Abstract
Free full text

A Novel Virulence Gene in Klebsiella pneumoniae Strains Causing Primary Liver Abscess and Septic Metastatic Complications
Abstract
Primary Klebsiella pneumoniae liver abscess complicated with metastatic meningitis or endophthalmitis is a globally emerging infectious disease. Its pathogenic mechanism remains unclear. The bacterial virulence factors were explored by comparing clinical isolates. Differences in mucoviscosity were observed between strains that caused primary liver abscess (invasive) and those that did not (noninvasive). Hypermucoviscosity correlated with a high serum resistance and was more prevalent in invasive strains (52/53 vs. 9/52; P < 0.0001). Transposon mutagenesis identified candidate virulence genes. A novel 1.2-kb locus, magA, which encoded a 43-kD outer membrane protein, was significantly more prevalent in invasive strains (52/53 vs. 14/52; P < 0.0001). The wild-type strain produced a mucoviscous exopolysaccharide web, actively proliferated in nonimmune human serum, resisted phagocytosis, and caused liver microabscess and meningitis in mice. However, magA − mutants lost the exopolysaccharide web and became extremely serum sensitive, phagocytosis susceptible, and avirulent to mice. Virulence was restored by complementation using a magA-containing plasmid. We conclude that magA fits molecular Koch's postulates as a virulence gene. Thus, this locus can be used as a marker for the rapid diagnosis and for tracing the source of this emerging infectious disease.
Introduction
Klebsiella pneumoniae is an enteric gram-negative bacillus (1, 2) causing hospital-acquired infections (1–3) and infections in debilitated or immunocompromised patients (1–3). In the past 15 yr, a new type of invasive K. pneumoniae disease has emerged in Taiwan (4–10) that typically presents a community-acquired primary liver abscess with sepsis and bacteremia (4, 5). Metastatic meningitis or endophthalmitis complicated the course in 10–12% of cases (6–9). Approximately 50% of the reported patients had diabetes mellitus (4–10), with the remainder displaying no apparent underlying diseases. The responsible K. pneumoniae strains were uniformly susceptible to all cephalosporins and aminoglycosides (5, 10). Nevertheless, mortality rates as high as 10% for primary liver abscess (4–9) and 30–40% among those with metastatic meningitis have been reported (11, 12). In addition to being particularly common in Taiwan (4), where >900 cases have been reported (4–10), this invasive K. pneumoniae disease has also been reported in North America (13, 14), Europe (15–19), and Japan (20, 21).
The mechanism of this new infectious disease remains unclear. Identification of the specific bacterial virulence factors would help spur the development of rapid molecular diagnosis methods and innovative drug therapies. Previous molecular epidemiology analyses (6, 10) have highlighted the genetic diversity among the involved K. pneumoniae strains, indicating that this disease is not caused by one or a few epidemic strains.
To identify factors that are unique, and so presumably vital to the virulence process, we compared strains causing primary liver abscess with strains not associated with liver abscess.
Materials and Methods
Bacterial Strains and Culture Conditions.
Clinical K. pneumoniae strains isolated from patients with septicemia were collected at National Taiwan University Hospital (NTUH) from 1996 to 2001. Identification of the isolates was according to standard clinical microbiologic methods (1). All strains were stored at −80°C before use.
Among the total of 1,352 isolates obtained from patients with septicemia, 101 strains were obtained from patients displaying primary liver abscess. The diagnosis of primary liver abscess was confirmed by sonography-guided aspiration or surgical drainage in 53 of these 101 patients. Of these 53 patients, 26 had diabetes mellitus, 25 were otherwise healthy before development of the abscess, one patient had nephrotic syndrome, and one patient displayed hepatic failure associated with advanced liver cirrhosis. The strains isolated from the 53 patients were designated as tissue-invasive (invasive) strains. In addition to displaying primary liver abscesses, four patients displayed metastatic endophthalmitis, whereas another displayed metastatic meningitis.
Of the remaining 1,251 patients who did not display clinical symptoms of liver abscess, meningitis, or endophthalmitis, 52 patients were confirmed to be free of abscess by either abdominal sonography or computed tomography. The K. pneumoniae strains from these patients were designated as nontissue invasive (noninvasive) strains.
For comparative purposes, we obtained 21 nonblood isolates from nonseptic patients at NTUH, and 101 strains from other facilities. These included 15 strains (6 of which were found capable of causing primary liver abscess) were obtained from the National Cheng Kung University Hospital (NCKUH; a gift from I.-J. Su, National Health Research Institute, Taipei, Taiwan). Another 13 strains (1 of which caused meningitis without liver abscess and 1 that caused abscess) were a gift from S.-S. Wang (ECK Hospital, Sansia, Taiwan). 34 strains, all of which caused nosocomial infections without liver abscess, meningitis, or endophthalmitis, were a gift from J.-T. Wang (Far Eastern Memorial Hospital, Banciao, Taiwan). 15 strains from Hong Kong were a gift from L.K. Siu (National Health Research Institute, Taipei, Taiwan). Finally, 24 strains, none of which caused liver abscess, were purchased from the American Type Culture Collection, including strain MGH-78578, which caused pneumonia and was selected for genome sequencing.
For general use, both K. pneumoniae and Escherichia coli were grown in Luria-Bertani (LB) broth or agar at 37°C. When necessary, 50 μg/ml of either kanamycin or chloramphenicol was added.
String Test for Hypermucoviscosity.
The tested strains were inoculated on 5% sheep blood agar plates and incubated at 37°C overnight. A standard bacteriologic loop was used to stretch a mucoviscous string from the colony. Hypermucoviscosity (HV) was defined by the formation of viscous strings >5 mm in length when a loop was used to stretch the colony on agar plate (positive string test).
Serum Resistance Assay.
The serum resistance of K. pneumoniae strains was determined by an established method (22). An inoculum of 2.5 × 104 CFU, prepared from the mid-log phase, was mixed at a 1:3 vol/vol ratio with nonimmune human serum donated by healthy volunteers. The final mixture was comprised of 75% nonimmune serum by volume. The mixture was incubated at 37°C. The colony count, determined by the serial dilution method, was checked at the baseline, and at 1, 2, and 3 h. The average survival percentage was plotted against the time to obtain a survival curve that characterized the serum susceptibility.
Phagocytosis Assay.
The phagocytosis assay of K. pneumoniae strains was performed by an established method (23). An inoculum of 107 CFU was mixed with 106 freshly isolated human neutrophils or mouse macrophage cell line RAW-264.7 in RPMI 1640 media (23). After 15 min of incubation, extracellular bacteria were washed out, and 100 μg/ml gentamicin was added to kill any remaining extracellular bacteria. Cell-associated bacteria were quantified at zero and 30 min after the end of incubation by plating on LB agar plates after lysis of the cells with 0.1% Triton X-100. Initial experiments established that the magA knockout mutants were no more sensitive to the detergent than the wild type. To determine whether cell-associated bacteria counted by the aforementioned method were indeed ingested by human neutrophils, fluorescence microscopy using confocal microscope was performed. Before fluorescence microscopy, plasmid GFPuv (CLONTECH Laboratories Inc.), which carries a gene that encodes green fluorescent protein, was electroporated into selected K. pneumoniae strains.
Complement C3 Deposition Assay.
Complement C3 deposition assay was performed by an established method (24). A bacterial suspension (2 × 108 CFU/ml) was opsonized with 25% serum at 37°C for 1–5 min. Bacteria were washed three times with PBS by pelleting and resuspended. The final pellet was resuspended in 50 mM carbonate-bicarbonate buffer, pH 9.0, containing 1 M NH4OH, to disrupt ester bonds between C3 fragments and the bacterial surface. After 2 h at 37°C, the C3 fragment suspension was reduced with 10 mM dithiothreitol in 1% SDS at 37°C for 1 h and alkylated with 22 mM iodoacetamide at 37°C for 1 h. Western blot analysis was performed. During the procedure, membranes were incubated sequentially with anti–human complement C3 (1:2,000; Biogenesis) and goat horseradish peroxidase–conjugated anti–human IgG antibodies, before development with an enhanced chemiluminescence system (Amersham Biosciences).
Animal Inoculation.
Female BALB/cByl 6-wk-old mice were used for inoculation. K. pneumoniae inocula consisting of 102–106 mid-logarithmic growth phase CFUs were diluted in 100 μl normal saline and injected intraperitoneally (25, 26). Four mice were used to test the effects of each inoculum. After inoculation, the mice were observed for 30 d. The LD50 was calculated using the method established by Reed and Muench (26).
Mini-Tn5 Transposon Mutagenesis.
A mini-Tn5 transposon was used for random transposon mutagenesis (27, 28). E. coli S17–1λpir, which carries the F plasmid and mini-Tn5 transposon on pUTKm1 plasmid, served as the donor. The conjugation between wild-type K. pneumoniae NTUH-K2044 and E. coli S17–1λpir was facilitated by mechanical force generated by vacuum suction. During the conjugation, the mini-Tn5 transposon was transposed into the NTUH-K2044 genome randomly. After insertion, the transposase was lost, thus ensuring the genetic stability of generated mutants. 5 μg/ml chlorhexidine was added to kanamycin-containing LB broth to select for the chlorhexidine-resistant K. pneumoniae NTUHK-2044 oversensitive donor E. coli S17–1λpir (29). The mutants were harvested and stored at −80°C before use.
Inverse PCR and DNA Sequencing.
DNA sequences adjacent to the transposon insertion sites were determined by inverse PCR and DNA sequencing. The genomic DNA of mutants was extracted and subjected to complete digestion by restriction endonuclease PstI (2 U of enzyme for 2.0 μg genomic DNA at 37°C overnight) followed by circularization. Inverse PCR was performed using the following primers: 5′-GTACCGAGCTCGAATTCGGC-3′ and 5′-ATGAGCCATATTCAACGGGA-3′. DNA sequencing was performed using the BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (PerkinElmer and Applied Biosystems) on an ABI PRISM 377 DNA Sequencer. Primers 5′-GTACCGAGCTCGAATTCGGC-3′ were used for sequencing of the inverse PCR products. The obtained sequences were further used in designing primers for the sequencing of the entire nearby region on the genome.
Dot-Blot Hybridization and PCR.
Standard procedure for dot-blot hybridization was used (30). For each strain, 100 ng of extracted genomic DNA was used. The nucleotide probe for magA, corresponding from +10 to +855, was prepared by PCR amplification followed by photolabeling using a psoralen–biotin system (Ambion). Hybridization was performed at 63°C overnight. After hybridization, the membrane was washed with 2 × SSC plus 0.1% SDS at room temperature for 10 min, washed 3 × 15 min with 0.1 × SSC plus 0.1% SDS in 58°C, followed by 2 × 5–min treatments with wash buffer (Ambion) at room temperature. Detection was conducted using a chemiluminescence system (Ambion) based on streptavidin and alkaline phosphatase. Two primer pairs derived from magA were as follows: outer, forward, 5′-GGTGCTCTTTACATCATTGC-3′, and reverse, 5′-GCAATGGCCATTTGCGTTAG-3′; and inner, forward, 5′-CGCCGCAAATACGAGA- AGTG-3′, and reverse, 5′-GCAATCGAAGTGAAGAGTGC-3′. These were used for nested PCR to detect the presence of magA. Ribosomal RNA gene was used as internal positive control for both dot-blot hybridization and PCR.
Complementation and Deletion Clones Construct.
The gene magA and its predicted promoter region were inserted into plasmid pCRII® (Invitrogen) by Taq-amplified cloning. A chloramphenicol resistance gene (31) was inserted as the selection marker. The constructed plasmid was sent into magA − mutant by electroporation (29, 32, 33). Deletion clones were constructed by sending plasmids carrying PCR- or inverse PCR–generated mutated magA, with various types of deletion, into magA − mutants. All deletion constructs were confirmed by DNA sequencing.
Cell Fractionation Procedure and Western Blot.
Cell fractionation was performed following a published protocol (34). The K. pneumoniae cells were harvested by centrifugation at 11,000 g for 20 min. The pellet was resuspended in water, and the cells were lysed by sonication. Unlysed cells were removed by centrifugation at 250 g for 5 min. The supernatant was collected and centrifuged at 11,000 g for 30 min. The pellet was resuspended in 25 μl PBS by vigorous pipetting, 25 μl of 4% Triton X-100 in PBS was added, and the mixture was incubated at 37°C for 30 min. Triton X-100–insoluble proteins (outer membrane protein-enriched fraction) were pelleted by centrifugation at 11,000 g for 30 min. The supernatant containing Triton X-100–soluble proteins (cytoplasmic membrane protein-enriched fraction) was also collected. Anti-MagA antibodies for Western blot were prepared by inoculating rabbits with a truncated MagA protein (amino acids 44–221) expressed in E. coli. During the procedure, membranes were incubated sequentially with anti-MagA antibodies and goat horseradish peroxidase–conjugated anti–rabbit IgG antibodies before development with an enhanced chemiluminescence system (Amersham Biosciences). Spheroplast preparation was performed as described previously (35). An antibody to NlpB in Serratia marcescens (a gift from H.C. Lai, National Taiwan University, Taipei, Taiwan), which has been shown to be an outer membrane protein in E. coli (36), was also used as a cell fractionation control.
Results
HV and Clinical Diseases.
Differences in the HV phenotype (formation of viscous strings >5 mm in length; Fig. 1 A shows a positive string test) were observed between 53 invasive strains and 52 noninvasive strains. In an experiment to test the reproducibility of these results, two independent blind readings of 50 clinical isolates yielded a consistency of 98%. The HV phenotype was identified in 52 out of 53 invasive strains (98%), but only in 9 out of 52 noninvasive strains (17%). The difference was highly significant (P < 0.0001; Fisher's exact test).
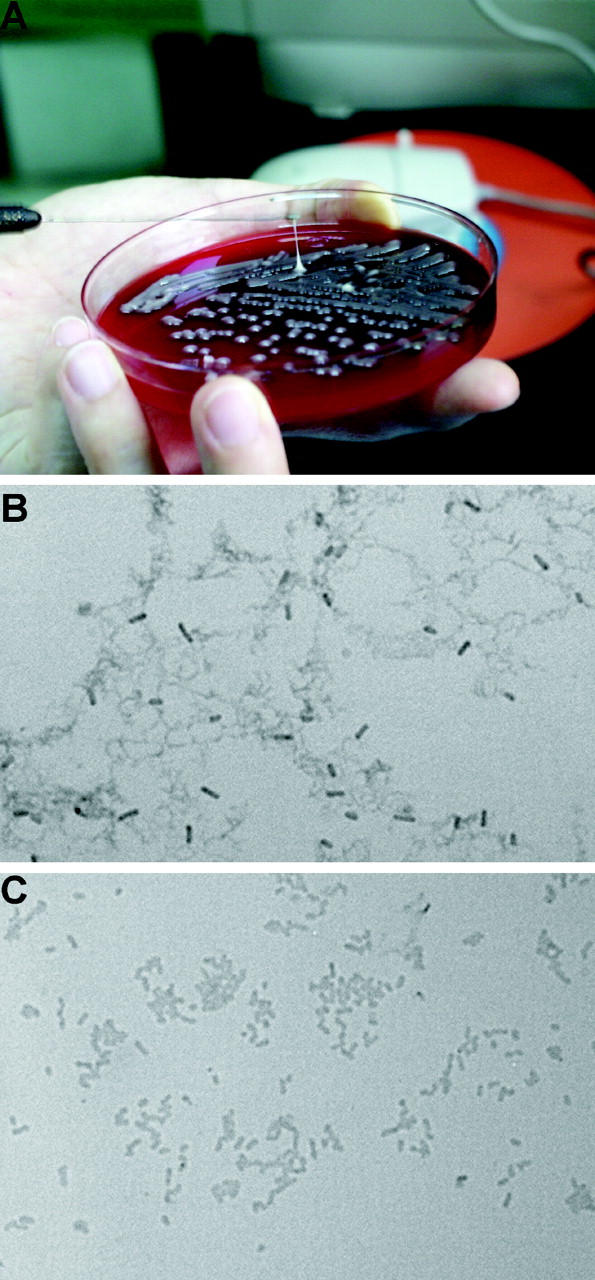
HV phenotype. (A) Demonstration of a positive string test; formation of a viscous string >0.5 cm in length stretched by the inoculation loop. (B) Under light microscopy, wild-type strain NTUH-K2044 produces an exopolysaccharide web attached to the capsule (Periodic Acid Schiff stain; original magnification, 1,000). (C) magA − mutant loses the capability of producing an exopolysaccharide web.
Virulence Assay In Vitro and In Vivo.
Three K. pneumoniae strains were selected for use in serum resistance assays and in vivo effect in mice. One was an invasive, HV phenotypic strain NTUH-K2044, isolated from the blood of a previously healthy 40-yr-old man suffering from community-acquired primary liver abscess and metastatic meningitis. The second was a noninvasive, non-HV phenotypic clinical strain NTUH-K9, and the third was the non-HV phenotypic strain MGH-78578.
The serum resistance assay showed that NTUH-K2044 was not only resistant to serum killing but actually actively proliferated in nonimmune human serum. In contrast, MGH-78578 was much less resistant to serum, and NTUH-K9 was extremely sensitive to serum (Fig. 2 A). This high serum resistance also correlated with HV phenotype in another eight randomly selected clinical strains, including four HV+ invasive strains and four non-HV noninvasive strains. The LD50 of NTUH-K2044 to BALB/cByl mice was <102 CFU. Intraperitoneal inoculation of 106 CFU in these mice resulted in rapid death in 12 h, without definite histopathology change. However, inoculation of 102–105 CFU resulted in death after 1 wk with meningitis and liver microabscess (Fig. 3, A and B). In contrast, inoculation of NTUH-K9 or MGH-78578 did not cause death, meningitis, or liver microabscess in mice (Fig. 3, C and D) at any doses (102–106 CFU). The animal experiments were repeated by different investigators and the results were reproducible.
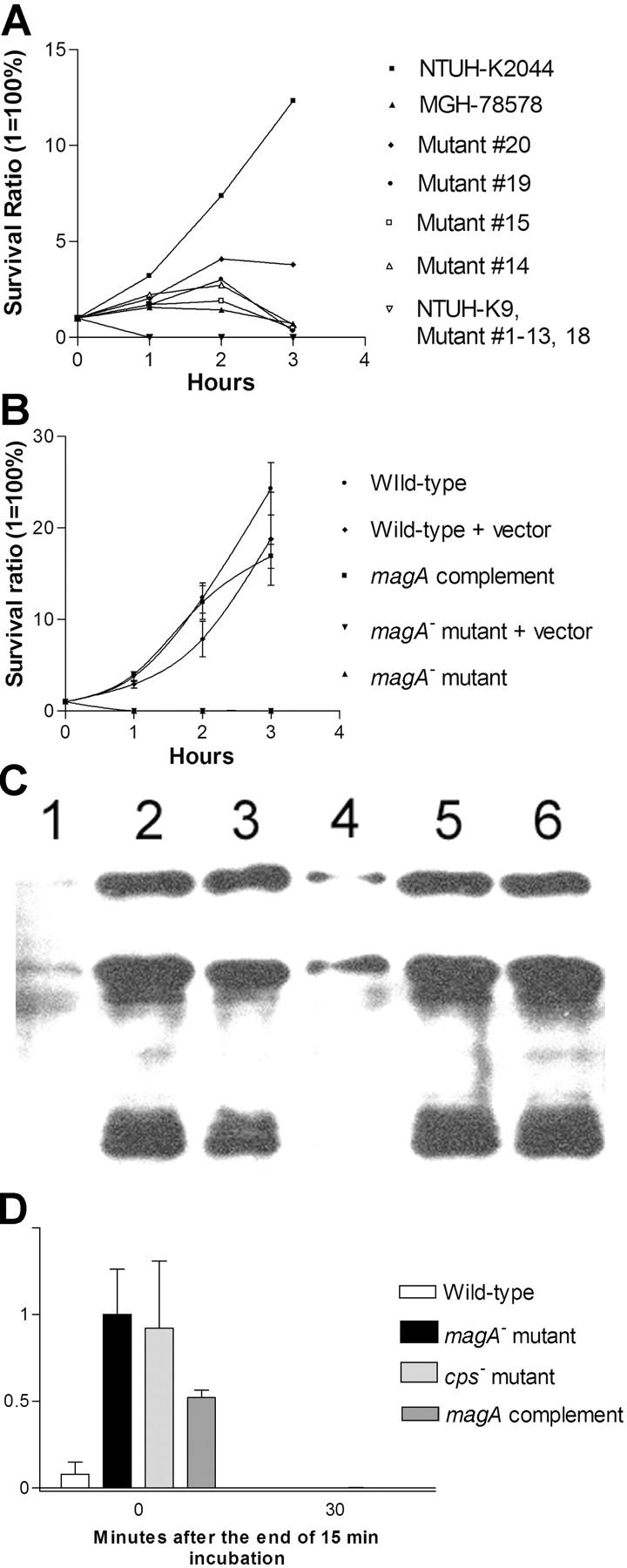
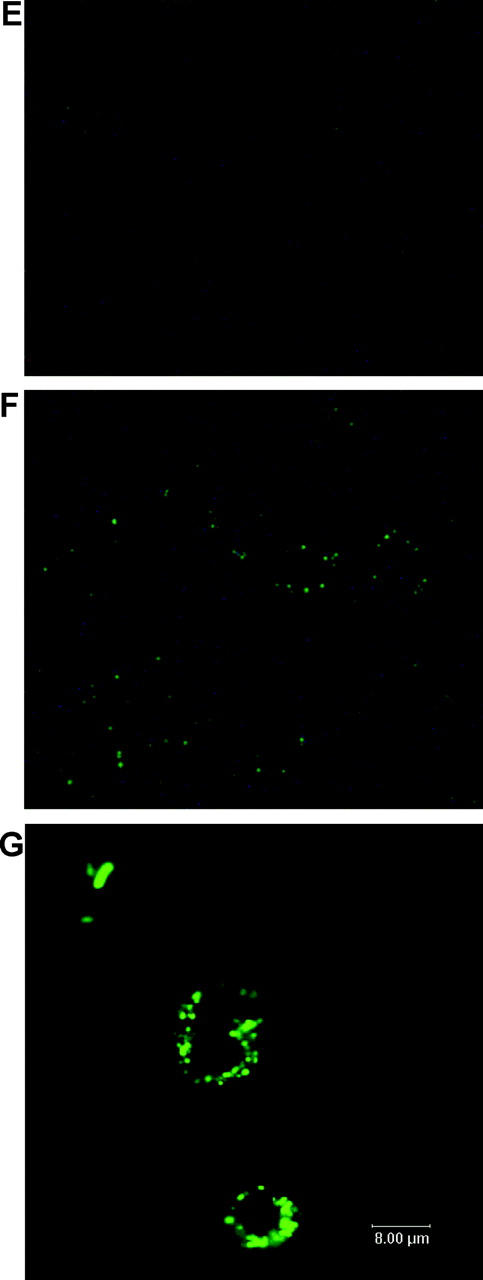
Serum and phagocytosis resistance assays. (A) Difference in serum resistance between NTUH-K2044, MGH-78578, NTUH-K9, and mutants 1–20. The assays were performed sequentially three times. A high degree of consistency was observed between experiments. (B) Restoration of serum resistance by magA complementation in a magA − mutant, in comparison with magA − mutant, magA − mutant–carrying vector only, wild-type–carrying vector only, and wild type. (C) Western blot analysis of complement C3 deposition showing that magA − mutant (lanes 2 and 5) and cps − mutant (lanes 3 and 6) have much higher levels of complement C3 deposition than the wild type (lanes 1 and 4). 1-min exposure to serum (lanes 1–3). 5-min exposure to serum (lanes 4–6). (D) Phagocytosis assay showing wild-type NTUH-K2044 is less associated with mouse macrophages than magA − mutant or cps − mutant. Complementation with magA-carrying plasmid partially restores the phenotype. (E and F) Fluorescence microscopy images (original magnification, 400). Wild-type NTUH-K2044 (E) is less likely to be ingested or attached by human neutrophils than a magA − mutant (F). (G) Confocal fluorescence microscopy showing that magA − mutants were ingested and disintegrated by human neutrophils.
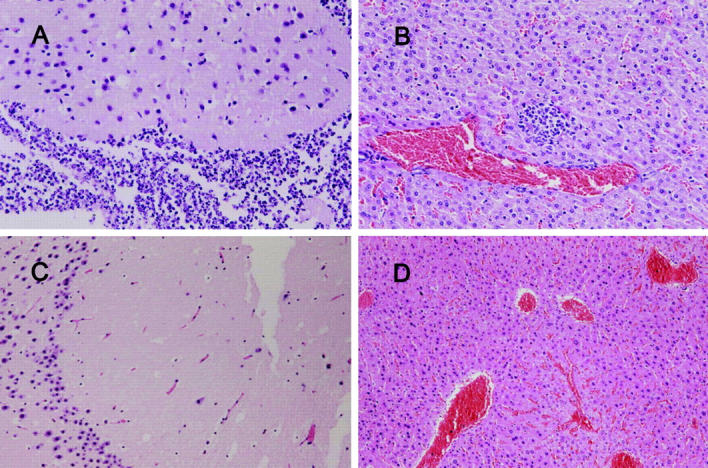
(A and B) Histopathology in the liver and brain of a mouse inoculated with 105 CFU of K. pneumoniae NTUH-K2044, examined on day 9 after inoculation (hematoxylin and eosin stain; original magnification, 200). (C and D) Histopathology in the liver and brain of a mouse inoculated with 105 CFU of K. pneumoniae MGH-78578, examined on day 9 after inoculation (hematoxylin and eosin stain; original magnification, 200). A and B show severe meningitis and liver microabscess. C and D show normal histology.
Loci Associated with HV.
To identify genetic loci associated with the HV phenotype, a mutant library of NTUH-K2044 was constructed using mini-Tn5 transposon mutagenesis. A total of 2,500 mutants were obtained. Southern blot hybridization and DNA sequencing (not depicted) in randomly selected mutants confirmed random transposon insertions. After screening all the mutants, it was determined that 20 had lost the HV phenotype. Among these mutants, mutants 1–11 were identified by their grossly nonmucoid colony appearance, whereas mutants 12–20 were identified by their presentation of a negative string test. All 20 mutants exhibited a marked decrease in serum resistance compared with the wild type (Fig. 2 A). By inverse PCR and DNA sequencing, the 20 loci were completely identified. The loci included K. pneumoniae cps (capsular polysaccharides) cluster (37), wb (lipopolysaccharide) cluster (38), rmpA (extracapsular polysaccharides synthesis regulator; references 39, 40), as well as four novel genetic loci (Table I).
Table I.
Genetic Loci Associated with Hypermucoviscosity in K. pneumoniae
| Mutant | Loci | Function | Reference |
|---|---|---|---|
| 1–11a | cps cluster | capsular polysaccharide synthesis | 37 |
| 12 | wb cluster | lipopolysaccharide synthesis | 38 |
| 13b | novel 1.2-kb ORF | identical to mutant 18 | – |
| 14 | novel 1.6-kb ORF | 57-kD hypothetical protein, putative thiosulfate sulfurtransferase | – |
| 15 | novel 0.9-kb ORF | 33-kD hypothetical protein | – |
| 16c | – | – | – |
| 17 | cps cluster ORF 6 | 80-kD hypothetical protein, function unknown | 37 |
| 18b | novel 1.2-kb ORF | 43-kD hypothetical protein, putative membrane lipoprotein | – |
| 19 | novel 1.6-kb ORF | 57-kD hypothetical protein, putative phosphotransferase | – |
| 20 | rmpA | extracapsular polysaccharide synthesis regulator | 39, 40 |
ORF, open reading frame.
Higher Prevalence of magA in Invasive Strains.
Genomic DNA were extracted from 53 invasive strains and 52 noninvasive strains. Dot-blot DNA hybridizations were performed using labeled PCR products of each locus as probes. We found only a novel 1.2-kb locus was significantly more prevalent in invasive strains, whereas the other six genetic loci were universally present in all tested strains. The novel locus was hit in both mutants 13 and 18 (transposon inserted in nucleotide +70 and +965, respectively). This locus, designated as magA (mucoviscosity-associated gene A), was present in 52 out of 53 invasive strains (98%; the sole strain negative for magA was from the patient with liver cirrhosis) and in 15 out of 52 noninvasive strains (29%; Fig. 4, A and B). The difference in frequencies of occurrence of the locus in the invasive versus noninvasive strains was statistically significant (P < 0.0001; Fisher's exact test). All of the five strains causing septic metastatic complications were strongly positive. The 15 dot-blot positive noninvasive strains included 8 out of 9 HV+ noninvasive strains. The invasive strain negative for string test was also magA positive and highly serum resistant. We further tested 21 nonblood isolates and 100 strains obtained outside NTUH. The result showed that magA was positive in six out of the eight invasive and abscess-causing strains (five out of the six strains were collected at the National Cheng Kung University Hospital, and one strain was collected at the ECK Hospital). However, magA was found to be positive in only three isolates (1 of the 21 nonseptic isolates from our hospital, 1 of the 15 Hong Kong strains, and ATCC8045) of the remaining 113 strains that did not cause apparent liver abscess. The correlation between the presence of magA and clinical tissue-invasive diseases remains significant (P < 0.0001; Fisher's exact test). The results of the dot-blot hybridizations were confirmed by PCR and DNA sequencing (Fig. 4 C). The two tests correlated very well, only one weakly positive dot (Fig. 4 A, dot 3G) was proven to be magA negative by PCR and sequencing.
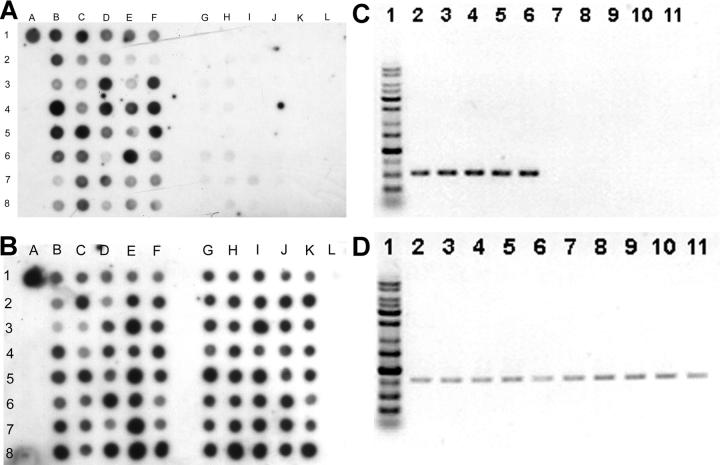
Dot-blot hybridization of genomic DNA extracted from 40 invasive strains (lanes B–F) and 40 noninvasive strains (lanes G–K). Dot A1 is NTUH-K2044 as the positive control. Dot L1 is the PCR product of cepA (reference 29) as the negative control. (A) Hybridization with magA probe. All the invasive strains shown here were positive. The noninvasive strains at dots 3G, 3H, 4H, 6G, 6H, 7G, 7H, 7I, and 8H were positive. (B) Hybridization with 23S rRNA gene probe as internal positive control. (C) PCR for magA. (lane 1) Marker. (lanes 2–6) 5 positive strains. (lanes 7–11) 5 negative strains. (D) PCR for 23S rRNA gene as control.
Characteristics of magA− Mutants.
The growth curves of magA − mutants were not significantly different from wild type (not depicted). Under light microscopy with Periodic Acid Schiff stain, wild-type NTUH-K2044 produced a mucoviscous exopolysaccharide web attached to the capsule (Fig. 1 B). The individual bacteria were interconnected to each other by this web. In addition to a high resistance to nonimmune human serum (Fig. 2 A), wild-type bacteria were also highly resistant to phagocytosis by macrophages (Fig. 2 D) or neutrophils (Fig. 2 E). The magA − mutants remained encapsulated, but did not produce the exopolysaccharide web (Fig. 1 C). magA − mutants were extremely sensitive to nonimmune human serum, which killed all of the mutants in 1 h (Fig. 2 A). After exposure to serum, both the magA − and cps − mutants had a much higher level of complement C3 deposition than wild-type bacteria at 1 min (Fig. 2 C). The cells of mutant bacteria began to degrade in 5 min. magA − mutants were also susceptible to phagocytosis. Phagocytosis assay using either mouse macrophage (Fig. 2 D) or human neutrophil (Fig. 2 F) showed that the amount of intracellular and cell-associated magA − mutants was much higher than that of the wild type. Confocal fluorescence microscopy confirmed that the cell-associated magA − mutants were ingested and disintegrated by human neutrophils (Fig. 2 G). In contrast, the microscopy technique failed to detect intracellular wild-type bacteria at any time point. Moreover, CFUs of the wild-type strain in nonphagocyte-associated washout was consistent with the noningestion of the bacteria. In the mouse experiments, all BALB/cByl mice remained healthy 6 wk after inoculation of 102–107 CFU of the magA − mutants. In mice that were killed 3 wk after inoculation of the bacteria, there was no microscopic evidence of liver microabscess or meningitis. Thus, magA was essential for NTUH-K2044 to cause invasive disease in vivo.
Complementation.
Complementation using a magA-containing plasmid restored the HV phenotype, strong serum resistance (Fig. 2 B), and virulence in the mouse model. The LD50 of magA complement strains to BALB/cByl mice was reduced to 103 CFU. The successful complementation excluded the possibility that mutations elsewhere were responsible for the phenotype change of magA − mutants.
Functional Study of MagA.
The magA gene was 1.2 kb in length and encodes a protein with 408 amino acids (GenBank/EMBL/DDBJ accession no. AB085741). Analysis using the Protein Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/BLAST) and the SWISS-PROT database showed that MagA was related to WaaL and a class of proteins involved in exopolysaccharide production. Analysis using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de; reference 41) showed that the predicted amino acid sequence of MagA started with a signal peptide (amino acids 1–18) followed by nine transmembrane domains (amino acids 44–63, 70–92, 107–126, 135–152, 172–189, 209–231, 246–268, 334–353, and 363–382). Western blotting detected a 43-kD protein (the predicted size of MagA) in the outer membrane fraction of wild-type NTUH-K2044 but not in that of magA − mutants (Fig. 5). Further bioinformatics analysis of motif function using the Protein Families Database of Alignments and Hidden Markov Models (Pfam 7.1) (http://www.sanger.ac.uk/Pfam; reference 42) suggested that amino acids 338–348 functioned as a prokaryotic membrane lipoprotein lipid attachment site (lipobox). Deletion constructs were prepared in a magA − mutant carrying the complementation plasmid. Deletion of the 3′ end up to amino acid 368 did not change the phenotype. However, an inframe deletion of the 20 amino acids (amino acids 338–357) of the lipid attachment site made the strain serum sensitive and resulted in the loss of the HV phenotype. This result was consistent with the identity of magA as a lipoprotein. Nevertheless, the lipobox was not located in the signal peptide as in a traditional prokaryotic lipoprotein.

Western blot of magA (lanes 1–7) and NlpB (lanes 8–10). (lanes 1–4) Cytoplasmic membrane, outer membrane, total proteins, and spheroplast of NTUH-K2044. (lanes 5–7) Cytoplasmic membrane, outer membrane, and spheroplast proteins of a magA − mutant. (lanes 8–10) Outer membrane, cytoplasmic membrane, and periplasmic proteins of NTUH-K2044. (arrow) 43 kD. (arrowhead) 37 kD.
Sequence of magA Flanking Regions.
We sequenced the nearby genomic region of magA, and found a 33-kb genomic segment (GenBank/EMBL/DDBJ accession no. AB117611), which was absent in the complete genome sequence of MGH-78578 (http://genome.wustl.edu/projects/bacterial/?kpneumoniae1). The 5′ flanking region of this fragment was a truncated open reading frame (ORF), whose amino acid sequence displayed 97% homology to that of E. coli OrfX (43). The 3′ flanking region of this fragment was hisI, a phosphoribosyl-AMP cyclohydrogenase gene involved in amino acid transport and metabolism in K. pneumoniae (44). This 33-kb DNA fragment contained 24 ORFs. magA was the fourth ORF from the 5′ end. Bioinformatics analysis suggested that the other 23 genes might function in exopolysaccharide biosynthesis export of the O-antigen of the lipopolysaccharides, glycosylation and other modification of cell envelope, and polysaccharides synthesis and transport.
Discussion
This paper shows that magA is an important virulence gene in invasive K. pneumoniae strains causing primary liver abscess and septic metastatic complications. Disruption of magA in NTUH-K2044 results in complete loss of resistance to serum and phagocytosis, along with a >105-fold increase in LD50 to mice. The virulence was significantly but not completely restored by complementation using a magA-containing plasmid, suggested that adjacent loci might also be contributing to virulence. Although other genetic loci, including the cps cluster (37), wb cluster (38), and rmpA (39, 40) might also be involved in bacterial virulence, only magA displayed a significantly higher prevalence rate in invasive strains (98 vs. 29%; P < 0.0001). Thus, magA meets the criteria of molecular Koch's postulates (45) for a virulence gene associated with this type of invasive K. pneumoniae disease. Thus, this locus can be used as a marker for rapid molecular diagnosis.
In the present work, our data provided supports for the hypothesis that MagA was an outer membrane protein essential for the formation of a protective exopolysaccharide web in invasive K. pneumoniae strains. Deletion construction experiments suggested that amino acids 338–348 might have a key role in MagA functionality; however, an indirect role (such as altered protein stability) cannot be excluded. In addition to magA, the 33-kb fragment contains genes involved in exopolysaccharide biosynthesis, export of the O-antigen of the lipopolysaccharides, glycosylation, and other modifications of cell envelope, and polysaccharide synthesis and transport. Further studies are needed to clarify the precise role of MagA in HV phenotype, and its interaction with components of bacterial surface envelopes such as exopolysaccharide, capsular polysaccharides, and lipopolysaccharides. Although the molecular details remain to be explored, the presence of MagA appears to alter the physicochemical properties of the bacterial surface such that serum complements cannot easily reach the bacterial cell membrane, and phagocytic ingestion also becomes more difficult. Being highly resistant to both serum killing and phagocytosis, these invasive enteric bacteria may reach the liver through the portal vein and cause primary liver abscess. If the invasion of K. pneumoniae cannot be contained within the liver, septic metastatic complications, such as metastatic meningitis or metastatic endophthalmitis, might occur. Our preliminary in vitro adherence assay found no evidence of adhesion between human hepatocyte-derived cell lines and these invasive K. pneumoniae strains. Thus, the predominant presentation of primary liver abscess in this disease may be the result of its enteric port of entry, rather than any real affinity between K. pneumoniae and liver. But further investigations are needed to make a definite conclusion.
The pathogenesis of bacterial diseases often involves both bacterial virulence and host factors (45). In the case of primary K. pneumoniae liver abscess, bacterial virulence is likely to play a major role because approximately half of the cases occur in otherwise healthy persons (4–10). In vitro serum resistance assays and in vivo animal inoculation experiments in the present work demonstrated the highly significant difference in virulence between clinical strains, and suggested a potential virulence of NTUH-K2044 to healthy subjects. There was a strong correlation between magA and clinical invasive diseases among tested clinical strains, including isolates from NTUH and other hospitals/sites outside Taiwan. Nevertheless, the host factors may still be important in some cases. The only magA-negative strain in the invasive strains group was isolated from the patient with liver cirrhosis and hepatic failure. Because of the unusual host factor in this case, this strain might not be a true invasive strain. The presence of magA in 29% of the strains that did not cause liver abscess should not be a surprise because the formation of a liver abscess might require an incubation time up to several days, as shown by the results of animal inoculation experiments in the present work. Prompt early antibiotic therapy could modify the clinical course and prevent the formation of liver abscess in some cases. Thus, these magA positive strains might not be truly “noninvasive.” Because patients with K. pneumoniae meningitis or endophthalmitis do not always have concomitant liver abscess (11, 46), there could be other bacterial as well as host and environmental factors involved. Further investigation of the involved host, and microbial and environmental factors is needed.
In the past two decades, primary K. pneumoniae liver abscess and its septic metastatic complications have emerged as one of the most common community-acquired bacterial diseases in Taiwan (4–12). Due to an increase in cases of metastatic K. pneumoniae meningitis secondary to primary liver abscess, K. pneumoniae has replaced Streptococcus pneumoniae as the leading pathogen of adult community-acquired bacterial meningitis in Taiwan (11, 47, 48). During this relatively short period, public sanitation in Taiwan has continued to improve, and there has been no major change in the population structure. The reason for this rapid increase of primary K. pneumoniae liver abscess remains unclear. One possible contributing factor is the documented overuse of amoxicillin and ampicillin in both agriculture and medical practice in Taiwan in the past 20 yr (49). K. pneumoniae is constitutively resistant to these two antibiotics (1, 2, 10). Their overuse will eliminate susceptible microorganisms and encourage the colonization of K. pneumoniae in both humans and animals. It is unknown whether this type of invasive K. pneumoniae strain has a survival advantage under antibiotic pressure, or whether they simply acquire their unique virulence genes from other bacteria. Identification of the virulence gene in K. pneumoniae strains causing primary liver abscess and septic metastatic complications will be helpful in tracing the origins of this emerging infectious disease.
In conclusion, magA meets the molecular Koch's postulates of a virulence gene for invasive K. pneumoniae strains causing primary liver abscess and septic metastatic complications. The gene is essential for invasive K. pneumoniae strains to demonstrate an extraordinarily high resistance to human serum and phagocytosis killing. This gene can be used as a marker for rapid molecular diagnosis, and might be also helpful in tracing the origins of this emerging infectious disease. The MagA protein could also become a good candidate for new drug targets in the future.
Acknowledgments
We thank Drs. Po-Ren Hsueh, Leung Kei Siu, Shih-Si Wang, Jann-Tay Wang, and Professor Ih-Jen Su for their gifts of the K. pneumoniae strains, and Dr. Arai (Sankyo Co. Ltd.) for providing us with globomycin.
This work was supported by grants from the National Science Council (NSC-91-2314-B-002-178 and NSC-92-3112-B-002-002) and from the National Taiwan University Hospital (NTUH-89-M009, NTUH-90-A09, and NTUH-92-021).
Notes
Chi-Tai Fang and Yi-Ping Chuang contributed equally to this work.
Abbreviations used in this paper: HV, hypermucoviscosity; LB, Luria-Bertani; ORF, open reading frame.
References
Articles from The Journal of Experimental Medicine are provided here courtesy of The Rockefeller University Press
Full text links
Read article at publisher's site: https://doi.org/10.1084/jem.20030857
Read article for free, from open access legal sources, via Unpaywall:
https://rupress.org/jem/article-pdf/199/5/697/1148733/jem1995697.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102684126
Article citations
Synergy of bacteriophage depolymerase with host immunity rescues sepsis mice infected with hypervirulent <i>Klebsiella pneumoniae</i> of capsule type K2.
Virulence, 15(1):2415945, 21 Oct 2024
Cited by: 0 articles | PMID: 39429194 | PMCID: PMC11497950
Isolation and characterization of lytic bacteriophage vB_KpnP_23: A promising antimicrobial candidate against carbapenem-resistant Klebsiella pneumoniae.
Virus Res, 350:199473, 01 Oct 2024
Cited by: 0 articles | PMID: 39332682 | PMCID: PMC11474366
RfaH contributes to maximal colonization and full virulence of hypervirulent <i>Klebsiella pneumoniae</i>.
Front Cell Infect Microbiol, 14:1454373, 17 Sep 2024
Cited by: 0 articles | PMID: 39364146 | PMCID: PMC11448354
Agnostic B cell selection approach identifies antibodies against K. pneumoniae that synergistically drive complement activation.
Nat Commun, 15(1):8100, 16 Sep 2024
Cited by: 0 articles | PMID: 39285158 | PMCID: PMC11405761
Genomic study substantiates the intensive care unit as a reservoir for carbapenem-resistant <i>Klebsiella pneumoniae</i> in a teaching hospital in China.
Microb Genom, 10(9), 01 Sep 2024
Cited by: 0 articles | PMID: 39325028 | PMCID: PMC11541224
Go to all (422) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (1 citation) ENA - AB117611
- (1 citation) ENA - AB085741
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection.
J Infect Dis, 192(1):117-128, 25 May 2005
Cited by: 102 articles | PMID: 15942901
magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of K. pneumoniae serotype K1.
J Med Microbiol, 55(pt 6):803-804, 01 Jun 2006
Cited by: 36 articles | PMID: 16687604
Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia.
J Intern Med, 259(6):606-614, 01 Jun 2006
Cited by: 121 articles | PMID: 16704562
Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed.
Virulence, 4(2):107-118, 09 Jan 2013
Cited by: 583 articles | PMID: 23302790 | PMCID: PMC3654609
Review Free full text in Europe PMC




