Abstract
Background
The aims of our study were to determine the correlation of the strain variation and degree of homogeneity of infecting Helicobacter pylori (H. pylori) with their disease outcomes, and the relevance of duodenal H. pylori expression of cagA and/or vacA gene to the development of duodenal ulcer in Korean patients.Methods
One hundred and twenty bacterial colonies isolated from different anatomical sites of the stomach and duodenum were used. The study population was consisted of 40 Korean patients, 21 with duodenal ulcer, 7 with gastric ulcer, 3 with combined gastric and duodenal ulcer, and 9 with chronic gastritis. Genomic characteristics of each strain were analyzed by random amplified polymorphic DNA (RAPD) fingerprinting. The cagA and vacA genes were detected by polymerase chain reaction (PCR).Results
PCR-based RAPD was proved to be a reliable method for the discrimination of individual bacterial genomic characteristics. Genomic fingerprinting showed a varying degree of inter- and intra-patient variation. Thirteen patients (32.5%) were colonized by a single strain throughout the corpus, antrum and duodenum, whereas the other 27 (67.5%) harbored multiple H. pylori strains. Thirty-six isolates (90.0%) each from the corpus and antrum, and 34 (85.0%) from the duodenum, expressed the cagA gene. The prevalence of duodenal H. pylori expression of the cagA gene was not different between patients with chronic gastritis and those with duodenal ulcer. All isolates were positive for both genes vacA s1 and vacA s1a.Conclusion
These results suggested that many of the H. pylori-infected Korean patients were actually colonized with mixed populations of different H. pylori strains and that the prevalence of duodenal H. pylori expression of the cagA and/or vacA gene was not correlated with the development of duodenal ulcer in Korean patients.Free full text

High Prevalence of Multiple Strain Colonization of Helicobacter pylori in Korean Patients: DNA Diversity Among Clinical Isolates from the Gastric Corpus, Antrum and Duodenum
Abstract
Background :
The aims of our study were to determine the correlation of the strain variation and degree of homogeneity of infecting Helicobacter pylori (H. pylori) with their disease outcomes, and the relevance of duodenal H. pylori expression of cagA and/or vacA gene to the development of duodenal ulcer in Korean patients.
Methods :
One hundred and twenty bacterial colonies isolated from different anatomical sites of the stomach and duodenum were used. The study population was consisted of 40 Korean patients, 21 with duodenal ulcer, 7 with gastric ulcer, 3 with combined gastric and duodenal ulcer, and 9 with chronic gastritis. Genomic characteristics of each strain were analyzed by random amplified polymorphic DNA (RAPD) fingerprinting. The cagA and vacA genes were detected by polymerase chain reaction (PCR).
Results :
PCR-based RAPD was proved to be a reliable method for the discrimination of individual bacterial genomic characteristics. Genomic fingerprinting showed a varying degree of inter- and intra-patient variation. Thirteen patients (32.5%) were colonized by a single strain throughout the corpus, antrum and duodenum, whereas the other 27 (67.5%) harbored multiple H. pylori strains. Thirty-six isolates (90.0%) each from the corpus and antrum, and 34 (85.0%) from the duodenum, expressed the cagA gene. The prevalence of duodenal H. pylori expression of the cagA gene was not different between patients with chronic gastritis and those with duodenal ulcer. All isolates were positive for both genes vacA s1 and vacA s1a.
Conclusion :
These results suggested that many of the H. pylori-infected Korean patients were actually colonized with mixed populations of different H. pylori strains and that the prevalence of duodenal H. pylori expression of the cagA and/or vacA gene was not correlated with the development of duodenal ulcer in Korean patients.
INTRODUCTION
Long-term colonization with genotypically single, homogeneous or multiple heterogeneous strains of Helicobacter pylori (H. pylori), causes chronic inflammation of the stomach and duodenum, although the magnitude of inflammation varies from strain to strain and from host to host. In 20 to 30% of infected patients, clinical end results range from chronic active gastritis to peptic ulceration, gastric adenocarcinoma and gastric MALToma1–3).
The data on the genomic characteristics of infecting H. pylori have shown that an individual person can be colonized by a strain with the same genomic DNA throughout the stomach and duodenum4–8), or by different strains with DNA heterogeneity in each organism found at different anatomical sites1, 9–14). Multiple strain infection of H. pylori occurs in both developed and developing countries. In addition, Campbell et al. suggested that separate bacterial lineages might have evolved in parallel with race-specific specialization15). Since the virulence differs between H. pylori strains, discrimination between the genomic characteristics of each isolate found in an individual with multi-strain infection may be important for diagnosis and treatment of infected patients16). Of the two representative techniques to discriminate between isolates of H. pylori, genotyping studies may be more useful since phenotypic expression of virulent determinants may be affected by storage and culture condition17). Among the most currently used techniques of typing systems based on polymerase chain reaction (PCR), random amplified polymorphic DNA (RAPD) analysis and repetitive extragenic palindromic DNA sequence-based PCR (REP-PCR) demonstrated optimal typeability (100%) and excellent discriminatory powers (0.99 to 1 and 0.99, respectively), while PCR-based restriction fragment length polymorphism (PCR-RFLP) analysis demonstrated a lower discriminatory power (0.70 to 0.97). Chromosome restriction-based typing methods, such as ribotyping and pulsed-field gel electrophoresis (PFGE), are limited by a low typeability (12.5 to 75%) that strongly decreases their discriminatory power (0.92 and 0.24 to 0.88, respectively)18).
Although there have been increasing reports opposed to the disease-specific significance of cagA and vacA19–23), several investigators still consider that these two genes are important risk factors in certain ethnic populations for developing more serious gastroduodenal diseases24–29).
The present study aimed to determine the strain variation of H. pylori by studying clinical isolates from four different patient groups and to determine the degree of homogeneity in organisms found at different anatomical sites within an individual patient. We also evaluated the significance of cagA, vacAs1 and vacAs1a as virulent determinants for developing peptic ulcer diseases.
MATERIALS AND METHODS
1. Population Studied
Among H. pylori-infected Korean patients with verified diagnoses, 40 patients were selected who simultaneously showed positive results for the culture of mucosal biopsies from all three of the corpus, antrum, and duodenal bulb.
The 40 patients consisted of 21 with endoscopically confirmed duodenal ulcer (DU, mean age 43.2 years; age range 13–69 years; 14 men and 7 women), 7 with benign gastric ulcer (GU, mean age 51.6 years; age range 30–66 years; 6 men and 1 woman), 3 with combined gastric and duodenal ulcer (GDU, mean age 53.7 years; age range 48–58 years; all men), and 9 with chronic gastritis (CG, mean age 49.7 years; age range, 30–73 years; 3 men and 6 women). Patients were excluded if they had a history of gastric surgery, were receiving steroids or NSAIDs, had taken H2-receptor antagonists, proton pump inhibitor, or antimicrobial agents within 30 days prior to study, or had active gastrointestinal bleeding. Patients with any other chronic illness were also excluded.
2. Clinical Isolation of H. pylori
A mucosal biopsy specimen was taken from each of the antrum, corpus and duodenal bulb in each patient. Antral and corpus biopsy specimens were taken at the greater curvature of the prepyloric region and the proximal corpus, respectively. Duodenal biopsy was taken at least 1 cm away from any ulcer margin. Each biopsy specimen was taken with a different pair of biopsy forceps. Between patients, the endoscope and biopsy forceps were washed automatically with a cycle including soaking in 2% glutaraldehyde for at least 4 minutes. In addition, the duodenal bulb was flushed several times with sterile water before sampling.
3. H. pylori Culture
One hundred and twenty clinical isolates were grown at 37°C in microaerophilic condition for 3 to 7 days. H. pylori were confirmed by typical Gram stain morphology and biochemical tests positive for urease, oxidase, and catalase.
4. Extraction of Genomic DNA from Clinical Isolates
The best-growing colony from each biopsy was incubated with STE buffer, 20% sodium dodecyl sulphate and proteinase K solution. An equal volume of phenol: chloroform solution and 1/10 of final volume of a 3M SDS solution, pH 5.3, was added. Double stranded DNA was precipitated with ethanol, the reaction mixture was allowed to stand at −80°C for 1 hour, and after centrifugation for 30 minutes at 12,000 g the resultant pellet was dissolved in 50 to 100 L of deionized water.
5. Detection of vacA and cagA Genes by PCR
PCR primers were designed on the basis of the published sequence of H. pylori vacA s1 (VA1-F/VA1-R ATGGAAATACAACAAACACAC and CTGCTTGAATGCGCCAAAC, nucleotide position 797–1055, size of product 259 bp), vacA s1a (SS1-F/VA1-R GTCAGCATCACACCGCAAC and CTGCTTGAATGCGCCAAAC, nucleotide position 866–1055, size of product 190 bp)26), and two different sets of cagA gene (set 1: primer cagA1, GATATAGCCACTACCACCACCG, nucleotide position 1249–1270; and primer cagA2, GGAAATCTTTAATCTCAGTTCGG, nucleotide position 1797–1819; size of product 570bp; set 2: cagA7, AGGAATCTCGCAATTAAGGG, nucleotide position 710–731; and cagA8 TTCTATGCCATTATGACTCCCC, nucleotide position 1446–1467; size of product 757bp)28). The cagA genes were amplified with 5 minutes at 94C; 40 cycles of 1 minute at 94°C, 1 minute at 55°C, and 1 minute at 68°C or 72°C; followed by 5 minutes at 72°C for extension. The reaction conditions for vacA s1 and s1a were as follows: 5 minutes at 94°C; 40 cycles of 1 minute at 52°C, 1 minute at 52°C, and 1 minute at 68C or 72°C; followed by 5 minutes at 72°C for extension.
6. H. pylori Genome Typing by RAPD-PCR
PCR-based RAPD fingerprinting was performed using the method described by Akopyanz et al1). The reaction was carried out in 50 L containing 20 ng of H. pylori genomic DNA, 3 mM MgCl2, 20 pM of primer, 2U of AmpliTaq DNA polymerase, 250 μM of each of dCTP, dGTP, dADP and dTTP in 1mM Tris-HCl, pH8.3, 50 mM KCl. For each primer, primer 1247 (AAGAGCCCGT), 1254 (CCGCAGCCAA), 1281 (AACGCGCAAC), or 1283 (GCGATCCCA), the following cycling program was used: 4 cycles of 94C for 5 minutes, 36°C for 5 minutes, and 72°C for 5 minutes; 30 cycles of 94°C for 1 minute, 36°C for 1 minute, and 72°C for 2 minutes; and then 72°C for 10 minutes. After PCR, 20 L aliquots of products were electrophoresed in 2% agarose gel containing 0.5 g/mL ethidium-bromide in the gel, and photographed under UV light.
7. Statistical Analysis
Chi square test and Fisher’s exact test were used to determine the significance of differences between the four groups. Unless stated otherwise, a p value less than 0.05 was taken to be significant. All statistical analyses were performed using SPSS for windows ver 9.0 (Chicago, Ill., U.S.A).
RESULTS
Genomic Fingerprinting of H. pylori Isolates
RAPD-PCR amplification resulted in 2 to 10 resolved DNA bands; 4 to 8 bands with primer 1281, 2 to 9 bands with primer 1283, 2 to 8 bands with primer 1254, and 2 to 10 bands with primer 1247. In this study, primers 1281 and 1283 yielded clearly distinct DNA fingerprints with multiple band differences but primers 1247 and 1254 did not (Figure 1A, ,1B).1B). Discrimination of individual isolates and their DNA typing was based on the characteristics of DNA fingerprints, and band differences and subtypes were defined when only one or two bands differed (Figure 2).
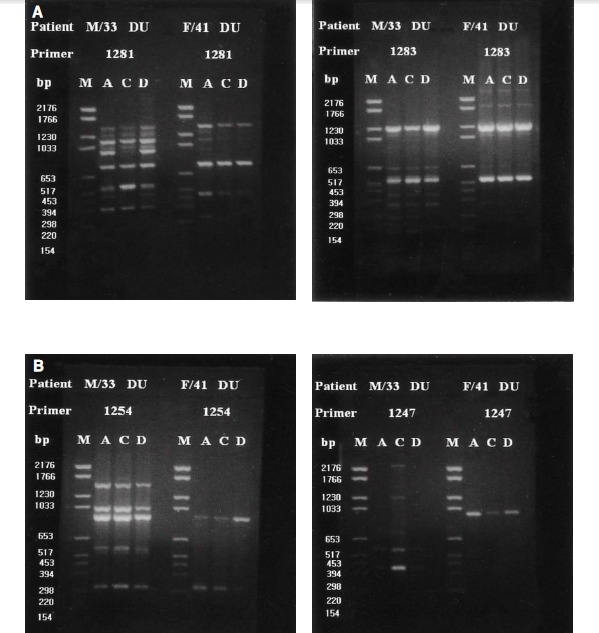
(A) Representative chromosomal DNA profiles of H. pylori isolated form 2 unrelated patients by RAPD-PCR DNA fingerprints obtained with primer 1281 and 1282. Column A, C, and D represent fingerprints of antral, corpus, and duodenal isolaeds of each patient, respectively. Primer 1281 and 1283 yielded clearly distinct DNA fingerprints with band differences.
(B) Representative chromosomal DNA profiles of H. pylori isolated form 2 unrelated patients by RAPD-PCR DNA fingerprints obtained with primer 1254 and 1247. Column A, C, and D represent fingerprints of antral, corpus, and duodenal isolaeds of each patient, respectively. DNA fingerprints obtained with primer 1254 and 1247 wrer less distinct than those of 1281 and 1283.
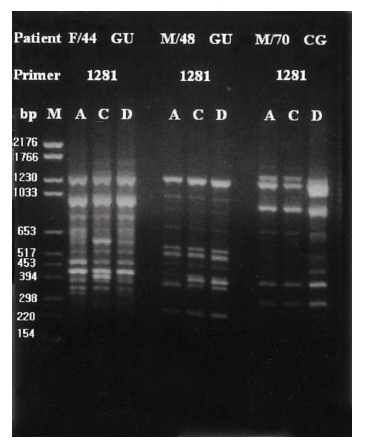
Representative chomosomal DNA subtypes of H. pylori isolated from 2 unrelated patients by RAPD-PCR DNA fingerprints obtained with primer 1281. Column A, C, and D represent fingerprints of antral, coupus, and duodenal isolates of each patient, respectively. DNA fingerprints showed slight variation in the banding pattern, i.e., bands missing or bands appearing with different intensity or at slightly different molecular weights.
RAPD-PCR fingerprintings of 120 isolates from 40 patients showed variable divergence of both inter-patient and intra-patient bacterial DNA characteristics. Many patients showed slight variations in the banding pattern, i.e., bands missing or bands appearing with different intensity or at slightly different molecular weights. On the basis of the fingerprint results, DNA patterns of the all isolates could be categorized into 5 different groups (Table 1).
Table 1.
Summary of RAPD-PCR fingerprints results according to disease status
| Group
| I | II | III | IV | V | Total |
|---|---|---|---|---|---|---|
| Disease Status | ||||||
| Chronic Gastritis | 4 | 1 | 2 | 2 | 0 | 9 |
| Duodenal Ulcer | 4 | 10 | 3 | 1 | 3 | 21 |
| Gastric Ulcer | 3 | 3 | 0 | 1 | 0 | 7 |
| Gastroduodenal Ulcer | 2 | 1 | 0 | 0 | 0 | 3 |
| Total | 13 | 15 | 5 | 4 | 3 | 40 |
Group I, H. pylori from the corpus, antrum and duodenum are all identical strains.
Group II, H. pylori from the corpus, antrum or duodenum are totally different strains from each other.
Group III, H. pylori only from the corpus and antrum are identical strains.
Group IV, H. pylori only from the antrum and duodenum are identical strains.
Group V, H. pylori only from the corpus and duodenum are identical strains.
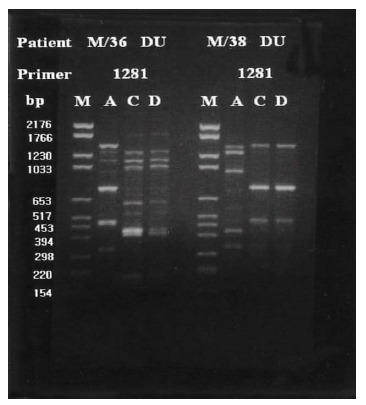
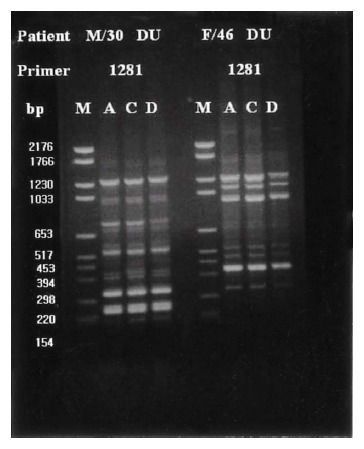
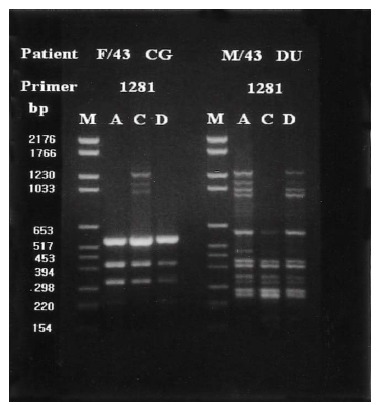
Group I patients (13; 4 DU, 3 GU, 2 GDU and 4 CG) were colonized by H. pylori with the same genomic DNA throughout the corpus, antrum and duodenum (Figure 3). Group II patients (15; 10 DU, 3 GU and 1 GDU and 1 CG) showed multi-strain infection, i.e., DNA patterns of H. pylori from the corpus, antrum and duodenum were totally different from each other within an individual patient (Figure 4). In five patients (Group III, 3 DU and 2 CG), only H. pylori from the corpus and antrum were an identical strain (Figure 5). Group IV patients (4; 1 DU, 1 GU and 2 CG) showed only infecting H. pylori from the antrum and duodenum were identical (Figure 6). Three DU patients (Group V) showed only H. pylori from the corpus and duodenum were identical (Figure 7).
Representative chromosomal DNA profiles of antral (column A), corpus (column C), and duodenal (column D) H. pylori isolated from 2 unrelated patients. RAPD-PCR DNA fingerprints from different anatomical sites of each patient showed homegeneous genomic patterns.
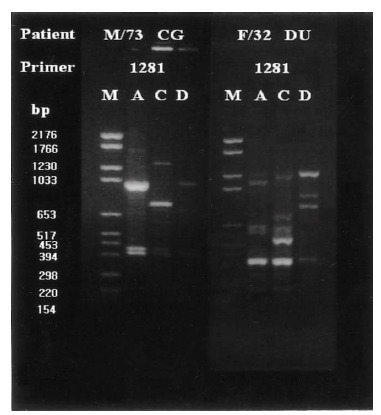
Representative chromosomal DNA profiles of antral (column A), corpus (column C), and duodenal (column D) H pylori isolated from 2 unrelated patients. RAPD-PCR DNA fingerprints from different anatomical sites of each patient showed totally different genomic patterns.
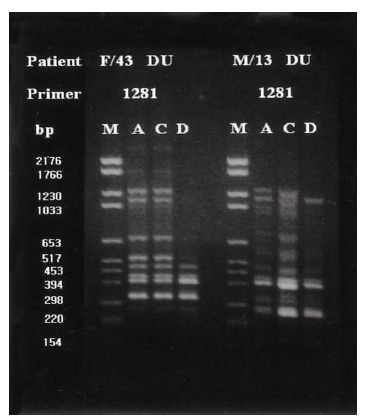
Representative chromosomal DNA profiles of antral (column A), corpus (column C), and duodenal (column D) H. pylori isolated from 2 unrelated patients. Isolates only from and corpus showed identical RAPD-PCR DNA fingerprints.
Representative chromosomal DNA profiles of antral (column A), corpus (column C), and duodenal (column D) H. pylori isolated from 2 unrelated patients. Isolates only from antrum and duodenum showed identical RAPD-PCR DNA fingerprints.
Detection of cagA and vacA Genes
Thirty-six isolates (90.0%) each from the corpus and antrum, and 34 (85.0%) from the duodenum, expressed cagA gene. H. pylori strains positive for cagA from the corpus were found in all of the patients with CG (100%), in 18 of 21 (85.7%) with DU, and in 6 (85.7%) of 7 with GU. Antral strains positive for cagA were found in 8 (88.9%) patients with CG, 18 (85.7%) with DU and 7 (100%) with GU. cagA+ duodenal strain were found in 8 CG (88.9%), 18 DU (85.7%) and 5 GU patients (71.4%) (Table 2). Out of thirty-six patients who were infected by cagA+ H. pylori, 11 (30.6%) were below the age of 29.
Table 2.
Relevance of cagA expression to clinical outcomes
| Disease | No. | cagA Expression | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Corpus | Antrum | Duodenum | |||||
|
|
|
| |||||
| Status | Patients | No. Positive | No. Negative | No. Positive | No. Negative | No. Positive | No. Negative |
| Chronic | |||||||
| Gastritis | 9 | 9 (100%) | 0 | 8 (88.9%) | 1 (11.1%) | 8 (88.9%) | 1 (11.1%) |
| Duodenal | |||||||
  Ulcer Ulcer | 21 | 18 (85.7%) | 3 (14.3%) | 18 (85.7%) | 3 (14.3%) | 18 (85.7%) | 3 (14.3%) |
| Gastric | |||||||
  Ulcer Ulcer | 7 | 6 (85.7%) | 1 (14.3%) | 7 (100%) | 0 | 5 (71.4%) | 2 (28.6%) |
| Gastroduodenal | |||||||
| Ulcer | 3 | 3 (100%) | 0 | 3 (100%) | 0 | 3 (100%) | 0 |
|
| |||||||
| Total | 40 | 36 (90.0%) | 4 (10.0%) | 36 (90.0%) | 4 (10.0%) | 34 (85.0%) | 6 (15.0%) |
All of the 120 strains isolated from the gastric corpus, antrum and duodenum were positive for both the vacA s1 and vacA s1a genes.
DISCUSSION
There have been two opposite opinions on the genomic characteristics of H. pylori strains colonizing the gastroduodenal mucosa. An individual patient can be colonized by either a single strain or a genetically predominant strain with the same genomic DNA, or by different strains which show the genomic heterogeneity of their DNA sequences found at different anatomical sites within an individual patient. Furthermore, some researchers have reported that infecting H. pylori in the duodenal mucosa in a particular gastroduodenal disease (i.e., DU) are genomically different strains from those of simple gastritis patients and that specific PCR products are associated with isolates from a particular disease and may contain genes encoding potential virulence factors9, 15, 28).
The principal objectives of our study were to determine the prevalence of inter- and intra-patient variation of genomic characteristics of H. pylori colonizing at different anatomical sites of the stomach and duodenum and their relevance to clinical outcomes. Along with this aim, we also evaluated the significance of cagA and vacA genes as virulent determinants for developing peptic ulcer diseases in Korean patients. We analyzed DNA patterns of 120 H. pylori strains cultured from different anatomical sites of the stomach and duodenum of patients with DU, GU, GDU, and CG by PCR-based RAPD fingerprints.
The results confirmed that the genotypic comparison by RAPD-PCR fingerprinting between isolates collected from different patients or different anatomic sites within the same patient is a reliable method for discrimination of their genomic characteristics since the PCR results obtained with the primers 1281 and 1283 yielded clearly distinct DNA patterns with multiple band differences. We speculated that H. pylori subtypes showing minor genomic variations might have resulted from the existence of a subclone that probably arose from the same original strain.
Our study revealed that there were variable degrees of genomic DNA diversity between clinical isolates. Thirteen (4 CG, 4 DU, 3GU &2GDU) out of 40patients (32.5%) harbored a single strain of H. pylori throughout their stomach and duodenum, while 27 (67.5%) showed heterogeneous DNA fingerprints, indicating multi-strain infection at different anatomical sites within an individual patient. Fifteen of the 40 (37.5%) harbored totally different strains of H. pylori, i.e., the genomic DNA fingerprints of the organisms isolated from the corpus, antrum and duodenum were all different from each other. In the remaining 12 patients (30.0%), one or two isolates from different biopsy sites showed genetically different fingerprints. Although the patient sample was small, our study showed an interesting result that duodenal H. pylori isolated from 13 out of 21 DU patients (61.9%) were genotypically different from those isolated either from the corpus or antrum. However, the prevalence of multi-strain infection in the duodenum and stomach of either CG (3 of 9, 33.3%) or GU patients (3 of 7, 42.9%) was not different from that of DU patients (p>0.1). These findings suggested that the strain variation of infecting H. pylori and the degree of homogeneity in organisms within an individual Korean patient were not correlated with their disease outcomes. Thoreson et al. also reported similar results of a mixed population of different H. pylori strains with marked variation, both genotypically and phenotypically, colonizing the same DU patient14).
The genetic diversity in the H. pylori population could be due to, i) the diversity of the species worldwide, ii)the large number of co-existing variants, or iii) the reports of natural transformation and genetic rearrangements or alterations in H. pylori within an individual host during colonization or adaptation to the host30). Furthermore, the case of maintaining the same DNA fingerprints may be a result of, i) the continuous evolution which occurs within the stomach of the infected person, because of nucleotide mutations, ii) excision of the cagA pathogenicity island (PAI), iii) transposition of insertion elements of cag PAI, iv) recombination with DNA from incoming strains that do not establish a chronic infection, and v) horizontal transfer of new genes3, 31–34).
The present study showed that the majority of patients irrespective of their disease states were infected with cagA+ H. pylori, and that the frequency of infection with cagA− strains was very low. Hamlet et al. suggested that a high density of cagA+ strains in the duodenum with severe duodenitis was an important determinant of DU disease, because their DU patients had a much higher prevalence of cagA+ strains in the duodenum than the asymptomatic subjects did (81% vs. 30%), despite a similar prevalence of cagA+ strains in the antrum (86% vs. 75%) [28]. In the present study, however, no association between the presence of cagA+ strain in the duodenum and the development of DU in Korean patients was evident since the prevalence of cagA+ duodenal H. pylori of DU patients was not significantly higher than that of CG patients (88.9% vs. 85.7%, p>0.05).
The reason for the low frequency of infection with cagA− strains in Korean patients is difficult to explain. We proposed two possibilities: the predominant strains naturally found in Korea might be cagA+ H. pylori, or, as seen in Covacci’s proposal, the decreased frequency of excision of cag PAI by the cagA+ strains with reduced generation of isogenic cagA− strains or to certain host factors that restricts the growth of cagA− strains35). Following from our previously reported results23), this study also showed that infection with H. pylori strains positive for vacA gene was not correlated with clinical outcomes since both vacA s1 and vacA s1a genes were also expressed in all of the 120 isolates.
On the basis of these observations, we concluded that the majority of H. pylori-infected Korean patients were actually colonized with mixed populations, but often with a single strain, and that the prevalence of duodenal H. pylori expression of cagA and/or vacA was not correlated with the development of DU disease in Korean patients.
Acknowledgments
Chung-Ang University Research Grants supported this research in 2002.
REFERENCES
Articles from The Korean Journal of Internal Medicine are provided here courtesy of Korean Association of Internal Medicine
Full text links
Read article at publisher's site: https://doi.org/10.3904/kjim.2004.19.1.1
Read article for free, from open access legal sources, via Unpaywall:
https://www.kjim.org/upload/kjim-19-1-1-1.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.3904/kjim.2004.19.1.1
Article citations
High prevalence of Helicobacter pylori mixed infections identified by multilocus sequence typing in Ningbo, China.
Front Microbiol, 14:1207878, 08 Aug 2023
Cited by: 1 article | PMID: 37614601 | PMCID: PMC10442550
Increased Risk of Severe Gastric Symptoms by Virulence Factors vacAs1c, alpA, babA2, and hopZ in Helicobacter pylori Infection.
J Microbiol Biotechnol, 31(3):368-379, 01 Mar 2021
Cited by: 6 articles | PMID: 33622995 | PMCID: PMC9705970
Helicobacter pylori cagA+ Is Associated with Milder Duodenal Histological Changes in Chilean Celiac Patients.
Front Cell Infect Microbiol, 7:376, 23 Aug 2017
Cited by: 6 articles | PMID: 28879170 | PMCID: PMC5572207
Genomic variability of Helicobacter pylori isolates of gastric regions from two Colombian populations.
World J Gastroenterol, 23(5):800-809, 01 Feb 2017
Cited by: 9 articles | PMID: 28223724 | PMCID: PMC5296196
Platelet Count Response to Helicobacter pylori Eradication in Iranian Patients with Idiopathic Thrombocytopenic Purpura.
Mediterr J Hematol Infect Dis, 4(1):e2012056, 10 Aug 2012
Cited by: 13 articles | PMID: 22973500 | PMCID: PMC3435127
Go to all (12) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Dominant cagA/vacA genotypes and coinfection frequency of H. pylori in peptic ulcer or chronic gastritis patients in Zhejiang Province and correlations among different genotypes, coinfection and severity of the diseases.
Chin Med J (Engl), 118(6):460-467, 01 Mar 2005
Cited by: 14 articles | PMID: 15788126
Clinical and histological associations of cagA and vacA genotypes in Helicobacter pylori gastritis.
J Clin Pathol, 51(1):55-61, 01 Jan 1998
Cited by: 64 articles | PMID: 9577374 | PMCID: PMC500433
Helicobacter pylori cagA, iceA and vacA status in Taiwanese patients with peptic ulcer and gastritis.
J Gastroenterol Hepatol, 18(11):1244-1249, 01 Nov 2003
Cited by: 15 articles | PMID: 14535980
Prevalence of cagA and vacA among Helicobacter pylori-infected patients in Iran: a systematic review and meta-analysis.
J Infect Dev Ctries, 9(7):686-696, 30 Jul 2015
Cited by: 15 articles | PMID: 26230117
Review