of MyoD by Twist2, it is also likely that Twist2-dependent transcriptional repression of ADD1/SREBP1c is, at least partially, mediated by the formation of transcriptionally inactive protein complex through recruiting co-repressors to ADD1/SREBP1c target gene promoter region (25). As described above, sodium butyrate, which is a well-known HDAC inhibitor, relieved the Twist2-induced transcriptional suppression of ADD1/SREBP1c, which implies that Twist2 represses the transactivity of ADD1/SREBP1c by recruiting HDACs. Although, the mechanism(s) by which Twist2 recruits co-repressors to the ADD1/SREBP1c target gene promoter region remains to be elucidated, it is plausible that Twist2 recruiting co-repressors to ADD1/SREBP1c might change protein complex or inactivate it by protein modifications. Recently, Giandomenico et al. reported that p300 binding to SREBP1a increases the DNA-binding ability of SREBP1a, and p300-dependent acetylation of SREBP1a and SREBP2 regulates their protein stability (38).
One may speculate that Twist2, as a repressor of ADD1/SREBP1c, affects several aspects of energy homeostasis regulated by ADD1/SREBP1c (39,40). For example, increased production of TNFα is observed in the obese state and has been implicated in obesity-associated insulin resistance. Also, TNFα has been shown to block the stimulatory effect of insulin on ADD1/SREBP1c in human adipocytes (41). Thus, it is likely that this TNFα-induced down-regulation of ADD1/SREBP1c in the adipocytes of obese or diabetic patients is, at least partially, mediated by increased levels of Twist2, because TNFα has been shown to stimulate expression of Twists (26). Consistent with this, we have observed elevated Twist protein levels in fat tissue from ob/ob and db/db mice that have an obese phenotype combined with severe insulin resistance and high serum TNFα (Y.S.Lee and J.B.Kim, unpublished data).
In summary, we have shown that Twist2 is a novel ADD1/SREBP1c interacting protein. Since the interaction between ADD1/SREBP1c and Twist2 suppressed the transcriptional activity of ADD1/SREBP1c, abnormal expression of Twist2 may affect energy homeostasis such as lipid and glucose metabolism, which are governed by ADD1/SREBP1c. However, future studies are required to define the role of Twist2 and ADD1/SREBP1c in whole body energy homeostasis.
ACKNOWLEDGEMENTS
This work was supported by a grant from the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (HMP-00-B-20900-0098). R.H.S., S.H.J., J.P. and J.B.K. were supported by a BK21 Research Fellowship from the Ministry of Education and Human Resources Development.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkg934
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc291873?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Mechanisms of Regulation of the CHRDL1 Gene by the TWIST2 and ADD1/SREBP1c Transcription Factors.
Genes (Basel), 14(9):1733, 30 Aug 2023
Cited by: 0 articles | PMID: 37761873 | PMCID: PMC10530651
Kinetic networks identify TWIST2 as a key regulatory node in adipogenesis.
Genome Res, 33(3):314-331, 21 Feb 2023
Cited by: 5 articles | PMID: 36810156 | PMCID: PMC10078291
Twist2 is NFkB-responsive when p120-catenin is inactivated and EGFR is overexpressed in esophageal keratinocytes.
Sci Rep, 10(1):18829, 02 Nov 2020
Cited by: 4 articles | PMID: 33139779 | PMCID: PMC7608670
Regulatory Roles of SREBF1 and SREBF2 in Lipid Metabolism and Deposition in Two Chinese Representative Fat-Tailed Sheep Breeds.
Animals (Basel), 10(8):E1317, 30 Jul 2020
Cited by: 7 articles | PMID: 32751718 | PMCID: PMC7460493
microRNAs targeting cellular cholesterol: implications for combating anticancer drug resistance.
Genes Cancer, 11(1-2):20-42, 01 Jan 2020
Cited by: 4 articles | PMID: 32577155 | PMCID: PMC7289906
Go to all (41) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Regulatory role of glycogen synthase kinase 3 for transcriptional activity of ADD1/SREBP1c.
J Biol Chem, 279(50):51999-52006, 04 Oct 2004
Cited by: 67 articles | PMID: 15466874
Adipocyte determination- and differentiation-dependent factor 1/sterol regulatory element-binding protein 1c regulates mouse adiponectin expression.
J Biol Chem, 279(21):22108-22117, 22 Mar 2004
Cited by: 83 articles | PMID: 15037635
Functional characterization of the human resistin promoter with adipocyte determination- and differentiation-dependent factor 1/sterol regulatory element binding protein 1c and CCAAT enhancer binding protein-alpha.
Mol Endocrinol, 17(8):1522-1533, 01 May 2003
Cited by: 41 articles | PMID: 12730330
SREBPs: transcriptional mediators of lipid homeostasis.
Cold Spring Harb Symp Quant Biol, 67:491-498, 01 Jan 2002
Cited by: 133 articles | PMID: 12858575
Review





