Abstract
Free full text

Practical Implementation of a Multiplex PCR for Acute Respiratory Tract Infections in Children
Abstract
Molecular testing for acute respiratory infections (ARIs) has documented value but limited implementation due to questions that typically slow the acceptance of new tests. This study sought to address these questions and achieve implementation. Rhinovirus was added to a nested multiplex PCR (M-PCR), increasing its diagnostic yield. Over one winter, three hospital pediatric departments used the M-PCR to complement their direct fluorescent-antibody assay (DFA) for respiratory syncytial virus (RSV). Clinicians recorded “pretest probability estimates” (using continuous scales for various pathogen groups) for comparison with test results; treatments and test turnaround times were also recorded. Transnasal and throat swabs, with or without nasopharyngeal aspirate (NPA), were M-PCR tested. NPA-containing sample sets found to be RSV positive by DFA were not further tested. Single PCR for human metapneumovirus (hMPV) was performed retrospectively. Of 178 ARI episodes representing 172 patients, NPA was included in 97 sample sets; 54 (56%) were determined to be RSV positive. The other NPA-containing sample sets (n = 43) yielded 27 findings (63%), and the swab-only sets (n = 81) yielded 47 findings (58%); rhinovirus was found most often. Testing for hMPV yielded seven positive results. M-PCR median turnaround times were 4 days in swab-only samples and 5 days with NPA. Antibiotics were prescribed in 50 episodes, at rates similar for RSV and rhinovirus. Pretest probability estimates of a viral cause were lower in episodes caused by rhinovirus than in episodes caused by RSV. The hospitals continued to use M-PCR for NPA-containing samples found to be RSV negative by DFA. Test implementation is more likely with higher diagnostic yield and a protocol that reflects day-to-day clinical and laboratory operations.
Acute respiratory tract infections (ARIs) in children are a frequent cause for parents to seek medical help, especially in the youngest children. Although the course of these infections is usually benign, hospital admission can be needed, and many clinical laboratories in The Netherlands routinely perform rapid testing for respiratory syncytial virus (RSV) by direct fluorescent-antibody assay (DFA) on nasopharygeal aspirate (NPA) (5). For the ARIs that remain undiagnosed, viral culture methods are used but fill the diagnostic gap only to a limited extent. Studies document the potential advantages of molecular testing in diagnosing ARI, especially if it is caused by rhinovirus, but give little information on its implementation in clinical practice (1, 11, 16, 20, 24). Laboratories considering introduction of molecular diagnosis of ARI face questions such as: which pathogens should be tested for, which methods should be used, should a commercial test be used, how often should the test be performed, and should it be restricted to specified categories of patients? As long as these questions remain, molecular tests for ARI will be introduced only very slowly into clinical practice. We therefore developed a noncommercial nested multiplex PCR (M-PCR), based on available sequence data and literature, to be implemented for both routine clinical use as well as public health requests. We conducted the present study to confirm its clinical usefulness, in cooperation with three hospital pediatric departments. The study was designed to match with day-to-day practices in the pediatric departments as well as in their shared clinical microbiology laboratory. For example, DFA for RSV usually gives a result within hours upon receipt of sample, so we intended our M-PCR to supplement that DFA, as suggested previously (L. Liolios and S. Wesselingh, Authors' Reply to Letter, J. Clin. Microbiol. 40:733-734, 2002). We included rhinovirus in our M-PCR to increase the number of diagnosed episodes of illness. Because rhinovirus is not commonly included in multiplex assays, we recorded clinical symptoms to document its clinical importance and used the prepresentation duration of illness as an indicator of severity (22). We decided to perform M-PCR testing twice weekly, a frequency the laboratory considered “affordable” for a prolonged period, although this choice would lengthen average test turnaround times (TATs) compared to daily testing. Because our M-PCR is based on well-described assays, our study was not designed to generate sensitivity and specificity figures, the classical indices of diagnostic accuracy (17). Instead, it was used to explore pretest probability estimates as a representation of physicians' judgement, borrowing a concept used in the evaluation of diagnostic decision rules (10). As is common in that field, these pretest estimates were recorded and linked with specific test results to indicate clinical usefulness. Thus, while evaluating M-PCR in our specific setting, using obtained numbers of diagnoses and test TATs, we also evaluated whether pretest probability estimates could help demonstrate the usefulness of a molecular test for ARI.
MATERIALS AND METHODS
Pediatric departments, patients, and questionnaires.
All pediatric ARI patients treated at three hospitals between 15 October 2001 and 1 April 2002 were eligible to the study if their attending physician judged that M-PCR could be useful. The three hospitals were Flevo Hospital at Almere, IJsselmeer Hospital at Lelystad, and IJsselmeer Hospital at Emmeloord. Their pediatric departments provide general, not intensive respiratory care for infants. Their shared laboratory for microbiology, based in Amsterdam, provides daily road transport to and from the hospitals, a one-way trip of 35, 65, or 95 km. After written informed consent was obtained from the parents, transnasal and throat swabs with or without NPA were obtained as samples from the children. Decisions regarding requests for routine bacteriology culture and PCR for Bordetella pertussis were left to the physician, as were all other laboratory and X-ray requests. Questionnaires on presenting symptoms, the starting day, risk factors, initial diagnosis, and treatment were completed by the pediatric department staff. In addition, both pediatricians and house staff gave a quantitative estimate of the cause of the illness (10), marking probability scales for the likelihood of various causes (Fig. (Fig.1).1). At the time of discharge, another questionnaire was completed as to the outcome and final diagnosis. The study was approved by the hospitals' ethical committees.
Conventional laboratory methods. (i) DFA.
The NPA samples were processed on all weekdays. They arrived in the laboratory with phosphate-buffered saline (PBS) and were shaken with glass beads for better homogenization. DFA to detect RSV was performed on 10 μl of homogenized sample with 25 μl of fluorescence-labeled anti-RSV antibodies (fluorescein isothiocyanate-labeled antibody MB120 [Imagen; Dako, Glostrup, Denmark]). The slides were read in a fluorescence microscope (Zeiss), and the slide was scored as positive if at least two cells showed specific cytoplasmic fluorescence.
(ii) Virus isolation in tubes and rapid shell vial culture.
If DFA for RSV was negative, both conventional culture and rapid shell vial culture were started and maintained basically as described previously (28). For conventional culture, duplicate aliquots of tertiary monkey kidney cells, human fetal lung fibroblast lung cells and HeLa cells were inoculated with 200 μl of sample fluid at 34°C. If after 4 weeks no cytopathic effect was seen and all staining efforts were negative, the cultures were considered negative. The rapid shell vial culture was performed in duplicate with 200-μl cell suspensions on tertiary monkey kidney and HeLa cells. One series was stained after 2 days, and the other was stained after 5 to 7 days. Direct staining was performed with pools of specific fluorescence-labeled mouse antibodies to influenza A virus, influenza B virus, RSV, and adenoviruses (Imagen; Dako). Indirect staining was performed for parainfluenza type 1, 2 or 3 virus with anti-mouse antibodies (Chemicon Brunschwig, Amsterdam, The Netherlands). Subsequent staining was performed with labeled rabbit anti-mouse antibodies (Imagen; Dako). As controls, virus cultures derived from the Dutch National Institute for Public Health were used (RIVM, Bilthoven, The Netherlands).
Nucleic acid extraction for M-PCR.
Samples from nasal and throat swabs were each suspended in 200 μl of PBS. Total nucleic acids were isolated by isopropanol precipitation as follows. First, 100 μl of PBS eluate was added to 600 μl of lysis buffer (bioMérieux, Boxtel, The Netherlands) plus 1 μl of glycogen (10 mg/ml [Roche Diagnostics, Almere, The Netherlands]). After an incubation of 60 min at 65°C to increase the lysis efficiency, the sample was cooled to room temperature for 2 min. After a short spin (10 s, 14,000 rpm), 1 volume of ice-cold isopropanol (700 μl) was added, followed by mixing. The precipitate was spun down in an Eppendorf centrifuge (type 5417C) for 20 min at 14,000 rpm. The supernatant was removed, and the pellet was washed twice with 70% ethanol. The pellet was dissolved in 50 μl of T10 (10 mM Tris-HCl [pH 8.0] plus 0.4 U of RNasin [Roche Diagnostics]/μl). The nucleic acid was left at 4°C to dissolve overnight and subsequently stored at −80°C until use in the PCR.
M-PCR.
M-PCR was performed twice weekly. Tests for DNA and RNA pathogens were performed separately by using an assay that was based on a previously described method (11). Adjustment to a nested format required outer and inner primers of our design as shown in Table Table1.1. For the DNA pathogens Chlamydia pneumoniae, Mycoplasma pneumoniae, and adenovirus, one nested M-PCR was performed. Both the outer and the inner PCR mixtures contained 20 ng of each of the six appropriate primers, 200 μM deoxynucleoside triphosphate, 2.0 mM MgCl2, 0.5 U of Taq polymerase, 2.5 μl of 10× Taq buffer (Silverstar; Eurogentec, Maastricht, The Netherlands), and water. To the outer PCR mixture, 5 μl of nucleic acid solution was added to obtain a final volume of 25 μl. The PCR program consisted of one step of 94°C for 3 min, followed by 30 cycles of 93°C for 30 s, 55°C for 30 s, and 72°C for 50 s, with a final extension step of 72°C for 7 min. After completion of the outer PCR, 2 μl was transferred to the inner PCR mixture, and the same PCR cycling program was followed. The inner PCR products were separated on a 10% acrylamide gel and stained with ethidium bromide. The length of the inner PCR fragments was indicative of the putative pathogen. Mixtures of culture-positive controls and a buffer sample as a negative control were added in each test run.
TABLE 1.
ARI M-PCR primers and hMPV primers
| Detection panel and microorganism | Target | Designation | Sequence (5′ to 3′) | Amplimer length (bp)
| Source or reference | |
|---|---|---|---|---|---|---|
| Outer | Inner | |||||
| DNA panel | ||||||
    Chlamydia pneumoniae Chlamydia pneumoniae | Omp1 gene | C pneu I3 | TTG GAG CGT AGC CGC TCG TGG A | 309 | 19 | |
| C pneu P3 | GCT CGA GAC CAT TGT ACT CCA | |||||
| C pneu K3 | ATG GGA ATG CGG TTG TGC AAC T | 223 | This study | |||
| C pneu U3 | GGC TCC TAC TTG CCA TTC ATG A | |||||
    Adenovirus Adenovirus | Hexon gene | Adeno3 out se | CCT TTG GCG CAT CCC ATT CT | 370 | 12 | |
| Adeno4 out as | GCG CTT GTC ATA GGT GCC CA | |||||
| Adeno5 inn se | CCA CGC GCT AGA CAT GAC TT | 142 | This study | |||
| Adeno6 inn as | GAA GGG CGT GCG CAG GTA CA | |||||
    Mycoplasma pneumoniae Mycoplasma pneumoniae | P1 gene (three internal repeats) | M pn F out se | CCC TCG ACC AAG CCA ACC TC | 340 | 31 | |
| M pn R out as | TGC GCG TTG TTC TTG TTG GTG | |||||
| P1-7 inn se | GTT TAA CCA CGC AGG ACG GCA A | 92 | This study | |||
| P1-7 inn as | TCA ATC AGC GGT GGG GGT GAG | |||||
| RNA panel 1 | ||||||
    Rhinovirus Rhinovirus | 5′NCR-VP4 | NCR 1 out se | CGG TAA TTT TGT ACG CCA GTT T | 501 | 1 | |
| NCR 2 out se | GAA ACA CGG ACA CCC AAA GTA GT | |||||
| NCR 3 inn se | CAA GCA CTT CTG TTT CCC CGG | 295 | This study | |||
| NCR 4 inn se | CAT TCA GGG GCC GGA GGA | |||||
    Influenza B virus Influenza B virus | NSP gene 8 | Infl B3 out se | GGG ATA TAC GTA ATG TGT TGT | 489 | This study | |
| Infl B4 out as | GCA CTG CCT GCT GTA CAC TT | |||||
| Infl B1 inn se | ATG GCC ATC GGA TCC TCA AC | 241 | 7 | |||
| Infl B2 inn as | TGT CAG CTA TTA TGG AGC TG | |||||
    Parainfluenza virus type 3 Parainfluenza virus type 3 | 5′ NCR-fusion gene | PIV 3C out se | CTG GGC TTC ATC AGT AGA GA | 585 | 15 | |
| PIV 3D out as | GAT CTG TTG GTC ACC ACA AGA | |||||
| PIV 3B inn se | AGA GGT CAA TAC CAA CAA CTA | 206 | This study | |||
| PIV 3A inn as | TAG CAG TAT TGA AGT TGG CA | |||||
    Influenza A virus Influenza A virus | Nonstructural protein gene | Infl A1 out se | AAG GGC TTT CAC CGA AGA GG | 171 | 7 | |
| Infl A2 out as | CCC ATT CTC ATT ACT GCT TC | |||||
| Infl A3 inn se | CCA GGA CAT ACT ATT GAG GA | 112 | This study | |||
| Infl A4 inn as | CCC AAG CGA ATC TCT GTA GA | |||||
| RNA panel 2 | ||||||
    RSV RSV | Nucleocapsid gene | N1A-B out se | GAR GTR GCT CCA GAA TAY AGG CAT | 343 | 20 | |
| Osi-2 out as | AGC ATC ACT TGC CCT GAA CCA TAG | |||||
| Osi N4 inn se | ACT AAG TTA GCA GCA GG | 197 | This study | |||
| Osi N3 inn as | CTG GRA GAA GAT TGT GC | |||||
    Parainfluenza virus type 1 Parainfluenza virus type 1 | Hemagglutinin-neur- minidase gene | PIV 1C out se | TGC AGA CGG CAT ATC TCC TCT GGA | 307 | 9 | |
| PIV-1D out as | GGT ATG AGA AAT TAC CGG GT | |||||
| PIV-1E inn se | CGT AGG AGA ACC CCT ACT GA | 173 | This study | |||
| PIV-1F inn as | CTA TAT CTG CAC ATC CTT GAG TGA | |||||
    Pan-entero virus Pan-entero virus | 5′ NCR | Fox AB out se | GGT GYG AAG AGY CTA YTG AGCT | 206 | 25 | |
| Srent2 out as | CAC CGG ATG GCC AAT CCA | |||||
| SRent3 inn se | CCC CTG AAT GCG GCT AAT | 146 | This study | |||
| SRent4 inn as | ATT GTC ACC ATA AGC AGC CA | |||||
| hMPV single PCR-hMPV | L gene | hMPV-3s out se | AAC ACT GGG GAA AAT GCT CA | 204 | This study | |
| hMPV-4a out se | ATC CGA TAT GAA CCC GTC | |||||
| hMPV-6 inn se | CAT GCC CAC TAT AAA AGG TCA G | 171 | -a | |||
| hMPV-7na inn as | GTC ACC CCA GTC TTT CTT GA | |||||
For the RNA pathogens (seven viruses [see Table Table1])1]) a combined reverse transcriptase (RT) outer PCR was performed by using the Access kit (Promega, Leiden, The Netherlands) according to the instructions of the manufacturer. The total mixture volume was 25 μl, of which 5 μl consisted of the nucleic acid solution. Thus, the outer Access RT-PCR contained a mixture of 12 primers (degenerate primers for the pan-enterovirus and RSV detection). The amplification program consisted of a first RT step at 48°C for 45 min, followed by a denaturation step of 94°C for 2 min and then 30 cycles of 93° for 30 s, 55°C for 30 s, and 72°C for 50 s, with a final extension step of 72°C for 7 min. After completion of the outer PCR, 2 μl was transferred to two inner PCR mixtures containing the same components as the DNA PCR mixtures, except for the primers. The two inner mixtures were selected to enable optimal discrimination by length separation of the inner PCR products. As shown in Table Table1,1, RNA panel 1 contained the primers (20 ng each) for detecting influenza A and B viruses, pan-rhinovirus, and parainfluenza type 3 virus, whereas RNA panel 2 contained the primers (20 ng each) for detecting pan-enterovirus, RSV, and parainfluenza type 1 virus. The PCR program used for the DNA PCR amplification steps described above was followed. Gel detection and inclusion of positive and negative controls were similar to the DNA PCR.
hMPV PCR.
For retrospective use, a nested PCR was designed by using sequences of the L gene of human metapneumovirus (hMPV) (Table (Table1).1). The outer and inner PCR mixtures and amplification conditions and the gel detection analysis were similar to the RT-PCR approach described above for the M-PCR. Each run contained, as a positive control sample, isolated RNA from cultured hMPV that was kindly provided by A. Osterhaus, Erasmus Medical Center, Rotterdam, The Netherlands.
Comparison of the sensitivity of M-PCR versus specific PCR.
To assess the analytical sensitivity, we compared the M-PCR to each pathogen-specific PCR by performing each test six times on endpoint titrations of control cultures for each pathogen. The specific PCR was done by performing the M-PCR but with only the primers for the pathogen under study, with identical conditions otherwise and with sterile water replacing the other primers. The dilution at which 50% of the PCRs was still positive was calculated by using formulas similar to those used to compute the 50% tissue culture infectious dose. The difference in results between the multiplex setting and the specific PCR were <0.7 log for all pathogens except for influenza B virus (1.3 log) and parainfluenza type 3 (1.3 log), an outcome considered acceptable for this diagnostic amplification system.
Clinical data analysis.
Clinical and laboratory data were entered in an Access database (Microsoft). Continuously recorded probability estimates were grouped into five intervals and, where appropriate, comparisons for the numbers in these intervals were made between specific test outcomes. Test TAT was defined as the day that the result was available minus the day that the sample was taken. The duration of illness prior to presentation to the hospital was figured from the day of presentation back to the day that the illness started. Where appropriate, chi-square tests were performed by using EPI-INFO version 6 (Centers for Disease Control and Prevention, Atlanta, Ga.).
RESULTS
Patients and samples.
During the study period, 188 ARI episodes were considered for our study. Of the sets received at the laboratory, 10 were excluded for one of the following reasons: (i) lack of informed consent (n = 4); (ii) samples were sent, but no questionnaires (n = 2); and (iii) questionnaires were sent, but there was not enough sample for an M-PCR (n = 4). The included 178 episodes represented 172 patients. Patient episode characteristics are summarized by site in Table Table2.2. Patients were seen by four pediatricians at site A, four pediatricians at site B, and two pediatricians at site C. In addition, six junior doctors saw patients at site A, whereas sites B and C each had one junior doctor working at their pediatric ward. Especially the youngest children were preferably sampled with both NPA and swabs. Severe illness was rare; only three patients required referral to other hospitals for intensive respiratory care. Risk factors were reported in 58 episodes; these factors varied from low birth weight to asthma and mental retardation. The largest number of samples was collected at site A, although the number of NPA samples was low compared to site B. The median length of stay of admitted children was shorter at site A compared to site B. Site C contributed the smallest number of samples.
TABLE 2.
Patient episode characteristics
| Episode category | Site A | Site B | Site C | All sites |
|---|---|---|---|---|
| Patient episodes (all) | 105 | 50 | 23 | 178 |
    Patients with a second episode Patients with a second episode | 5 | 1 | 0 | 6 |
    Age groups Age groups | ||||
        0-2 mo 0-2 mo | 18 | 19 | 1 | 38 |
        3-5 mo 3-5 mo | 26 | 5 | 5 | 36 |
        6-11 mo 6-11 mo | 20 | 14 | 4 | 38 |
        12-23 mo 12-23 mo | 13 | 5 | 6 | 24 |
        24-47 mo 24-47 mo | 10 | 3 | 4 | 17 |
        48 mo to 16 yr 48 mo to 16 yr | 18 | 4 | 3 | 25 |
    Transfers for intensive respiratory care Transfers for intensive respiratory care | 2 | 1 | 0 | 3 |
    Median length of stay (days) (range) Median length of stay (days) (range) | 4 (2-9) | 5 (2-17) | 5 (1-8) | 5 (1-17) |
| Inpatient episodes | 24 (23) | 42 (84) | 20 (87) | 86 (48) |
    NPA and swabs NPA and swabs | ||||
        No. RSV positive/no. tested for rapid RSVa (%) No. RSV positive/no. tested for rapid RSVa (%) | 7/18 (39) | 26/41 (63) | 8/14 (57) | 41/73 (56) |
        No. any pathogen positive/no. tested by M-PCRb (%) No. any pathogen positive/no. tested by M-PCRb (%) | 8/11 (73) | 8/15 (53) | 5/6 (83) | 21/32 (66) |
        No. positive in rapid RSV or M-PCR/no. tested (%) No. positive in rapid RSV or M-PCR/no. tested (%) | 15/18 (78) | 34/41 (83) | 13/14 (93) | 62/73 (85) |
    Swabs only (no. any pathogen positive/no. tested by M-PCR) (%) Swabs only (no. any pathogen positive/no. tested by M-PCR) (%) | 3/6 (50) | 1/1 (100) | 5/6 (80) | 9/13 (69) |
| No. of M-PCR results (positive or negative) available at least 1 day before discharge/no. of all M-PCR tests performed for admitted patients (%) | 4/17 (23) | 5/16 (31) | 5/12 (42) | 14/45 (31) |
| Outpatient episodes | 81 (77) | 8 (16) | 3 (13) | 92 (52) |
    NPA and swabs NPA and swabs | ||||
        No. RSV positive/no. tested for rapid RSV (%) No. RSV positive/no. tested for rapid RSV (%) | 11/18 (61) | 1/5 (20) | 1/1 (100) | 13/24 (54) |
        No. any pathogen positive/no. tested by M-PCR (%) No. any pathogen positive/no. tested by M-PCR (%) | 4/7 (57) | 2/4 (50) | 0/0 | 6/11 (55) |
        No. positive in rapid RSV or M-PCR/no. tested (%) No. positive in rapid RSV or M-PCR/no. tested (%) | 15/18 (83) | 3/5 (60) | 1/1 (100) | 19/24 (79) |
    Swabs only (no. any pathogen positive/no. tested by M-PCR) (%) Swabs only (no. any pathogen positive/no. tested by M-PCR) (%) | 35/63 (56) | 2/3 (67) | 1/2 (50) | 38/68 (56) |
RSV DFA results and viral culture results.
Table Table22 shows that 97 sample sets (73 inpatients and 24 outpatients) included an NPA. Of these, 54 (41 inpatients and 13 outpatients) were determined to be positive by RSV DFA. Thus, in 56% of the patients sampled by NPA, the first test yielded a positive RSV result. Since admission policies differed among hospitals, less than half of the RSV-positive patients (7 of 18) were admitted at site A, whereas most of the RSV-positive patients were admitted at sites B (26 of 27) and C (8 of 9). Of the 43 NPA samples with a negative or equivocal DFA result, one was insufficient for all remaining tests, so 42 samples were cultured for respiratory viruses, yielding nine positive results (21%): adenovirus (n = 3), rhinovirus (n = 4), influenza virus A (n = 1), and RSV (n = 1).
M-PCR results.
Table Table33 summarizes PCR results of the sample sets with (n = 43) or without (n = 81) NPA. The upper part of Table Table33 shows the results of the M-PCR that became available to the clinicians during our study. The percentage of diagnosed episodes reached 84% or, conversely, the diagnostic deficit was reduced to 16% if NPA was sent to the laboratory, and M-PCR results were added to the DFA results. If only swabs were sent, M-PCR detected a possible pathogen in 58% of the cases. Double infections were common and, with three exceptions, these were all rhinovirus combinations: infection with adenovirus (n = 6), infection with C. pneumoniae (n = 1), infection with M. pneumoniae (n = 4), infection with enterovirus (n = 2), infection with influenza virus A (n = 2), and infection with RSV (n = 3). The three other combinations were influenza virus A and C. pneumoniae, adenovirus and RSV, and influenza virus B and RSV.
TABLE 3.
Amplification test results of 124 patient episodes by type of sample set
| Analysis | No. of sample sets (%)
| |
|---|---|---|
| Sample sets including NPA, negative by DFA for RSV (n = 43) | Swab-only sample sets (n = 81)        | |
| M-PCR (prospectively) | ||
    Adenovirus Adenovirus | 2 (5) | 4 (5) |
    Chlamydia pneumoniae Chlamydia pneumoniae | 0 | 1 (1) |
    M. pneumoniae M. pneumoniae | 1 (2) | 2 (2) |
    Influenza A virus Influenza A virus | 3 (7) | 7 (9) |
    Influenza B virus Influenza B virus | 1 (2) | 1 (1) |
    Parainfluenza 1 Parainfluenza 1 | 0 | 0 |
    Parainfluenza 3 Parainfluenza 3 | 1 (2) | 2 (2) |
    Enterovirus Enterovirus | 0 | 1 (1) |
    Rhinovirus Rhinovirus | 13 (30) | 12 (15) |
    RSV (PCR) RSV (PCR) | 0 | 2 (2) |
    Double infection Double infection | 6 (14) | 15 (19) |
    Negative Negative | 16 (37) | 34 (42) |
| Bordetella-PCR (if requested)a | 1 (2) | 0 |
| hMPV-PCR (retrospectively)b | 4 (9) | 3 (4) |
| Negative after hMPV-, Bordetella-, and M-PCR | 11 (26) | 31 (38) |
Of the 66 cases considered to be “bronchiolitis” at presentation, 33 yielded RSV as single pathogen, whereas 9 (7 with NPA plus 2 swab only) yielded a rhinovirus as single pathogen (Table (Table4).4). Four bronchiolitis double infections sampled with NPA all involved rhinovirus: in combination with RSV (n = 3) and C. pneumoniae (n = 1). In bronchiolitis episodes, influenza virus A was found only once. In episodes regarded to be pneumonia on presentation (17 patients, of whom 8 were admitted), rhinovirus was found slightly more frequently than RSV. There were three episodes with rhinovirus alone, one in combination with M. pneumoniae, and one in combination with adenovirus; RSV alone was found twice in such patients, and the RSV-rhinovirus combination was found once. The combination of rhinovirus and M. pneumoniae was found in a 6-year-old child. In three episodes considered to be pneumonia, influenza virus A was detected. Of the three children referred for intensive respiratory care, RSV was detected in one, rhinovirus was detected in another, and no pathogen was detected in the third.
TABLE 4.
Symptoms by sample type
| Symptom or syndrome at presentationa | Outcome (all tests combined)b | No. of sample sets (%)
| |
|---|---|---|---|
| NPA included in sample sets (n = 97) | Swab-only sample sets (n = 81) | ||
| Upper respiratory tract infection (undifferentiated) | 25 (26) | 49 (60) | |
    Pharyngitis Pharyngitis | 2 (2) | 29 (36) | |
    Tonsilitis Tonsilitis | 3 (3) | 9 (11) | |
    Sinusitis Sinusitis | 0 | 1 (1) | |
    Otitis media Otitis media | 4 (4) | 7 (9) | |
| Lower respiratory tract infection (undifferentiated) | 76 (78) | 58 (72) | |
    Bronchitis Bronchitis | 10 (10) | 32 (40) | |
    Bronchiolitis Bronchiolitis | Total | 50 (52) | 16 (20) |
| RSV | 33 | 0 | |
| Rhinovirus | 7 | 2 | |
| Influenza A virus | 0 | 1 | |
| hMPV | 2 | 1 | |
| Other and double | 4 | 5 | |
| Negative | 4 | 7 | |
    Pneumonia Pneumonia | Total | 6 (6) | 11 (14) |
| RSV | 1 | 1 | |
| Rhinovirus | 1 | 2 | |
| Influenza A virus | 1 | 2 | |
| hMPV | 1 | 1 | |
| Other and double | 2 | 2 | |
| Negative | 0 | 3 | |
| None stated | 7 (7) | 4 (5) | |
hMPV- and Bordetella-PCR and bacteriology results.
The lower part of Table Table33 shows that 7 (5.6%) more diagnoses were retrospectively added by performing the hMPV-PCR on all M-PCR processed samples. Most of the hMPV-infected children had lower respiratory tract infections, and five of the seven children were younger than 6 months, four of whom were admitted for 4 to 8 days. B. pertussis was not a target pathogen in the present study, so only samples with a specific request were tested by PCR for Bordetella. Of 51 such requests, only one was PCR positive for B. pertussis. This case of whooping cough was diagnosed in a 4-week-old baby admitted at ward B who persisted for almost 2 weeks. The infection was not immediately suspected on admission; the probability estimate for viral infection was 90%, and the probability of whooping cough was not stated. Blood cultures (n = 10) and/or lower respiratory tract samples (n = 4) for bacteriology were send in 13 episodes of illness. Blood cultures remained negative. Potential pathogenic bacteria were detected in 3 lower respiratory tract samples. Streptococcus pneumoniae and Haemophilus influenzae were detected in a 10-month-old child known to have previous asthma-like symptoms. Staphylococcus aureus was found in low numbers in a 2-month-old child with Downs' syndrome, and M-PCR detected rhinovirus. At almost 4 months of age, this child had a second episode of illness with a readily recognized RSV bronchiolitis. Finally, Pseudomonas aeruginosa was detected in a 15-year-old with cystic fibrosis; this was a repeated finding.
TATs.
TATs of NPA containing sample sets are graphically represented in Fig. Fig.2.2. The results that were obtained most rapidly are shown; thus, the largest number of positive results was obtained by DFA. Two rapid shell vial results were obtained the same day as the M-PCR result became available, and one conventional culture result (i.e., RSV) was not detected by M-PCR. In the latter sample, a rhinovirus was detected by M-PCR, and in the breakdown columns in Tables Tables44 and and5,5, this sample was designated a “double pathogen.” All other results were obtained only by M-PCR or more rapidly than by viral culture methods. As can be seen in Fig. Fig.2,2, the median TAT for positive M-PCR results was 5 days in the NPA-containing sample sets. Samples in the swab-only group had a median TAT of 4 days, since processing of these samples did not wait for a DFA result. We also calculated the number of admitted patients in whom the M-PCR result (both positive or negative) was available at least 1 day before discharge (Table (Table2),2), finding an average of 31%. The percentage was lowest at site A, which had the lowest admission rate and the shortest length of stay.
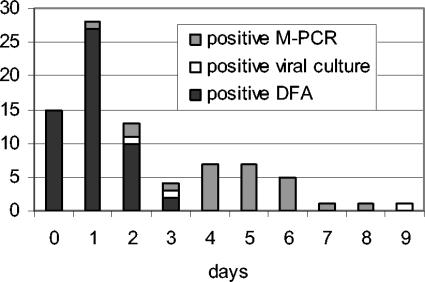
TATs obtained in NPA-containing sample sets (n = 97) for different laboratory tests. DFA for RSV was performed first and, depending on the result, M-PCR processing followed. Bars represent the numbers of first positive test results and the time (in days) required to produce the result. If a conventionally obtained result was available on the same day as an M-PCR result, the conventional TAT is represented. One adenovirus and one influenza virus A test result were obtained, both with rapid shell vial culture and M-PCR after 2 and 3 days. One RSV result was obtained with conventional culture only after 9 days.
TABLE 5.
Clinician probability estimates for causes of presenting illnesses
| Suspected cause of illnessa | Outcome (all tests combined)b | No. of samples with clinician pretest probability estimate of:
| |||||
|---|---|---|---|---|---|---|---|
| Not stated | <5% | 6-30% | 31-69% | 70-94% | ≥95% | ||
| Viral cause (any) | Total | 4 | 3 | 9 | 24 | 71 | 67 |
| RSV | 7 | 17 | 32 | ||||
| Rhinovirus | 3 | 4 | 15 | 3 | |||
| Influenza A virus | 2 | 2 | 4 | 2 | |||
| hMPV | 1 | 3 | 3 | ||||
| Other and double | 1 | 2 | 3 | 4 | 14 | 14 | |
| Negative | 1 | 3 | 7 | 18 | 13 | ||
| Bacterial cause (any) | Total | 21 | 78 | 52 | 23 | 4 | 0 |
| RSV | 3 | 33 | 14 | 4 | 2 | ||
| Rhinovirus | 5 | 7 | 10 | 3 | |||
| Influenza A virus | 1 | 3 | 3 | 2 | 1 | ||
| hMPV | 1 | 3 | 3 | ||||
| Other and double | 3 | 15 | 12 | 8 | |||
| Negative | 8 | 17 | 10 | 6 | 1 | ||
| “Atypical” cause | 24 | 103 | 20 | 23 | 8 | 0 | |
| Pertussis | 19 | 93 | 41 | 13 | 11 | 1 | |
| Asthma as a co-cause | 31 | 70 | 40 | 26 | 11 | 0 | |
Probability estimates, test outcomes, and antibiotic treatment.
Clinicians were asked to estimate the likelihood of various causes of the presented illness, as a representation of physicians' judgement (Fig. (Fig.1).1). In the majority of cases, there was a high level of suspicion for a viral cause of the presented symptoms (Table (Table5).5). Low to moderately low probability estimates were given for pertussis, “atypical” agents, and bacterial pathogens. Many physicians did not address the question on the probability of asthma as a co-cause and, of those who did, many indicated their uncertainty by marking the center range of the probability scale. Comparing diagnostic probability estimates with testing results (Table (Table5),5), we noted that doctors were more likely to suspect viral origin in cases of RSV infections (as diagnosed by both DFA and M-PCR) than in cases of rhinovirus infections (M-PCR, single finding). Taking the 95% probability estimate as a threshold in 54 RSV-positive patients, 32 estimates were at or above threshold, whereas only 3 estimates were above threshold in 22 rhinovirus-positive patients (P < 0.01 [chi-square analysis]). Repeating this procedure with inclusion of only the sample sets including NPA or with exclusion of the data of clinicians contributing more than 20 cases yielded similar results, although the numbers were lower (P < 0.05 [chi-square analysis]). Conversely, clinicians were more certain that children did not have a bacterial infection if RSV was detected compared to rhinovirus. Antibiotics were prescribed in 50 episodes of illness. As expected, such prescriptions were strongly related to the clinicians' estimate of bacterial cause. However, even among the 78 episodes in which a bacterial cause was estimated to be very unlikely (Table (Table5),5), antibiotics were prescribed in 10. Among the patients sampled via NPA, those with reported risk factors were more likely to receive an antibiotic prescription than those without risk factors (P < 0.01 [chi-square analysis]). RSV was detected in 16 and rhinovirus as a single pathogen was detected in 7 of the antibiotic-prescribed episodes. Thus, rhinovirus-infected patients were not more likely to have antibiotics prescribed than RSV-infected patients.
Duration of illness prior to presentation with RSV or rhinovirus infection.
Calculation of the duration of illness prior to presentation was possible in 152 of the 178 episodes, and the duration ranged from 0 to 122 days. In cases with the longest duration, we assume that parents and/or clinicians did not always consider only the current pathogen episode when indicating the starting date of the illness. However, taking 2 days as a threshold, 11 of 22 rhinovirus episodes had a prepresentation illness of ≤2 days, whereas only 8 of 45 RSV episodes had such a short prepresentation duration of illness (P < 0.01 [chi-square analysis]).
DISCUSSION
Our study in three pediatric departments was intended to enable their clinicians and microbiologists to decide whether or not to establish M-PCR as a test that is routinely available within the existing collaboration of the hospitals and shared laboratory. We focused on the number of results, test TATs, and the clinicians' pretest probability estimates in relation to test outcomes as indicators of clinical usefulness. Our noncommercial M-PCR was based on a previously described assay (11). We added rhinovirus to the pathogen set, expecting and finding a substantial increase of diagnosed episodes of illness. Primers were redesigned or supplemented with new primers to obtain a nested multiplex test format. Decreased analytical sensitivity of the multiplex PCR compared to single-pathogen PCR tests did not prove to be a problem; in our hands, differences were small and did not outweigh the advantages in both ease and decreased reagent costs. The M-PCR obtained far more diagnoses than conventional viral culture, at reasonable test TATs. Our finding that clinicians recorded lower pretest probabilities of viral causation in rhinovirus infections, compared to RSV, underscored the usefulness of M-PCR as an additional diagnostic tool.
While designing the present study, we decided to perform M-PCR on NPA only if DFA for RSV was negative. DFA usually gives a reliable result within hours upon receipt, and the associated costs are modest, making it unlikely that molecular methods will replace DFA for RSV detection in the next few years. Figure Figure22 illustrates that in our setting, with the laboratory distant from the hospitals and offering once-daily transport, less than half of the DFA results are available on the day of sampling. Performing a test for RSV in cases of bronchiolitis is considered standard in The Netherlands (5), but the typical bronchiolitis can be managed without it (23). Due to the sequential testing in our study, M-PCR results in the NPA-containing sample sets were delayed compared to the swab-only samples. Most M-PCR results were not received in time to affect decisions during hospital stays. Still, clinicians appreciated that the results could be reported to the parents at the next outpatient visit.
Our hypothesis was that the M-PCR test results would be more appreciated if clinicians were uncertain about the diagnosis, in other words, when the estimated pretest probability was low. We therefore recorded clinicians' diagnostic probability estimates before any microbiological test was done, thus introducing physicians' judgement into evaluating a new diagnostic procedure in clinical microbiology. Although used in other fields, to our knowledge this approach has not been used previously in diagnostic studies in infectious disease, perhaps due to certain limitations. For instance, clinicians may hesitate to estimate pretest probabilities in an individual case (3). However, sufficient numbers of observations made by a sizeable group of clinicians should allow inferences. The fact that clinicians frequently recorded very high levels of pretest probability of a viral cause in RSV infections (Table (Table5)5) but not in rhinovirus infections suggests that M-PCR indeed adds useful diagnostic information, particularly regarding that pathogen. The relative diagnostic uncertainty in rhinovirus infections could result in more antibiotic prescriptions, but this was not seen in our data set.
Rhinovirus was the single most frequently detected virus in the present study after RSV (Table (Table3).3). Appreciation of this virus as a potential pathogen is vital in evaluating the usefulness of M-PCR as an additional test. As shown in Table Table4,4, rhinovirus came second to RSV in episodes considered bronchiolitis at presentation and, in cases considered to be pneumonia, rhinovirus was slightly more common than RSV. Distinguishing pneumonia clinically may be difficult in children (30). These findings are in line with reports that rhinovirus does cause a number of lower respiratory tract infections (21). In hematologic cancer patients requiring bronchoalveolar lavage, in bronchopulmonary dysplasia patients, and in adults and children with exacerbated asthma, rhinovirus proved to be a serious pathogen (4, 6, 8, 14, 24, 29). Similar to a previous report, we found that more children with infected with rhinovirus had been ill no longer than 2 days before presentation compared to the RSV-infected children (P < 0.01 [chi-square analysis]), indicating more serious disease (22). We conclude that it is worthwhile to test for rhinovirus and that the few manufacturers of multiplex amplification tests for ARI should include rhinovirus in their assays.
Influenza A virus was also commonly found, but mainly during the high-incidence season in The Netherlands. Few of these children required admission. M-PCR did not add much speed because in NPA samples influenza viruses are readily detected with conventional rapid methods (2). M-PCR performed on swabs, however, made outbreaks caused by influenza easier to investigate. Our collected set of samples was used to determine the frequency of hMPV (27). Of the 124 samples tested, 7 (5.6%) were positive for hMPV, a percentage that is comparable to the findings of other reports (26, 27). Especially the youngest children were affected; they had lower respiratory tract symptoms, and all recovered uneventfully. Retrospective testing for hMPV thus reduced the diagnostic deficit to 11% (11 of 97) in the NPA-containing sample sets and to 38% (31 of 81) in the swab-only sample sets.
We recognize that there must be some selection bias in our data set, given the relatively low number of samples from sites B and C compared to site A (Table (Table2).2). A partial explanation may lie in the different numbers of junior doctors. In addition, pediatricians at locations B and C had more space to admit children presenting with RSV bronchiolitis. At location A such children were often sent home with equipment and instructions for the parents; these children were told to return the next day to the pediatric department to be seen by the clinician. Variation in resource utilization in hospitalized children in the United States has been studied by Willson et al., who found that the severity of illness did not explain the variation (32).
Costs have been considered throughout the planning and performing of the present study, and future economic assessments of molecular tests are warranted. However, we did not perform a formal economic assessment, mainly because molecular methods to diagnose ARI are still being improved and thus have no widely accepted standard. At present, introducing a new in-house molecular test is largely a matter of affordability. As for future assessment, ARI in children usually has a benign course, so outcome measures need to be chosen with this in mind. Improved care may reduce length of stay and save hospital costs, as found in a study of rapid viral culture methods (2); however, having seen differences in admission and discharge policies among our sites, we doubt that length of stay will be significantly influenced just by rapid diagnostic methods. Improved antibiotic stewardship and avoidance of resistance development are important possible effects of molecular testing, but economic evaluation of resistance is complicated (18), and the availability of M-PCR results may not automatically reduce the number of antibiotic prescriptions. As we noted above, patients may have an antibiotic prescribed even if the clinician is fairly certain that the illness is caused by a virus. To reduce such prescriptions, a combination of individual M-PCR test results with repeated feedback of aggregated results is probably required. Finally, we must stress the appreciation of the test results by the children's parents. Diagnostic certainty or “knowing for the sake of knowing” should not be ignored, in our opinion, and may be addressed with “willingness to pay” questionnaires (13). Within the present study, we attempted to determine the parents' “willingness to pay” for M-PCR but failed to collect sufficient data. We think that especially parents of infants at risk for developing asthma may value diagnostic information on an episode of wheezing.
At our study sites, pediatricians and laboratory staff decided to continue M-PCR testing of NPA samples. Swab-only samples, however, were discontinued since there was no consensus among the hospitals on the indication of these samples as routine requests. After approval by the hospitals' administrators, M-PCR testing was thus implemented. We hope that laboratories already capable of noncommercial molecular tests will be encouraged by the number of diagnoses we achieved and proceed to offer molecular tests for ARI at hospitals comparable to ours. Of course, newer molecular methods should be used as soon as they are available and affordable, with cheap and accurate point-of-care diagnosis being the ultimate goal. Future studies should confirm that both individual and aggregated feedback of molecular ARI test results indeed contribute to more appropriate antibiotic use.
Acknowledgments
We thank Patrick M. M. Bossuyt and Henri A. Verbrugh for critical reading of the manuscript and Lucy D. Phillips for editorial review.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.42.12.5596-5603.2004
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc535307?pdf=render
Citations & impact
Impact metrics
Article citations
Next Generation Sequencing Approaches to Characterize the Respiratory Tract Virome.
Microorganisms, 10(12):2327, 24 Nov 2022
Cited by: 3 articles | PMID: 36557580 | PMCID: PMC9785614
Review Free full text in Europe PMC
Diversity of respiratory viruses detected among hospitalized children with acute lower respiratory tract infections at Hospital Serdang, Malaysia.
J Virol Methods, 269:1-6, 22 Mar 2019
Cited by: 4 articles | PMID: 30910688 | PMCID: PMC7172173
The use of multiplex PCR for the diagnosis of viral severe acute respiratory infection in children: a high rate of co-detection during the winter season.
Eur J Clin Microbiol Infect Dis, 35(10):1607-1613, 10 Jun 2016
Cited by: 18 articles | PMID: 27287764 | PMCID: PMC7088036
Is there still room for novel viral pathogens in pediatric respiratory tract infections?
PLoS One, 9(11):e113570, 20 Nov 2014
Cited by: 24 articles | PMID: 25412469 | PMCID: PMC4239085
Incidence of Alpha-Herpes virus induced ocular disease in Suriname.
J Med Virol, 84(12):1937-1942, 01 Dec 2012
Cited by: 3 articles | PMID: 23080500
Go to all (42) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences
- (1 citation) ENA - AF371337
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
[Investigation of respiratory syncytial virus by three different methods in children with lower respiratory tract infection].
Mikrobiyol Bul, 43(3):433-438, 01 Jul 2009
Cited by: 2 articles | PMID: 19795618
Frequent detection of viral coinfection in children hospitalized with acute respiratory tract infection using a real-time polymerase chain reaction.
Pediatr Infect Dis J, 27(7):589-594, 01 Jul 2008
Cited by: 103 articles | PMID: 18520973
Excretion patterns of human metapneumovirus and respiratory syncytial virus among young children.
Eur J Med Res, 11(8):329-335, 01 Aug 2006
Cited by: 28 articles | PMID: 17052968
[Human metapneumovirus--virologic and diagnostic aspects].
Tidsskr Nor Laegeforen, 125(20):2766-2768, 01 Oct 2005
Cited by: 0 articles | PMID: 16244676
Review






