Abstract
Free full text

Detection of Inducible Clindamycin Resistance of Staphylococci in Conjunction with Performance of Automated Broth Susceptibility Testing
Abstract
This study has shown that inducible clindamycin resistance in staphylococci can be detected by disk testing on sheep blood agar inoculum purity plates used with the bioMerieux VITEK 2. Tests of 150 erythromycin-resistant isolates correlated with standard D-zone tests on Mueller-Hinton agar and with PCR for erm(A), erm(C), and msr(A).
Many clinical microbiology laboratories perform routine antimicrobial susceptibility testing either with a rapid automated instrument method or by broth microdilution. While broth-based test methods work well for the detection of the majority of antimicrobial resistance mechanisms, there has been increasing awareness that inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative Staphylococcus species may not be detected by standard tests (1, 4, 9, 10). Macrolide resistance in staphylococci may be due to ribosomal target modification that affects the activities of both macrolides and clindamycin, called macrolide-lincosamide-streptogramin B (MLSB) resistance, which is encoded by either erm(A) or erm(C) (2, 5, 6). While strains that demonstrate constitutive resistance to clindamycin can normally be detected by standard susceptibility testing methods, inducible resistance (MLSBi) present in some strains is not routinely detected by standard broth- or agar-based susceptibility test methods (9, 10). It is important to distinguish the MLSBi strains from macrolide-resistant strains that contain the gene msr(A), encoding an efflux pump that affects only macrolides, not clindamycin (2, 10).
Recently, we described a practical method for detection of MLSBi strains that involves simply placing standard erythromycin and clindamycin disks in adjacent positions on a Mueller-Hinton agar plate when the National Committee for Clinical Laboratory Standards disk diffusion test is performed (3). While this is a simple maneuver for laboratories that perform disk diffusion as their standard method for testing staphylococcal clinical isolates, it would constitute an additional test for laboratories that routinely use a broth-based method. The purpose of this study was to devise a simple procedure for performing disk induction testing in conjunction with broth-based susceptibility test methods.
A group of 150 erythromycin-resistant clinical isolates (75 S. aureus and 75 coagulase-negative staphylococci) were selected from among isolates previously characterized by disk induction testing and by PCR for the erm(A), erm(C), and msr(A) genes (3). These included 48 S. epidermidis, 15 S. haemolyticus, 3 S. auricularis, 3 S. hominis, 2 S. simulans, 2 S. warneri, and 2 S. capitis strains. Isolates were tested with the bioMerieux VITEK 2 instrument and standard AST-GP susceptibility cards in accordance with the manufacturer's protocol (bioMerieux, Durham, N.C.). A 0.5 McFarland standard suspension of each isolate was prepared and transferred to the VITEK 2 instrument for further dilution and card filling. After card inoculation, the tube containing the autodiluted (~1:10) inoculum suspension was streaked for confluent growth across the first one-third of the surface of a standard tryptic soy-5% sheep blood agar plate (Becton Dickinson, Cockeysville, Md.) and then streaked for isolated colonies on the remaining agar surface with the larger of the two VITEK 2 straws left in the inoculum tube. A 15-μg erythromycin disk (Becton Dickinson) and 2-μg clindamycin disk (Becton Dickinson) were placed on the plate in the area streaked for confluent growth, with a distance from disk edge to disk edge of 15 mm. After incubation for 16 to 20 h at 35°C in ambient air, the zones of inhibition were examined to detect any flattening of the shape of the clindamycin zone (D-zone), indicating inducible resistance. Examples of positive and negative induction tests with the inoculum purity plates are shown in Fig. Fig.11.
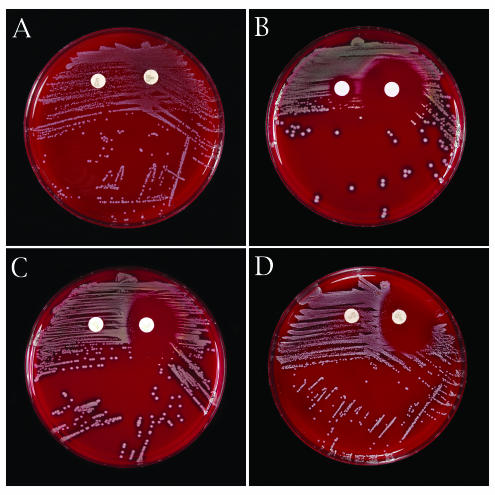
Purity plate disk induction testing. (A) Constitutive clindamycin resistance by purity plate testing. Growth extends to the edge of the clindamycin disk with an S. auricularis strain carrying erm(A). (B) Negative disk induction test indicating the absence of inducible clindamycin resistance, i.e., no blunting of the clindamycin zone adjacent to the erythromycin disk with an S. aureus carrying msr(A). (C) Positive disk induction test indicating inducible clindamycin resistance. Blunting of the clindamycin inhibition zone adjacent to the erythromycin disk (D-zone effect) with an S. epidermidis carrying erm(C). (D) Positive disk induction test indicating inducible clindamycin resistance with an S. aureus carrying erm(C).
A comparison of the purity plate induction tests, standard disk induction tests on Mueller-Hinton agar, and the VITEK 2 susceptibility results is shown in Table Table1.1. All isolates with inducible clindamycin resistance by the standard method and possessing either erm(A) or erm(C) were readily detected in the purity plate induction test. These included 22 S. aureus [4 with erm(A) and 18 with erm(C)] and 26 coagulase-negative staphylococci [5 with erm(A), 19 with erm(C), one with both erm(A) and msr(A), and one with erm(C) and msr(A)]. All 48 isolates had clindamycin MICs of ≤0.5 μg/ml by VITEK 2 but showed flattening of the clindamycin zone on the purity plate. All 62 isolates with constitutive clindamycin resistance (25 S. aureus and 37 coagulase-negative staphylococci) were detected by VITEK 2 (61 had MICs of ≥4 μg/ml) and demonstrated either no zone of inhibition on the purity plates or a heavy ingrowth pattern around the clindamycin disks. Eight isolates showed a double zone of growth, with flattening of the outer zone and heavy inner growth up to the edge of the disk; all of these isolates contained erm(A). All eight isolates were reported as clindamycin resistant by VITEK 2. One S. simulans isolate contained only msr(A) but showed constitutive clindamycin resistance by disk diffusion testing by an unknown mechanism and yielded a MIC of 1 μg/ml in the VITEK 2. None of the isolates carrying only msr(A) showed flattening of the clindamycin zone on the purity plate tests.
TABLE 1.
Purity plate disk induction testing for clindamycin resistance compared to standard disk induction testing on Mueller-Hinton agar and results of VITEK 2 susceptibility testing
| Species (no. of isolates) | Phenotypea | No. of strains | No. (%) of strains detected by purity plate induction testb | No. (%) of strains detected by VITEK 2 aloneb |
|---|---|---|---|---|
| S. aureus (75) | CS | 28 | 0 | 0 |
| CR-Ind | 22c | 22 (100) | 0 | |
| CR-Const | 25 | 25d (100) | 25 (100) | |
| Coagulase-negative staphylococci (75) | CS | 12 | 0 | 0e |
| CR-Ind | 26 | 26 (100) | 0e | |
| CR-Const | 37 | 37 (100) | 37f (100) |
This study demonstrated that inducible clindamycin resistance can be easily detected by disk induction testing on standard sheep blood agar plates used for verification of inoculum purity in conjunction with an automated susceptibility test system. It is important to note that clindamycin zone flattening, not zone size, is assessed on the purity plate. Thus, the use of standard sheep blood agar rather than Mueller-Hinton agar suffices for recognition of inducible resistance. While we evaluated purity plate induction testing with the bioMerieux VITEK 2 system in this study, any broth-based antimicrobial susceptibility test method, manual or automated, should be amenable to this approach. We evaluated an approximately 1:10 dilution of a 0.5 McFarland standard as the inoculum source. We also found that direct plating of the 0.5 McFarland standard or a dilution of up to 1:250 of the McFarland standard worked equally well (data not depicted).
The goal of routine detection of clindamycin resistance among clinically significant staphylococcal isolates is twofold. First, prior investigations have demonstrated the potential for clinical failures when patients infected with MLSBi strains are treated with clindamycin for various types of infections (1, 4, 10). However, to categorically regard all macrolide-resistant staphylococci as clindamycin resistant would deny potentially safe and effective therapy for patients infected with isolates that carry only the macrolide efflux mechanism. The percentage of clinical staphylococcal isolates that demonstrate macrolide efflux compared to MLSB resistance may vary widely by geographic location or patient group (2, 3, 4, 5, 6, 8, 9, 10). Therefore, the second benefit of routine testing for inducible clindamycin resistance is to clearly identify those strains that remain susceptible to clindamycin despite macrolide resistance. For these reasons, routine testing of significant staphylococcal isolates for inducible clindamycin resistance is now advocated by the National Committee for Clinical Laboratory Standards (7).
Acknowledgments
This study was supported in part by a grant from bioMerieux, Inc., Durham, N.C.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.42.4.1800-1802.2004
Read article for free, from open access legal sources, via Unpaywall:
https://jcm.asm.org/content/jcm/42/4/1800.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Article citations
Comparative Performances of Vitek-2, Disk Diffusion, and Broth Microdilution for Antimicrobial Susceptibility Testing of Canine Staphylococcus pseudintermedius.
J Clin Microbiol, 59(9):e0034921, 18 Aug 2021
Cited by: 6 articles | PMID: 34132581 | PMCID: PMC8373018
Bacterial Antibiotic Resistance: The Most Critical Pathogens.
Pathogens, 10(10):1310, 12 Oct 2021
Cited by: 297 articles | PMID: 34684258 | PMCID: PMC8541462
Review Free full text in Europe PMC
Ceftriaxone- and ceftazidime-resistant Klebsiella species, Escherichia coli, and methicillin-resistant Staphylococcus aureus dominate caesarean surgical site infections at Mulago Hospital, Kampala, Uganda.
SAGE Open Med, 8:2050312120970719, 10 Nov 2020
Cited by: 8 articles | PMID: 35154757 | PMCID: PMC8826261
Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Benin.
Ann Clin Microbiol Antimicrob, 19(1):54, 25 Nov 2020
Cited by: 7 articles | PMID: 33239061 | PMCID: PMC7687776
Detection of inducible and constitutive clindamycin resistance among Staphylococcus aureus isolates in a tertiary care hospital, Eastern India.
Avicenna J Med, 6(3):75-80, 01 Jul 2016
Cited by: 7 articles | PMID: 27390669 | PMCID: PMC4922212
Go to all (40) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci.
J Clin Microbiol, 41(10):4740-4744, 01 Oct 2003
Cited by: 216 articles | PMID: 14532213 | PMCID: PMC254362
Selection of strains for quality assessment of the disk induction method for detection of inducible clindamycin resistance in Staphylococci: a CLSI collaborative study.
J Clin Microbiol, 43(6):2613-2615, 01 Jun 2005
Cited by: 20 articles | PMID: 15956373 | PMCID: PMC1151953
Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus.
J Clin Microbiol, 43(4):1716-1721, 01 Apr 2005
Cited by: 81 articles | PMID: 15814990 | PMCID: PMC1081368
Agar dilution method for detection of inducible clindamycin resistance in Staphylococcus spp.
J Clin Microbiol, 45(12):4018-4020, 17 Oct 2007
Cited by: 8 articles | PMID: 17942656 | PMCID: PMC2168581




