Abstract
Free full text

Maintaining cholesterol homeostasis: Sterol regulatory element-binding proteins
Abstract
The molecular mechanism of how hepatocytes maintain cholesterol homeostasis has become much more transparent with the discovery of sterol regulatory element binding proteins (SREBPs) in recent years. These membrane proteins are members of the basic helix-loop-helix-leucine zipper (bHLH-Zip) family of transcription factors. They activate the expression of at least 30 genes involved in the synthesis of cholesterol and lipids. SREBPs are synthesized as precursor proteins in the endoplasmic reticulum (ER), where they form a complex with another protein, SREBP cleavage activating protein (SCAP). The SCAP molecule contains a sterol sensory domain. In the presence of high cellular sterol concentrations SCAP confines SREBP to the ER. With low cellular concentrations, SCAP escorts SREBP to activation in the Golgi. There, SREBP undergoes two proteolytic cleavage steps to release the mature, biologically active transcription factor, nuclear SREBP (nSREBP). nSREBP translocates to the nucleus and binds to sterol response elements (SRE) in the promoter/enhancer regions of target genes. Additional transcription factors are required to activate transcription of these genes. Three different SREBPs are known, SREBPs-1a, -1c and -2. SREBP-1a and -1c are isoforms produced from a single gene by alternate splicing. SREBP-2 is encoded by a different gene and does not display any isoforms. It appears that SREBPs alone, in the sequence described above, can exert complete control over cholesterol synthesis, whereas many additional factors (hormones, cytokines, etc.) are required for complete control of lipid metabolism. Medicinal manipulation of the SREBP/SCAP system is expected to prove highly beneficial in the management of cholesterol-related disease.
INTRODUCTION
The view of cholesterol as a nasty substance which clogs arteries and causes heart disease is wide-spread, but it does not do the molecule justice. Not only is it a vital component of cell membranes without which the cell cannot function, but it is also the precursor to all steroid hormones, bile acids, and oxysterols, which by themselves are important regulatory molecules in many metabolic pathways.
Cholesterol and fatty acids as building blocks of cell membranes are synthesized via regulated pathways. All cells must control these pathways in order to maintain levels within physiological boundaries. Excessive amounts of cholesterol in cells can destroy membrane function, precipitate as crystals which will kill the cell or result in atherosclerotic damage if spread to blood[1]. However, the original view of random distribution of cholesterol and lipids in the cell membrane no longer holds: Not only differs the lipid composition of the outer leaflet of the plasma membrane from the inner one, but the distribution of lipids and cholesterol in the outer leaflet is organized into domains, with so-called rafts[2] and caveolae[3] being rich in cholesterol and sphingomyelin. These structures play intricate roles in cholesterol trafficking to maintain cellular homeostasis, and they are also components of the cellular signalling system. The membranes of endoplasmic reticulum (ER) and Golgi, on the other hand, contain comparatively little cholesterol, a factor important in its own homeostasis, and one objective of this overview.
The understanding of cholesterol regulation has come a long way from the initial recognition of cholesterol feedback inhibition of its rate-limiting synthetic enzyme, 3-hydroxy-3-methylglutaryl coenzyme A (HMGCoA) reductase, through the role of lipoproteins in maintaining plasma cholesterol levels, to the recent discoveries of regulation of cholesterol synthesis via sterol-sensitive response elements (SREs), and degradation via liver X receptor (LXR) - or bile acid receptor (BAR) - regulated pathways.
Lipid homeostasis via SREs in animal cells is achieved by a family of transcription factors called SRE-binding proteins (SREBPs). SREBPs activate directly the expression of some 30-plus genes participating in the metabolism mostly of lipids, but also glucose. Activation of these originally membrane-bound transcription factors involves a proteolytic cascade through which the SREBP molecule is released from the membrane and obtains its mature form as a transcription factor. The active SREBP enters the nucleus and binds to those special DNA sequences, the SREs, in the promoter regions of many different genes.
In the health arena, SREBPs stand at a crucial point: They regulate expression of the LDL receptor, the molecule which enables the hepatocytes to remove cholesterol contained in LDL particles from the bloodstream. High (dietary) cholesterol prevents maturation of SREBPs and not only cuts off cholesterol synthesis, but also LDL receptor synthesis, resulting in high blood cholesterol and the imminent danger of atherosclerotic plaque formation. At this point in time the so-called statins, drugs which block HMGCoA reductase, another target of SREBP-mediated gene expression, are the most effective way to interrupt this vicious circle[4].
This brief overview is concerned with the SREBP-mediated control of cholesterol and lipid synthesis. Lipid synthesis is subject to many other regulatory influences which cannot be addressed here, nor can the degradation of cholesterol via hormone or bile acid synthesis be covered, which again commands its own set of regulatory substances. The focus of this minireview is to present the latest data on the SREBP-induced mechanism. Recent in-depth reviews have been available[5-9].
OXYSTEROLS AS NEGATIVE AND POSITIVE REGULATORY MOLECULES
Cholesterol is not the only sterol inside the cell to act as a regulatory substance, many of its hydroxylated derivatives, the oxysterols, share these important functions. Oxysterols exert a feed-back effect and down-regulate cholesterol synthesis, to be detailed in the following sections of this overview, but they also up-regulate their own metabolism and elimination via a feed-forward effect which will be briefly described in this section.
Cholesterol directly affects two enzymes which are vital in its own removal. It activates acyl-CoA: Cholesterol acyl transferase (ACAT), the enzyme required to synthesize cholesteryl esters, its major storage form, and it also activates cholesterol 7α - hydroxylase (CYP7A1), the initial and rate-limiting enzyme of bile acid synthesis, the leading pathway of cholesterol elimination[10].
The side chain-hydroxylated 27-hydroxycholesterol is presumed to function in cholesterol homeostasis by sustaining HDL-mediated reverse cholesterol transport, the process by which cholesterol is brought back to the liver from peripheral tissues[11]. The most important action of oxysterols, however, is to serve as ligands for nuclear orphan receptors. This family of proteins has been recognized as nuclear hormone receptors based on their molecular properties, but since ligands were initially not known, they were named orphan receptors. Most important among these is liver X receptor (LXR), which controls expression of CYP7A1[12] as the rate-limiting step of bile acid synthesis[13,14], reverse cholesterol transport[15], trafficking of cholesteryl esters, and aids in lipid metabolism[16] and the intestinal absorption of cholesterol. This receptor displays specific requirements for the position of additional hydroxy group (s) on the cholesterol molecule, the most potent ligands being 20 (S) -OH-, 22 (R) -OH-, 20, 22-di-OH-, and 24-OH-cholesterol[17,18]. An overview of oxysterol actions is shown in Figure Figure11.
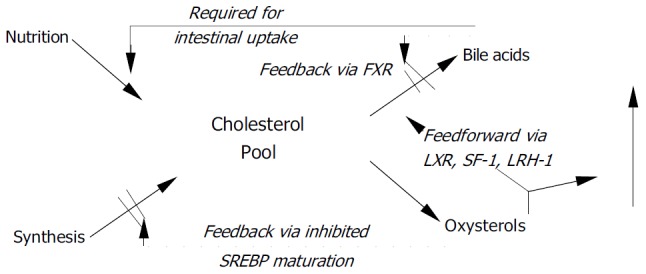
Feedforward and feedback effects of oxysterols FXR = farnesoid × receptor; LRH-1 = liver receptor homologue-1; LXR = liver × receptor; SF-1 = steroidogenic factor-1; SREBP = sterol responsive element binding protein.
Furthermore, oxysterols are ligands for steroidogenic factor-1 (SF-1) and liver receptor-homologue 1 (LRH-1), in which they regulate steroid hormone synthesis and sexual differentiation during prenatal development. SF-1 is limited to steroidogenic tissues, whereas LRH-1 occurs in liver and other tissues derived from the gut endoderm[7]. Oxysterols also play a role in meiosis[19]. Finally, these bile acids activate the farnesol X receptor (FXR)/retinoid X receptor (RXR)/receptor-interacting protein 14 (Rip14) system, FXR is also known as bile acid receptor (BAR). Binding of bile acids to FXR inhibits bile acids synthesis.
SREBPS, MEMBRANE-DERIVED TRANSCRIPTION FACTORS
SREBPs belong to the large family of basic helix-loop-helix-leucine zipper (bHLH-Zip) transcription factors. Three members of the SREBP family, SREBPs-1a, -1c, and -2, have been identified[20,21]. They are synthesized as inactive precursors. Isoforms -1a and -1c are produced from a single gene on human chromosome 17p11.2 by use of alternate promoters and splicing, resulting in different forms of exon-1[21-23]. SREBP-2 is encoded by a separate gene on human chromosome 22q13[20,24]. It has about 50% sequence identity with SREBP-1. Adipocyte determination differentiation factor (ADD)-1, a transcription factor which binds to E-boxes and promotes adipocyte differentiation in rats[25] is the homologue of human SREBP-1c[23].
Each nascent SREBP protein has a molecular size of about 125 ku and consists of about 1 150 amino acids (aa) in 3 functional domains. The first, NH2-terminal domain of SREBP contains the bHLH-Zip and an acidic domain, located at the very NH2 terminus. The acidic domain has to bind a transcription coactivator for function, it is shorter in SREBP-1c than in SREBP-1a, making SREBP-1c a weaker transcription activator[26]. The acidic domain is essential for function. When removed, the bHLH-Zip portion of SREBP still binds to DNA, but no longer activates transcription, thus acting as an inhibitor[27].
A djacent to the acidic DNA-binding domain is a variable area rich in serine, proline, glutamine, and glycine. Next follows the bHLH-Zip sequence, whose basic region mediates DNA binding. The rest of the bHLH-Zip motif imparts the ability to dimerize. Other bHLH-Zips tend to homo-dimerize, whereas SREBP needs other transcription factors such as SP-1 or NF-Y for full function. The remainder of the SREBP molecule has no analogy with other bHLH-Zip transcription factors[5].
SREBPs are embedded in membranes of the endoplasmic reticulum (ER) and the nuclear envelope, forming what has been called a hairpin shape[28,29]. The NH2-terminal portion of the SREBP molecule through the bHLH-Zip region protrudes into the cytosol. The following central portion of SREBP, the membrane anchoring region, is about 90 aa in length. It consists of two hydrophobic, membrane-spanning segments separated by a hydrophilic loop which extends into the lumen of the ER. The COOH terminal segment of about 590 aa again extends into the cytosol and serves as the regulatory domain for transformation into the mature, a transcriptionally active form also known as nuclear SREBP (nSREBP). Neither the membrane-spanning, nor the regulatory regions, are found in other bHLH-Zip transcription factors.
The NH2-terminal domain of SREBP binds to a sterol binding element (SRE) which must contain a direct, or tandem, repeat of the recognition sequence[30]. This is another evident difference to bHLH-Zip transcription factors, which will bind only to palindromic (i.e., tail-to-tail connected) repeats. SREBPs display no in vivo activity with palindromic sequences[5,19].
SREBP CLEAVAGE ACTIVATION PROTEIN (SCAP), AN ESCORT AND STEROL SENSOR PROTEIN
SREBPs attain biological activity only after being transferred to the membrane of the Golgi complex, where they are cleaved at the lumenal loop and the NH2-end is released from the membrane. They are confined to the membranes of the ER unless they are paired up with another protein which serves two functions: To sense the levels of sterols in the cell, and in response to low sterol levels, to escort SREBP to its place of activation. This is achieved by SREBP cleavage activating protein (SCAP), a protein originally cloned from a mutant cell line of Chinese hamster ovary cells which will not suppress SREBP cleavage even with very high sterol levels[31]. The mutation affects one specific codon, 443, where a C (ytosine) to G (uanine) transition on the DNA side replaces an aspartic acid by an asparagine in the amino acid sequence, rendering SCAP unresponsive to sterols[32].
SCAP has 1 276 amino acids[31] in two domains[33]: A membrane-spanning NH2 terminal domain of 730 aa, and a COOH terminal domain of 546 aa, which extends into the cytosol. The NH2-terminus contains eight hydrophobic sequences separated by short hydrophilic loops[31], the hydrophobic sequences are thought to span the membrane with the hydrophilic loops protruding at either side. SCAP shares this feature with HMGCoA reductase[34,35]. The membrane-spanning stretches comprise the sterol-sensing area, whereas the COOH-terminus contains all of the remaining biological activity[31]. The COOH-terminus is organized in five repeat sequences characteristic of the WD family. Wherever this sequence occurs, it mediates protein-protein interactions[36].
Newly synthesized SREBP forms a tight complex with the WD repeat domains of SCAP[37], but in the presence of oxysterols this complex is confined to the ER, establishing a feedback loop to control cholesterol synthesis. With oxysterol depletion, the SCAP/SREBP complex appears in vesicles budding from ER membranes[38] and translocates to another subcellular compartment, the Golgi[39-43]. Transfer of SREBP and proteolytic cleavage to release nSREBP cannot occur unless this complex has formed[42]. Oxysterols induce a change of the conformation of SCAP, and an additional protein has been postulated to retain SCAP in the ER once in its sterol-induced conformation[44]. Not only cholesterol, but also several other oxysterols display full activity in suppressing SCAP translocation into vesicles, but it has been suggested that cholesterol alone exerts the effect, while high levels of oxysterols simply force translocation of cholesterol from the plasma membrane to the ER[44]. Complete understanding could be of immense benefit in the management of hyperlipidemias.
SREBP ACTIVATION VIA TWO-STEP PROTEOLYTIC CLEAVAGE
The SREBP/SCAP-containing vesicles from the ER also contain a membrane-anchored serine protease of the subtilisin family, Site-1 protease (S1P), in an inactive form which becomes activated only during its transport to the Golgi[45]. The SREBP/SCAP complex and S1P now incorporate into the Golgi membrane. As the next step, the activated S1P attaches to the SCAP/SREBP complex and cuts the SREBP molecule right in the middle of its lumenal hydrophilic loop[29]. When active S1P is inserted into the ER, it will cleave SREBP without a need for SCAP[43].
To release active SREBP, another enzyme is required, Site-2 protease (S2P, a trans-membrane zinc metalloprotease). The cellular location of this enzyme is as yet unclear, but likely resides in the Golgi. S2P cuts the still membrane-anchored SREBP in a rather unusual place, viz., three amino acids into the membrane-spanning portion on the cytoplasmic side[46,47]. This process is known to regulate intra-membrane proteolysis (Rip)[48], and typically produces proteins which are transcriptionally active and participate in the control of various cellular processes[6]. S2P action is not directly affected by cellular oxysterol levels, since this enzyme cannot act unless S1P has separated the bHLH-Zip portion of SREBP from the regulatory COOH terminus[49]. S2P action results in the release of a mature, 68 kDa nSREBP, consisting of the bHLH-zipper domain with the first three membrane-spanning amino acids attached, which now can migrate to the nucleus and bind to a sterol-responsive element (SRE). SCAP is recycled back to the ER to chaperone another 125 kDa SREBP molecule to the Golgi[50], whereas the 68 kDa SREBP is degraded. Interestingly, the nuclear action of nSREBP induces new SREBP mRNA synthesis by way of SREs located in the promoter regions of their own genes[51]. A synopsis is shown in Figure Figure22.
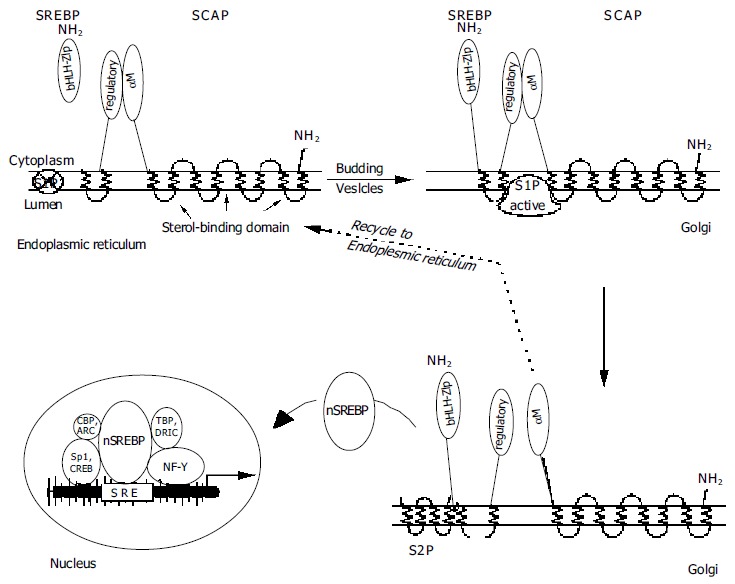
Maturation of SREBPs -NH2 = amino-terminal ends of SREBP or SCAP. S1P = Site-1 protease (crossed-out = inactive); S2P = Site-2 protease. SRE = sterol-responsive element; nSREBP = nuclear SREBP. Arrangement of the additional transcription factors NF-Y, Sp1, CREB, ARC, CBP, TBP, and DRIC (see text) is tentative as their requirements are not exactly known.
The process of SREBP cleavage and activation thus comprises 4 components which, upon mutation, can result in loss of oxysterol-sensitive regulation of cholesterol homeostasis. As a matter of fact, much of the knowledge of the SREBP regulatory system stems from experimentally mutated cell lines and subsequent selection for oxysterol resistance or cholesterol auxotrophy[8]. Oxysterol resistance has aided in cloning the genes for SREBP and SCAP, respectively. One type, class 1 mutation, produces an NH2-terminal bHLH-Zip portion of SREBP which is truncated even before the S2P cleavage site[52,53]. This molecule is therefore not membrane bound and can access the nucleus immediately after its synthesis, turning on the complete SREBP gene battery regardless of cellular oxysterol levels. Class 2 mutants contain a mutated SCAP molecule[54] which holds the SREBP/SCAP/S1P complex in a permanently active configuration, no matter how high the oxysterol level in the cell, resulting in permanent release of nSREBP with concomitant overproduction of cholesterol[31].
Cholesterol auxotrophy has been used as a selection criterion to clone the genes for S1P and S2P. With S1P[55], S2P[41,46,56] or SCAP[57] rendered non-functional, cells become dependent on exogenous cholesterol because they cannot produce the enzymes necessary for its biosynthesis.
It is interesting to know that the proteolytic activation of SREBP-1 and SREBP-2, respectively, can be regulated individually. In rodents, treatment with cholesterol suppressing drugs (statins) or with cholesterol-sequestering agents results in up-regulation and increased activation of SREBP-2 while reducing activation of SREBP-1[58]. Another interesting feature is that polyunsaturated fatty acids inhibit the proteolytic activation of SREBP-1, but contrary to the action of oxysterols, they have no effect on SREBP-2 maturation[59]. In addition, glucose metabolites such as glucose-6-phosphate may serve to accelerate maturation of SREBP-1c only[60], but mechanistic details have not been elucidated.
GENE ACTIVATION BY SREBPs, TARGET GENES
Immediately after the second cleavage of the SREBPs the now mature protein enters the nucleus where it binds to SREs in the promoters of the target genes and activates transcription. The SRE nucleotide sequence displays considerable variation among the promoters of the many genes activated by an SRE, a commonly found sequence is represented by 5’-nTCACnCCA Cn-3’ (cf.[6]). However, SREBP alone cannot activate transcription, but must act co-ordinately with additional transcription factors to obtain full activation of target genes.
Transcription factors known to activate SREs in conjunction with SREBP are Sp1, nuclear factor (NF)-Y, and cAMP response element binding protein (CREB)[10,61-64]. NF-Y interacts directly with SREBP[62], and Sp1 can stabilize the complex[61]. It appears that the promoters of different SRE-activated genes respond to different combinations of these transcription factors. Once a stable complex has formed in a promoter region, additional factors such as CREB binding protein (CBP)[65,66], activated recruited cofactor (ARC)[67], vitamin D receptor interacting protein (DRIC)[67] , or TATA box-binding protein (TBP)-associated factors may be recruited to initiate transcription. Histone acetylation is also required for full transcriptional activity[68]. Such complexes have been shown to exist with SREBP-1a and -2, but not with SREBP-1c. This may explain its rather weak potency to activate the SRE gene battery.
SREBP-activated genes predominantly belong to lipid metabolism pathways, viz., cholesterogenesis, fatty acid synthesis, lipogenesis, triglyceride and phospholipid synthesis, but also glucose metabolism. Yet, the three SREBPs do not activate identical gene batteries. Using transgenic mice over-expressing just one type of nSREBP it has been discovered that SREBPs-1a and -1c actions favour fatty acid synthesis, whereas SREBP-2 action favours cholesterol synthesis[5,69-71], but in vitro both SREBPs-1a and -2 can trigger expression of the complete set of enzymes required for cholesterol synthesis with similar potency[72]. While SREBPs-1a and -2 predominate in cultured cells, intact liver and most other tissues primarily express SREBP-1c and -2[9]. An overview of SREBP pathways is given in Figure Figure33.
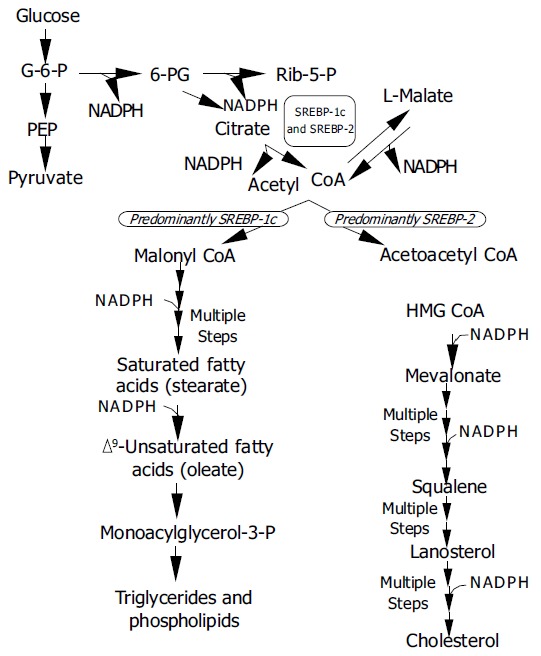
Metabolic pathways regulated by SREBPs G-6-P = glucose-6-phosphate; 6-PG = 6-phosphogluconate; Rib-5-P = ribulose-5-phosphate; PEP = phosphoenol pyruvate; CoA = coenzyme A; HMGCoA = 3-hydroxy-3-methylglutaryl coen-zyme A. SREBP-1c and SREBP-2 activate genes for the genera-tion of NADPH (ATP citrate lyase, malic enzyme, G-6-P dehydrogenase, 6-PG dehydrogenase) required in various steps of lipid synthesis.
SREBP-2 over-expression induces all 12 enzymes of the cholesterol biosynthetic pathway[72], most notably the mRNA for HMGCoA reductase, which may increase as many as 75-fold[73]. Overall cholesterol synthesis in such animals is up 28-fold, whereas fatty acid synthesis is increased only 4-fold. In contrast, expression of enzymes does not involve cholesterol synthesis but is related to cholesterol metabolism. Cholesterol 7α -hydroxylase (rate-limiting enzyme for bile acid synthesis) and acyl-CoA:Cholesterol acyltransferase (ACAT, catalyses cholesterol ester formation), are not activated[72].
SREBP-1a appears to be constitutively expressed in most tissues, with as yet no known factor to stimulate its low expression[26]. Over-expression in adult rats also resulted in over-stimulation of lipid synthesis, but in this case fatty acid synthesis was increased 26-fold, and cholesterol synthesis 5-fold. Since SREBP-1a and -1c (see below) also induce enzymes for fatty acid elongation and desaturation[73,74], SREBP-1a over-expression resulted in elevated hepatic levels of oleate[75].
SREBP-1c is predominantly involved in the regulation of adipogenesis and also in the regulation of insulin-responsive genes which control lipogenesis and glucose metabolism[76,77], but in vitro it does not stimulate cholesterol synthesis[75]. SREBP-1c mRNA synthesis responds to nutritional changes in parallel to insulin levels[78]. Insulin heightens the expression of SREBP-1c and its battery of genes involved in the synthesis of saturated and unsaturated fatty acids[75,79] as well as glucose metabolism genes[77,78]. The effect is opposed by glucagon and cyclic AMP. In short, available experimental data indicate that SREBP-1c mediates all effects of insulin on lipogenesis. In response to nutritional stimuli SREBP-1c also triggers expression of genes of enzymes required for fatty acid elongation[74], and of glycerol 3-phosphate acyltransferase required for triglyceride and phospholipid synthesis[6]. Last but not least SREBPs activate 3 genes necessary for the generation of NADPH, which is needed in fatty acid and cholesterol synthesis.
The promoter of the SREBP-1c gene contains response elements for insulin, glucagon, as well as liver X-activated receptors (LXR) α and LXRβ, the latter is activated by sterols[17,43]. This regulatory pathway, among others, results in increased synthesis of oleate[75], the major fatty acid used for cholesterol esterification, to ensure its removal from the liver.
KNOCK-OUT ANIMALS AND HEALTH IMPLICATIONS OF SREBPS
Knock-out mice which lack all SREBPs, or S1P to activate them, die at an early stage of embryonic development[80,81]. Specific knock-out of SREBP-2 also results in embryonic lethality. Deletion of SREBP-1a allows some foetuses to survive, whereas lack of SREBP-1c appears to be of no consequence. The survivors are found to compensate by higher SREBP-2 levels, and consequently, these animals have elevated hepatic cholesterol, but lower fatty acid levels[9].
In order to study SREBP knock-outs in adulthood, a gene manipulation was used in Brown and Goldstein’s laboratory which allows to turn specific genes off at will by stimulating interferon production. Disruption of the SCAP or S1P gene, respectively, almost abolished nSREBPs-1 and -2 in liver and diminished expression of all target genes of cholesterol and fatty acid synthesis. The result was a reduction in cholesterol and fatty acid levels in livers by 70%-80%[9].
The SREBP-related links between fatty acid and glucose metabolism draw immediate attention to the wide-spread disease, diabetes. It would appear that the fatty liver frequently observed in insulin-resistant diabetics is a result of high SREBP-1c levels caused by high insulin levels. Leptin, the organism’s response to fat accumulation in adipocytes, opposes SREBP-1c action[82], and consequently can heal the derailment of fat metabolism in diabetic animals[83].
One of the genes activated by nSREBP-1c is that for the hepatic LDL receptor, an effect clearly targeting at maintaining plasma lipid homeostasis. At first sight this would appear beneficial for arteriosclerosis since it should reduce elevated LDL-cholesterol levels in blood. However, since nSREBP-1c also induces lipogenesis, its effect is ambiguous, and other factors will decide whether the net result fights arteriosclerosis, or rather exacerbates it[7]. It is at this point where the HMGCoA reductase inhibitors or statins work, but they are effective only in one third of cases[84].
It must be understood that fatty acid synthesis is not only subject to SREBP regulation, but a host of other factors, whereas cholesterol synthesis appears to be exclusively controlled by SREPBs. Additional unknowns are the exact regulation of system by which HDL via the scavenger receptor B-1 mediates efflux of cholesterol from the liver, and the ATP-binding cassette-1 (ABC-1) system which mediates transfer of cholesterol to HDL[84]. Profound knowledge of these pathways will open a perspective beyond the statin drugs to specifically lower cholesterol levels in disease conditions.
References
Articles from World Journal of Gastroenterology are provided here courtesy of Baishideng Publishing Group Inc
Full text links
Read article at publisher's site: https://doi.org/10.3748/wjg.v10.i21.3081
Free to read at www.wjgnet.com
http://www.wjgnet.com/1007-9327/10/3081.asp
Free to read at www.wjgnet.com
http://www.wjgnet.com/abstract.asp?url=/1007-9327/10/3081
Free to read at www.wjgnet.com
http://www.wjgnet.com/downpdf.asp?url=/1007-9327/10/3081
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.3748/wjg.v10.i21.3081
Article citations
Unveiling Endogenous Serum Peptides as Potential Biomarkers for Hepatocellular Carcinoma in Patients with Liver Cirrhosis.
J Proteome Res, 23(9):3974-3983, 23 Aug 2024
Cited by: 0 articles | PMID: 39177206 | PMCID: PMC11385380
Lipid Metabolism Modulatory Cisplatin Prodrug Sensitizes Resistant Prostate Cancer toward Androgen Deprivation Therapy.
ACS Pharmacol Transl Sci, 7(9):2820-2826, 12 Aug 2024
Cited by: 0 articles | PMID: 39296252
SREBPs as the potential target for solving the polypharmacy dilemma.
Front Physiol, 14:1272540, 10 Jan 2024
Cited by: 0 articles | PMID: 38269061
Review
Photodynamic Therapy Supported by Antitumor Lipids.
Pharmaceutics, 15(12):2723, 03 Dec 2023
Cited by: 0 articles | PMID: 38140064 | PMCID: PMC10747669
Role of EGFR and FASN in breast cancer progression.
J Cell Commun Signal, 17(4):1249-1282, 25 Jul 2023
Cited by: 1 article | PMID: 37490191 | PMCID: PMC10713975
Review Free full text in Europe PMC
Go to all (91) article citations
Other citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Sterol regulatory element-binding protein family as global regulators of lipid synthetic genes in energy metabolism.
Vitam Horm, 65:167-194, 01 Jan 2002
Cited by: 81 articles | PMID: 12481547
Review
Sterol regulatory element-binding proteins (SREBPs) as regulators of lipid metabolism: polyunsaturated fatty acids oppose cholesterol-mediated induction of SREBP-1 maturation.
Ann N Y Acad Sci, 967:34-42, 01 Jun 2002
Cited by: 33 articles | PMID: 12079833
Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes.
Prog Lipid Res, 40(6):439-452, 01 Nov 2001
Cited by: 408 articles | PMID: 11591434
Review




