Abstract
Free full text

Cerebral hypoplasia and craniofacial defects in mice lacking heparan sulfate Ndst1 gene function
Summary
Mutant mice bearing a targeted disruption of the heparan sulfate (HS) modifying enzyme GlcNAc N-deacetylase/N-sulfotransferase 1 (Ndst1) exhibit severe developmental defects of the forebrain and forebrain-derived structures, including cerebral hypoplasia, lack of olfactory bulbs, eye defects and axon guidance errors. Neural crest-derived facial structures are also severely affected. We show that properly synthesized heparan sulfate is required for the normal development of the brain and face, and that Ndst1 is a modifier of heparan sulfate-dependent growth factor/morphogen signalling in those tissues. Among the multiple heparan sulfate-binding factors potentially affected in Ndst1 mutant embryos, the facial phenotypes are consistent with impaired sonic hedgehog (Shh) and fibroblast growth factor (Fgf) interaction with mutant heparan sulfate. Most importantly, the data suggest the possibility that defects in heparan sulfate synthesis could give rise to or contribute to a number of developmental brain and facial defects in humans.
Introduction
Heparan sulfate (HS) is produced by most mammalian cells as part of membrane and extracellular matrix proteoglycans (Esko and Lindahl, 2001). The chain grows by the copolymerization of GlcAβ1,4 and GlcNAcα1,4 residues, and undergoes modification by one or more of the four Ndst isozymes, which remove acetyl groups from subsets of GlcNAc residues and add sulfate to the free amino groups. In vertebrates, Ndst1 and Ndst2 mRNA are expressed in all embryonic and adult tissues examined, whereas Ndst3 and Ndst4 transcripts are predominantly expressed during embryonic development and in the adult brain (Aikawa et al., 2001). Subsequent modifications of the HS chain by O-sulfotransferases and a GlcA C5-epimerase depend on the presence of GlcNS residues, making the Ndsts responsible for the generation of sulfated ligand-binding sites in HS (Esko and Selleck, 2002; Lindahl et al., 1998).
Many growth factors and morphogens bind to HS. In some cases, HS proteoglycans are thought to act as co-receptors for these ligands. Studies in Drosophila melanogaster demonstrated that HS is crucial for embryonic development (Perrimon and Bernfield, 2000) and that the fly Ndst ortholog, Sulfateless, affects signalling mediated by Wingless (Wg), Hedgehog (Hh) and Fibroblast Growth Factor (Fgf) (Lin et al., 1999; Lin and Perrimon, 1999; The et al., 1999). The ability of HS to regulate the activity of morphogens and growth factors is currently best understood for the Fgfs. HS was found to be a necessary component of Fgf-Fgf receptor binding and assembly (Schlessinger et al., 2000), and global changes in HS expression regulate Fgf and Fgf receptor assembly during mouse development (Allen and Rapraeger, 2003). Owing to the multiple developmental processes regulated by the 22 Fgfs, including those of the lung, limbs, heart, skeleton and brain (reviewed by Ornitz and Itoh, 2001), perturbed Fgf-HS interactions can be expected to result in impaired morphogen signalling, resulting in a variety of developmental defects.
Sonic hedgehog (Shh), a member of the multi-gene hedgehog family in vertebrates, also binds HS (Rubin et al., 2002). Mice that lack Shh function show defective axial patterning of the brain and eye, skeleton and limbs (Chiang et al., 1996a). Shh has been demonstrated to be essential in craniofacial morphogenesis, survival of craniofacial neural crest cells, and growth and patterning of the cerebellum (for a review, see Ingham and McMahon, 2001). Members of the Tgfβ family and Tgfβ-binding proteins also bind HS. Additionally, HS proteoglycans in the extracellular matrix and on cell surfaces can affect gradients of these various factors in tissues.
In this paper, we ask which developmental processes are impaired by undermodified heparan sulfate in the mouse. Surprisingly, we found that decreased sulfation of HS due to Ndst1 deficiency leads to very specific developmental defects of the head and forebrain. We also found that Ndst1 is a modifier of Fgf- and Shh-dependent signalling in those tissues.
Materials and methods
Targeted recombination of the Ndst1 gene
The thymidine-kinase/neomycin containing targeting vector was constructed by insertion of loxP-sites in intron-sequences surrounding exon 2 (the first coding exon) of Ndst1. The final targeting vector was linearized using SalI before transfection of ES cells. R1 ES cells were grown, transfected and subjected to neomycin G418 selection. Homologous recombinants were identified by Southern blotting and PCR and transfected with a Cre-expressing vector, followed by ganciclovir selection. Four type II recombinants were chosen and injected in C57Bl/6J blastocysts. Two mouse lines, derived from two different ES cell clones, were obtained, and backcrossed into a C57Bl/6 background for more than 10 generations. The primers employed for genotyping were: P1, 5′-CCCAGATGGCGAGACTGAGG-3′; P2, 5′-CCAGGGCGTCAGGGCCTCCTG-3′; P3, 5′-CATCCTCTGAGGTGACCGC-3′; P4, 5′-GGTACCCGGGGATCAATTCG-3′; P5, 5′-CCAGAAGGCTAACACTGTAAAG-3′; P6, 5′-GAAAGTGAAGTCTCTGGGCGG-3′; P7, 5′-GCTTGGATGATTTGGTCACACT-3′.
Histology and in situ detection of RNA and protein expression
Embryos were fixed in 4% paraformaldehyde overnight, dehydrated, embedded in paraffin and sectioned. Sections were stained with Haematoxylin and Eosin for histological analysis. Cartilage and bone were stained with Alcian Blue and Alizarin Red in whole embryos. For whole-mount in situ hybridization a 700 bp riboprobe against the most variable N-terminal region of Ndsts was employed (DIG RNA Labeling Kit, Roche, Mannheim, Germany). Immunohistochemical analysis of Ndst1 expression and western blotting was performed using an anti-Ndst1 antiserum (Grobe and Esko, 2002) followed by detection with goat anti-rabbit HRP-conjugated antibodies (Zymed, San Francisco, USA).
Quantitation of apoptosis was performed on paraffin sections of three mutant and three wild-type E15.5 embryos, using the TUNEL Assay Kit (Roche, Mannheim, Germany). BrdU-labelled nuclei were quantified using anti-BrdU antibodies (Zymed) on three mutant and wild-type E15.5 and E17.5 embryos. BrdU (70 μg/g mouse) was injected intraperitoneally and mothers were sacrificed after 1 hour. The number of BrdU-labelled cells relative to nonlabelled cells in the VZ was determined. Two different horizontal levels of the embryonic brains, 250 μm apart, were assessed in each embryo. Eight serial sections per level, each of three wild type and three mutant E17.5 embryos, were analyzed in anterior, posterior, median and lateral forebrain positions.
Patched expression was detected using anti-Ptch1 antiserum (Acris Antibodies, Hiddenhausen, Germany) and secondary FITC-labelled goat anti rabbit antibodies (Dianova, Hamburg, Germany) on three mutant and wild-type embryos. Images were taken on a Zeiss Axiophot microscope employing a 10×/0.3, a 20×/0.5 and a 63×/1.25 Zeiss objective, and a Leica DFC280 camera. Leica software was used for image capturing and Photoshop 7 software run on Macintosh computers for the generation of figures. Contrast and brightness were adjusted for whole images during figure assembly.
Preparation of HS
Three mutant, heterozygous and wild-type E15.5 embryos were pooled, digested with 2 mg/ml pronase in 320 mM NaCl, 100 mM sodium acetate (pH 5.5) overnight at 40°C, diluted 1:3 in water and applied to a 2.5 ml column of DEAE Sephacel. After washing the column with 0.3 M NaCl, the glycosaminoglycans were eluted with 1 M NaCl. For disaccharide analysis, the GAG pool was β-eliminated overnight at 4°C (0.5 M NaOH, 1 M NaBH4), neutralized with acetic acid until the pH was ~6 and applied to a PD-10 (Sephadex G25) column (Pharmacia, Uppsala, Sweden). Glycosaminoglycans eluting in the void volume were lyophilized, purified on DEAE as described above, again applied to a PD-10 column and lyophilized. The sample was digested using Heparin-lyase I, II and III and the resulting disaccharides were separated from undigested CS using a 3 kDa spin-column (Centricon, Bedford, USA) followed by HPLC analysis using Carbopac PA1 columns (Dionex, Sunnyvale, USA). HS preparations from pooled embryos were independently analyzed twice and statistical analysis was performed using Student’s t-test in Microsoft Excel.
Analysis of Fgf2-dependent Mapk pathway activation was performed using anti-Erk1/2 and anti-phospho-Erk1/2 polyclonal antibodies (Promega, Madison, USA). Fibroblasts derived from the heads of E14.5 wild-type and mutant embryos (n=4) were cultured in DMEM+10% FBS, starved for 20 hours in DMEM without FBS, incubated in complete medium, DMEM without FBS or 10 ng/ml Fgf2 in DMEM without FBS for 5 minutes, and lysed. Analyses were carried out in duplicate.
To generate soluble alkaline phosphatase-Shh fusion proteins, the N-terminal sequence of Shh (amino acids 25–198, Shh-N) was produced by PCR (sense primer, 5′AGATATCAATGTGGGCCCGGCAGGGGGTTTG3′; antisense primer, 5′ATCTAGAAGCCGCCGGATTTGGCCGCC3′), ligated into pGEM (Promega) and sequenced. Shh-N was ligated into pWIZ (Gene Therapy Systems, San Diego, USA) after limited HpaI and subsequent XbaI restriction of the vector, and EcoRV and XbaI restriction of the Shh PCR product. After transfection of B16/F1 cells, the secreted protein was bound to heparin and eluted as a single-peak with 0.6 M NaCl (Pharmacia, Uppsala, Sweden). The biological activity of the recombinant AP-Shh was confirmed by Shh-dependent alkaline phosphatase induction in C3H10T1/2 cells using the method described below. About 200 μg of purified lyophilized glycosaminoglycans (see above) derived from E14.5 mutant and wild-type embryos were covalently coupled to Affi-Gel 10 (BioRad) according to the manufacturers instructions, and the extent of coupling was determined by carbazole reaction. The supernatant of AP-Shh transfected B16/F1 cells was applied to the columns at a final concentration of 0.5 M salt to prevent unspecific binding and bound material was eluted with a NaCl gradient from 0–1 M in 0.1 M sodium acetate buffer (pH 6.0). AP activity was measured at 405 nm by addition of 120 mM p-nitrophenyl phosphate (pNPP, Sigma) in 0.1 M glycine buffer, pH 10.4. Integration of elution peaks was performed employing Origin software (Origin Lab, Northampton, USA).
Results
Targeted disruption of Ndst1
To study the role of HS in mammalian biology, conditional knockout mice for Ndst1 were generated using the cre-loxP system and homologous recombination in embryonic stem (ES) cells (Fig. 1). In the targeting vector, Cre-recombination sequences (loxP-sites) were positioned in intron sequences surrounding the second exon of Ndst1, which includes most of the 5′ untranslated region and 171 of 882 amino acids of the ORF, containing the signal peptide, cytoplasmic tail, transmembrane region and part of the catalytic domain (Fig. 1A). Chimaeric mice were generated by blastocyst injections of four ES-cell clones. Two resulting type II Ndst1 mouse lines derived from independent ES cell clones were crossed with ZP3 Cre mice, deleting the ‘floxed’ allele in the oocyte and generating mice with a systemic deletion of Ndst1 (Type I) (Fig. 1A). The distribution of genotypes in the newborn floxed mice showed no deviation from the expected Mendelian distribution (n=36). No surviving Ndst1−/− pups could be detected from crosses of Ndst1+/− mice (n=31), confirming the finding that Ndst1−/− pups die at or just before birth (Fan et al., 2000; Ringvall et al., 2000). Genotypic analysis of embryos from E10-E18.5 indicated that some of the Ndst1−/− mice arrested their development in utero (27.5% Ndst1+/+, 54% Ndst1+/−, 18.5% Ndst1−/−; n=378).
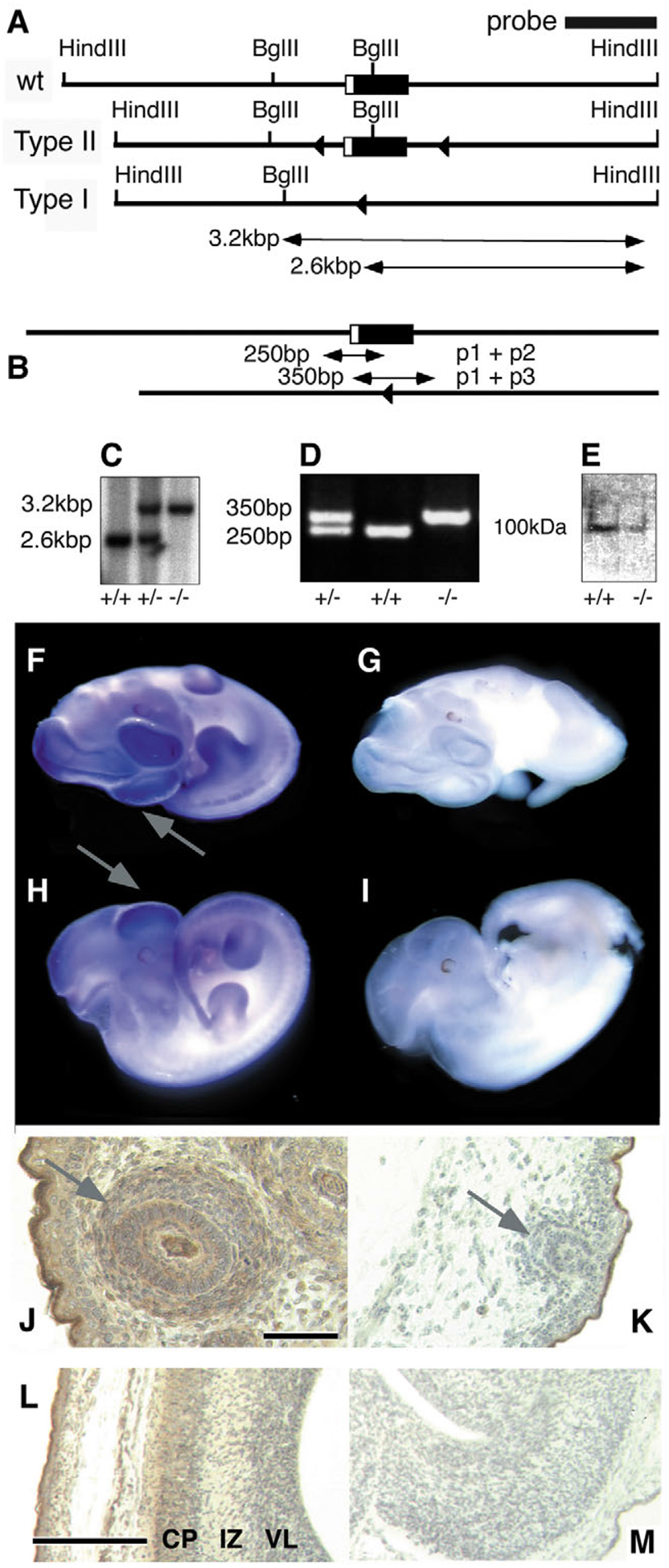
Disruption of the Ndst1 gene by targeted recombination. (A) Maps of the wild-type Ndst1 locus, the Type II ‘floxed’ allele and a Type I deletion allele, obtained after breeding of Type II mice with ZP3-Cre mice. Lox-sites located in intron sequences are shown as triangles. (B) PCR analysis. Deletion of exon 2 in Ndst1−/− mice yields a 350 bp product, whereas wild-type mice produce a 250 bp amplification product. Heterozygous mice yield both amplification products. (C) Southern blot analysis of DNA. DNA digestion using restriction endonucleases HindIII and BglII yields a 2.6 kbp (wild type) and a 3.2 kbp (Ndst1−/−) band. (D) PCR analysis of the same samples as in (C). (E) Western blotting of total embryo extract after SDS-PAGE. Samples were detected by an affinity-purified rabbit-anti-mouse Ndst1 antiserum (Grobe and Esko, 2002). The weak signal seen in the knockout lane is due to cross-reactivity with Ndst2–4, all of which are expressed in the developing embryo (Aikawa et al., 2001). (F-I) Detection of Ndst1 expression in the developing embryo. (F,H) Whole-mount in situ hybridization in E11.5 wild-type embryo showed strongest expression (blue stain) in the forebrain and frontonasal/maxillary processes (arrows). (G,I) Ndst1−/− embryo probed with an antisense riboprobe showed no reactivity. (J-M) Immunohistochemical detection of Ndst1 expression in E16.5 embryos using the affinity-purified rabbit-anti-mouse Ndst1 antiserum (Grobe and Esko, 2002). Ndst1 protein is strongly expressed (brown) in frontonasal mesenchyme (J) and telencephalic walls (L). In the developing face, hair follicles show the highest expression levels (arrow). (K,M) The hair follicles and telencephalic walls are not stained in Ndst1−/− tissues. CP, cortical plate; IZ, intermediate zone; VL, ventricular layer. (M) There is a lack of stratification in the Ndst1 mutant cortex. Scale bar: 10 μm in J; 100 μm in L.
Ndst1 expression and HS composition in the mouse embryo
Whole-mount in situ hybridization was performed to elucidate expression of Ndst1 in the developing embryo. By E11.5, Ndst1 expression was strongest in the developing brain and frontonasal process, as well as distal limb structures (Fig. 1F,,H).H). As Ndst proteins are known to be translationally regulated (Grobe and Esko, 2002), we also investigated Ndst1 expression on the protein level in E16.5 mice. Strong expression was observed in the frontonasal process (FNP) and maxillary prominences of the face, especially in hair follicles (Fig. 1J). Ndst1 protein expression was also detected in the forebrain, with highest levels in the cortical plate and the ventricular layer (Fig. 1L). Specificity of the Ndst1 antiserum was confirmed by using Ndst1 mutant embryo sections as controls (Fig. 1K,,MM).
Disaccharide analysis of purified E14.5 mouse embryo HS revealed that the relative amount of N-sulfated disaccharides decreased in the mutant relative to the wild type (13% versus 30%, respectively), whereas the relative amount of N-acetylated disaccharide increased proportionately (n=2, P<0.001) (Table 1). The relative amount of 6-O-sulfated disaccharides was only mildly reduced in the mutant (16.7% versus 19.2%, respectively), whereas the amount of 2-O-sulfated disaccharides was more dramatically affected (3.8% versus 9.7%). Notably, heterozygous mice for Ndst1 showed only minor changes in HS composition, with small reductions in the amount of sulfated disaccharides. Analysis of the amount of galactosamine relative to glucosamine in the glycosaminoglycan fraction, which provides a relative estimate of chondroitin sulfate (CS) relative to HS, showed that in E14.5 embryos, the CS/HS ratio was reduced by only 7% and in E15.5 embryos by 12%. These data suggest that the phenotype detected in Ndst1 mutant mice is related to the altered fine structure of the HS chains, rather than to a change in the ratio of these two glycosaminoglycans.
Table 1.
Reduced amount of sulfated disaccharides isolated from HS of mutant embryos
| IV–A | IV–S | II–A | II–S | III–S | I–S | |
|---|---|---|---|---|---|---|
| Wild type | 59.1±0.8 | 16.5±0.8 | 11.5±0.8 | 3.3±0.1 | 5.3±0.2 | 4.4±0.8 |
| Ndst1−/− | 75.7±0.7 | 6.5±0.2 | 11.2±0.9 | 2.9±0.14 | 1.2±0.07 | 2.6±0.14 |
| Ndst1+/− | 62.3 | 13.9 | 12.2 | 3.1 | 4.4 | 4.1 |
HS was isolated from wild-type, Ndst1−/− and Ndst1+/− embryos, and samples were digested with heparin lyases. The resulting disaccharides were analyzed by anion exchange HPLC. HS from Ndst1−/− embryos was undersulfated, resulting in a decrease in N-sulfated and 2-O-sulfated disaccharides, especially III-S and I-S. HS derived from heterozygous embryos showed only mild effects on sulfation. Values indicate the mean % of total disaccharide. Three pooled mutant, wild-type and heterozygous embryos were investigated in one experiment. Results for the wild-type and mutant embryos are the mean±s.d. from two independent experiments. The abbreviations used indicate the following disaccharides: IV-A (ΔUA1,4GlcNAc), II-A (ΔUA1,4GlcNAc-6S), IV-S (ΔUA1,4GlcNS), II-S (ΔUA1,4GlcNS-6S), III-S (ΔUA2S1,4GlcNS) and I-S (ΔUA2S1,4GlcNS-6S). Students t-test revealed statistical significance for the differences observed (P<0.001, P<0.002, P<0.045, P<0.001 and P<0.05 for the change in IV-A, IV-S, II-S, III-S and I-S between wild-type and mutant HS, respectively).
Abnormal anatomy and histology of Ndst1−/− mice
Ndst1−/− embryos displayed developmental defects of varying severity. Of 71 Ndst1−/− mutants, 61 (86% of total mutants) had relatively mild developmental defects, such as lack of the eye lens/iris coloboma (n=57), uni or bilateral microophthalmia (small eyes, n=11), complete lack of eyes (n=4), median cleft lip (n=4), and underdeveloped frontonasal and mandibular/maxillary prominences (n=8) (Fig. 2B,,D).D). The cerebral cortex was significantly smaller in null embryos (70% of wild-type size, n=6, P<0.001). Another group of Ndst1−/− embryos (n=10, 14% of total mutants) showed very severe phenotypic defects, such as the lack of a defined skull, including the complete lack of eyes, partial lack of the frontonasal process, mandibular/maxillary prominences and agnathia (lack of lower jaw) (Fig. 3B). Strikingly, the remaining body of these mutants showed no dysmorphology.
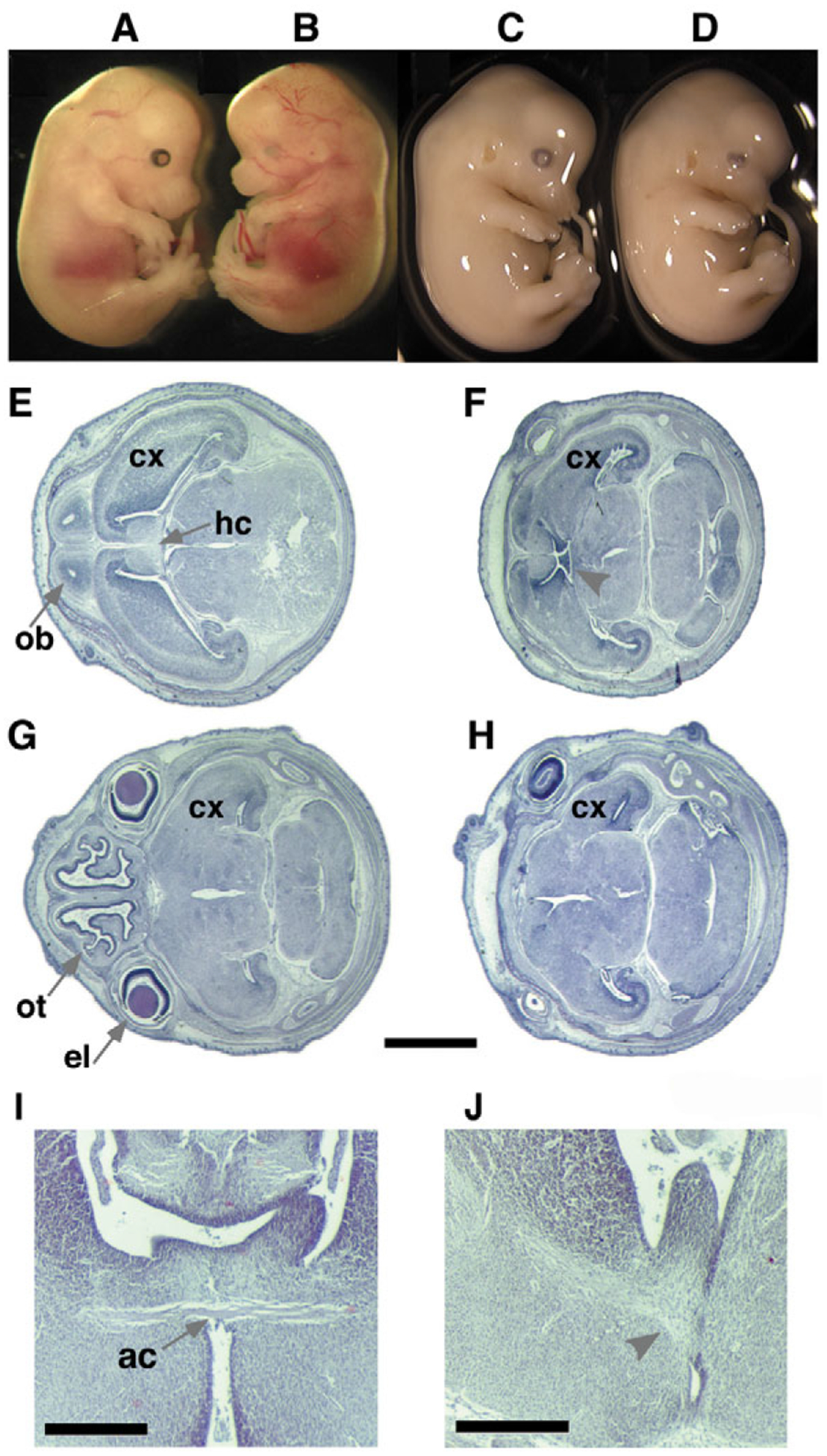
Phenotypes of mildly affected Ndst1 mutant E13.5 embryos. (A,C) Wild-type littermate controls. (B,D) Mutant embryos show lack of eyes and reduced frontonasal process (B) and (D) lack of eye lens. (E,G) Wild-type littermate controls. (F,H) Mutant embryos with mild defects in external features show severe defects in the frontonasal process and forebrain (E16.5, horizontal sections). Lack of olfactory tracts (ot), olfactory bulbs (ob), the eye lens (el) and the hippocampal commissure (hc, arrowhead) is evident. The cortex (cx) is reduced in size and stratification and the cleavage along transverse, sagittal and horizontal axes is defective. Scale bar: 1 mm. (I,J) Coronal sections of a E18.5 wild-type (I) and Ndst1−/− (J) brain show the lack of the anterior commissure (ac) in the mutant embryo (arrowhead). Axons disperse instead of crossing the midline. Scale bar: 500 μm.
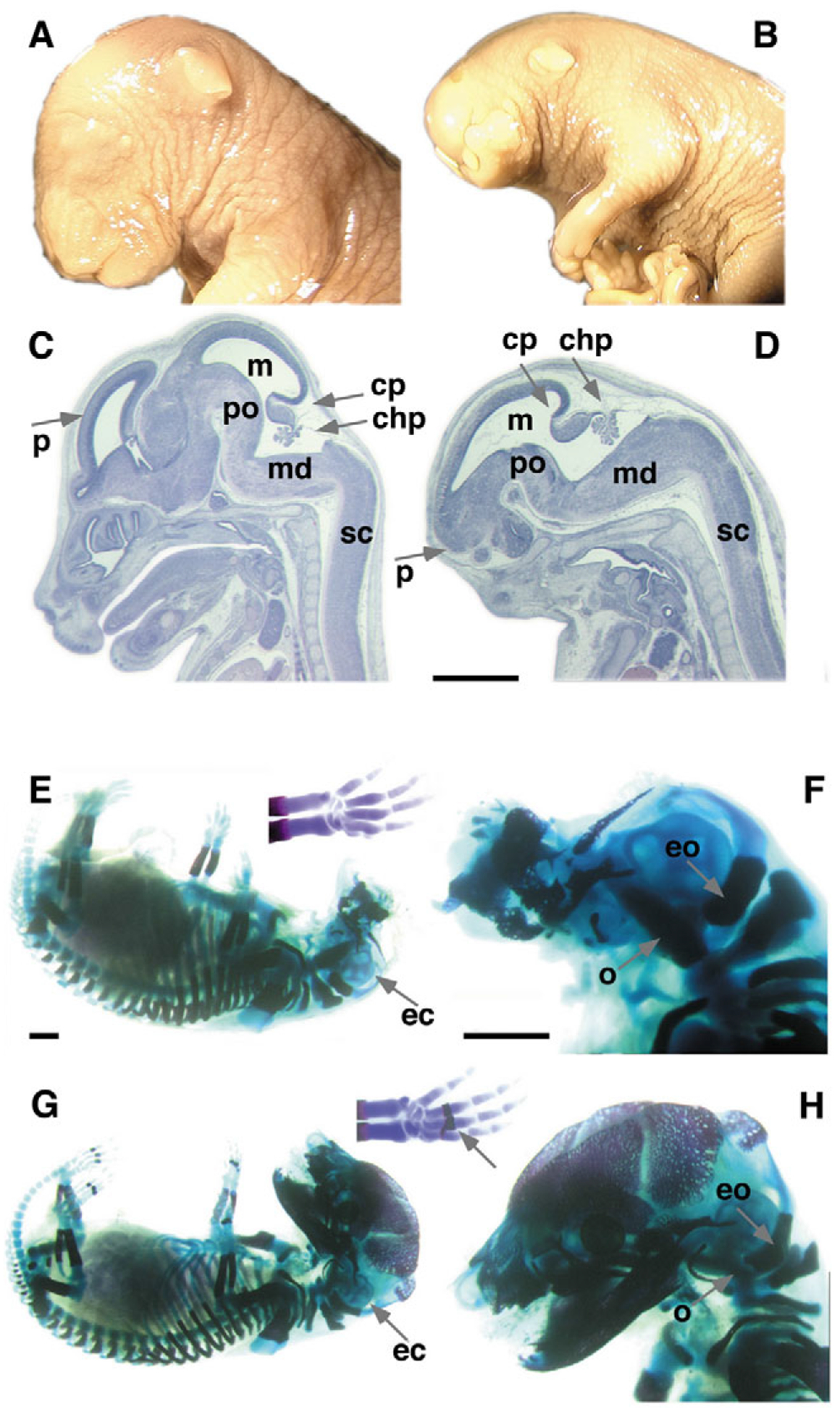
Anatomy and histology of strongly affected E18.5 Ndst1 mutant embryos. (A,C) Wild-type embryo, (B,D) Ndst1 mutant. Gross examination revealed severe frontonasal dysmorphism, including the lack of eyes but normal outer ear. (D) Hypoplastic prosencephalon (p). The midbrain (m), pons (po), medulla (md), the cerebellar primordium (cp), choroid plexus (chp) and spinal cord (sc) appear normal (sagittal sections). (E-H) Bone (red) and cartilage (blue) stain of E18.5 embryos. (E,F) Severely affected Ndst1 mutant embryos show lack of all neural crest cell-derived bones of the viscerocranium and neurocranium. Some lack or delay of ossification occurs, especially in the vertebrae and (shortened) limb digits (compare with wild type, arrow in G), which may be fused (E). The ear capsule (ec) is intact in these embryos (E,F), as are the occipital (o) and exoccipital (eo) bones, which are derived from paraxial mesoderm. (G,H) Wild-type littermate controls. Scale bars: 1 mm.
Histological analysis revealed that the development of the brain was affected in Ndst1−/− embryos (Figs 2F,,HH and and3D).3D). One group of embryos had an only mildly affected external appearance, but the forebrain showed patterning defects and typically lacked olfactory bulbs. Moreover, the anterior and hippocampal commissures were absent or hypoplastic in all cases (arrowheads, Fig. 2F,,J).J). Coronal sections revealed a hypoplastic pituitary in mutant embryos (not shown). A second group were severely affected (Fig. 3D). The size of the diencephalon and telencephalon was extremely reduced, whereas other brain regions surprisingly showed no strong dysmorphology. Forebrain-derived structures were also missing in these embryos. The neural crest-derived neurocranium and viscerocranium were almost completely absent, whereas the mesoderm-derived skeleton of the body appeared to be normal, with the exception of delayed ossification in the digits and vertebrae (Fig. 3E,,F).F). Ndst1 heterozygous mice were normal.
Ndst1 is a regulator of Shh and Fgf signalling in the developing frontonasal process and maxillary prominences
The phenotype of severely affected Ndst1−/− embryos strongly resembles chick embryos deficient in Fgf8 and Shh signalling (Schneider et al., 2001), and mouse embryos carrying neural crest specific deletions of Shh (Jeong et al., 2004) and Fgf8 (Trumpp et al., 1999). Thus, we decided to test Ndst1 mutant mice for impaired Shh and Fgf signalling as possible causes for the observed developmental defects.
First, Ndst1; Shh compound mutant embryos were produced and analysed (n=28) to test for genetic interactions. All compound heterozygous mice showed the expected Mendelian frequency and no brain abnormalities, which can be explained by the relatively low reduction of sulfation in Ndst1 heterozygous embryos (Table 1). However, four out of eight compound heterozygous E15.5 embryos showed hypoplastic frontonasal/maxillary processes as well as a hypoplastic lower jaw (Fig. 4C), whereas the remaining four embryos were normal. Histological analysis of the four affected embryos revealed a normal brain morphology including the olfactory bulbs, but absent olfactory epithelium, an absent tongue, severe developmental defects of the eyes and absent/hypoplastic eye lenses (Fig. 4E). Surviving compound heterozygous mice generally had smaller eyes and snouts. Shh heterozygous mice, however, did not manifest any defects, suggesting that Ndst1 deficiency sensitizes the animals to changes in Shh expression.
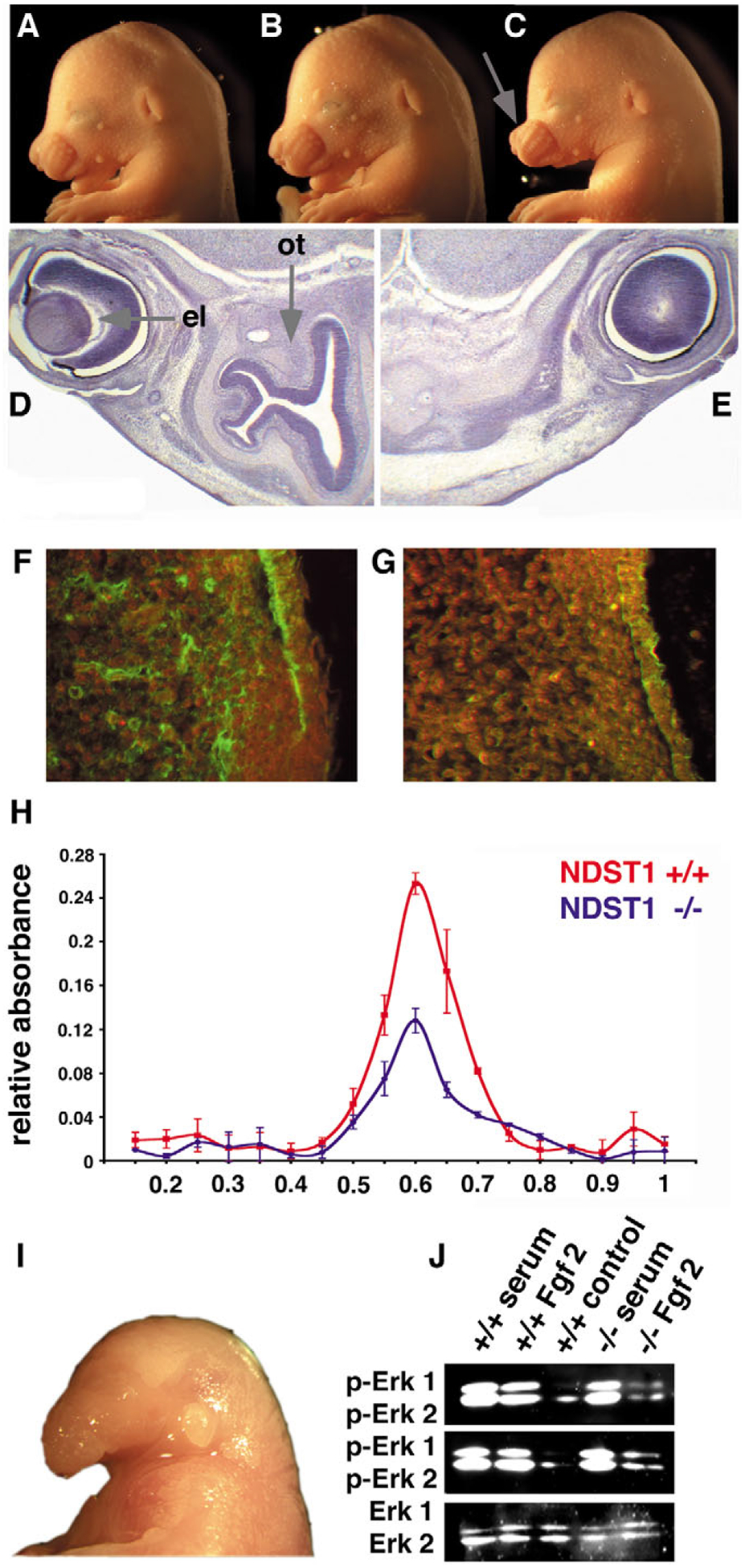
Ndst1; Shh interact genetically and biochemically. Ptch1 protein expression is reduced in the developing face of Ndst1−/− embryos and Fgf signalling is also affected. (A-C) Craniofacial morphology in E15.5 embryos. (A) Ndst+/− and (B) Shh+/− littermate controls. (C) Some Ndst1; Shh compound heterozygous mice showed hypoplastic maxillary processes (arrow). (D,E) Haematoxylin/Eosin staining of horizontal sections of embryos in A,C, respectively. The olfactory epithelium (ot) is absent in the Ndst1, Shh compound heterozygous mouse, and development of the eye lens (el) is also affected, both resembling features found in the Ndst1−/− mutant embryos (Fig. 2F,,H).H). (F) Wild-type embryo, (G) Ndst1−/− embryo. (F,G) Reduced expression of the Shh receptor Ptch1 in the frontonasal process of Ndst1−/− mutant E15.5 embryos, as assessed by immunohistochemical staining. Three wt and mutant embryos were analyzed, representative sections are shown. Green fluorescence indicates Ptch1 expression. (H) More AP-Shh binds to E14.5 wild-type GAGs (red line) than to GAGs isolated from mutant embryos (blue line). Equal amounts of GAGs isolated from these embryos were coupled with Affi-Gel, loaded with recombinant, soluble alkaline phosphatase fused to Shh (AP-Shh) followed by elution with increasing NaCl concentrations ranging from 0–1 M in 50 mM increments. (I,J) Impaired Fgf signalling in Ndst1 mutant embryos. (I) Twenty-five percent of all Ndst1−/− embryos showed developmental defects of the first branchial arch, including agnathia strongly reminiscent of neural- and neural-crest specific Fgf8 mutant mice (Trumpp et al., 1999). One representative E18.5 Ndst1 mutant embryo is shown. (J) Fgf-dependent Erk1/2 phosphorylation is significantly reduced in Ndst1 mutant mesenchymal cells isolated from two mutant embryos (top and middle). Cultured cells derived from facial mesenchyme of two E14.5 wild-type and two mutant littermates were stimulated with 10% serum in DMEM or 10 ng/ml Fgf2 in DMEM for 5 minutes before analysis. Phosphorylation of Erk1 and Erk2 (p-Erk1 and p-Erk2) were observed in the wild type after addition of serum or Fgf2; however, Fgf2 addition alone failed to stimulate Erk1/2 phosphorylation in the Ndst1 mutant cells. Control refers to no stimulation. (Bottom) Detection of total Erk1 and Erk2 with anti-Erk antibodies.
This effect was enhanced in Ndst1−/−; Shh+/− embryos, which were produced three times less than expected (n=5, 4%). Four out of the five embryos showed very dramatic developmental defects, resembling phenotypes found only in the second group of most strongly affected Ndst1−/− embryos (14% of total Ndst1−/− mutants). In both Ndst1−/− and Ndst1−/−; Shh+/− animals, the eyes and the frontonasal and maxillary prominences were severely hypoplastic. Brain morphology was also affected in Ndst1−/−; Shh+/− embryos, showing a collapsed fore- and midbrain in all cases (not shown). Only one extremely small and malformed E10.5 embryo with genotype Ndst1−/−; Shh−/− was produced (six times less than expected), showing that additional developmental pathways were also affected. The phenotypes of compound mutant mice demonstrate that Shh signalling is decreased in the developing FNP/maxillary prominences and may also be affected to a lesser degree in the forebrain of Ndst1-deficient mice. Based on those experiments it is not clear, however, whether Shh signalling was affected directly because of reduced Shh-HS-interaction or indirectly via an unknown, HS-binding protein influencing Shh signalling.
Next, we investigated impaired Shh signalling by assessing the expression of the Shh receptor patched (Ptch1) immunohistochemically in E15.5 embryos (Fig. 4F,,G).G). Ptch1-expression was strongly reduced in the developing face (n=3), an area that was strongly affected in the mutant and was previously found to express Ndst1 at high levels (Fig. 1F,,HH,,J).J). Last, binding of a soluble fusion protein consisting of alkaline phosphatase and Shh (AP-Shh) to Ndst1−/− derived HS was performed to demonstrate whether reduced Ptch1 expression in the mutant embryo, as well as the enhanced phenotypic defects in Ndst1; Shh compound mutant mice, could possibly be explained by impaired Shh-HS interactions. Indeed, binding of AP-Shh was reduced to 39% and 45% compared with binding to wild-type HS (n=2, Fig. 4H). The AP-Shh protein was expressed in a functional form, because it induced expression of alkaline phosphatase in C3H10T1/2 cells (not shown). Moreover, AP-Shh bound to heparin-sepharose and required 0.6 M NaCl to elute (not shown). Similar results were described for binding and elution of recombinant Shh consisting of residues 25–198 expressed in E. coli (Roelink et al., 1995).
Taken together, the genetic interaction between Ndst1 and Shh, the reduced binding of recombinant AP-Shh to mutant HS and the alteration in Ptch1 expression in Ndst1 mutant embryos all suggest that impaired signalling via Shh contributes to the observed phenotypes in Ndst1−/− embryos.
We next investigated if impaired Fgf signalling contributed to the observed phenotypes. About 25% of Ndst1−/− mice showed agnathia and other facial defects (Fig. 4I) strongly reminiscent of mice carrying a conditional, neural crest and neuron-specific Fgf8 deletion (Trumpp et al., 1999). To test whether impaired Fgf receptor signalling and mitogen activated protein (Map) kinase activity contributed to the phenotype, fibroblasts were derived from facial mesenchyme of two wild-type and two mutant E14.5 embryos. The cells were starved from serum for 20 hours and then stimulated with 10 ng Fgf2 or complete growth medium containing serum (Fig. 4J). Erk1/2 phosphorylation was strongly stimulated in fibroblasts derived from both wild-type embryos in response to Fgf2 or serum. By contrast, Erk1/2 phosphorylation in Ndst1−/− fibroblasts was unchanged in response to Fgf2. Erk1/2 was not affected per se, as the same level of Erk protein was present and serum stimulation activated phosphorylation in the mutant. These findings indicate that mutant HS in cells derived from the developing face was less effective as a Fgf co-receptor. Thus, the craniofacial phenotype in the mutant could also result from defective Fgf signalling.
Neural precursor cells show enhanced apoptosis and moderately decreased proliferation
We next investigated the functional basis for the observed morphological defects of the brain by assessing proliferation and apoptosis in the brains of mutant animals and wild-type littermate controls. The number of cells undergoing apoptosis was significantly enhanced in the neopallial cortex of E15.5 Ndst1−/− embryos (three mutant embryos and wild-type littermates were investigated, Fig. 5A,,BB,,I),I), as well as in the subventricular zone (Fig. 5C,,DD,,I).I). Cell proliferation in the ventricular zone of the prosencephalon, however, was found to be comparable in most brain regions of E15.5 and E17.5 embryos (Fig. 5J). Reduced proliferation could be detected only in lateral areas of the developing E17.5 cortex (Fig. 5E,,FF,,J).J). Immunohistochemical staining using Gfap and S100 antibodies, which detect glia-derived cell types, revealed that the number of glial cells in the mutant embryonic E18.5 brain was reduced (Fig. 5H).
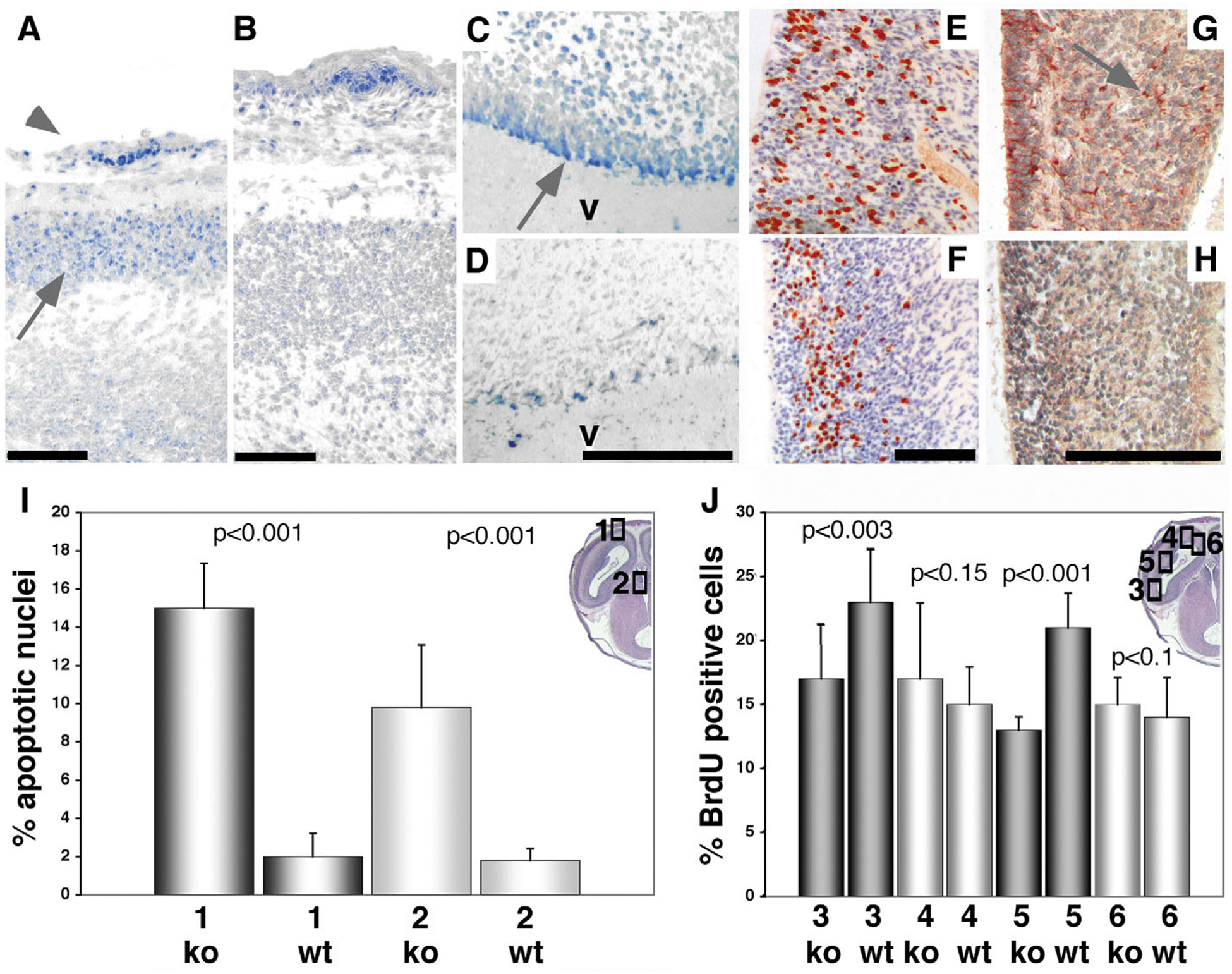
Enhanced apoptosis, decreased proliferation and impaired glia development in Ndst1−/− embryos. (A,B) Apoptosis is significantly enhanced in the forebrains of mutant embryos. Skin (arrowhead) serves as a positive control. (A) TUNEL staining reveals very high levels of cell death (blue nuclei) in the neopallial cortex (arrow) of E15.5 Ndst1−/− embryos, a region expressing Ndst1 at high levels (Fig. 1L). (B) Wild-type littermate controls. (C) Cells residing in the ependymal and ventricular layers of the diencephalon in Ndst1−/− embryos also undergo apoptosis. (D) Wild-type littermate control. v, ventricle. Horizontal sections. (E,F) Proliferation (brown stain) in a E17.5 mutant (F) embryo compared with the wild type (E) lateral telencephalic wall. In mutant mice, the proliferative rate is reduced in the lateral telencephalic wall of the brain (F), and dividing cells are more clustered within the VZ if compared with the wild type. Horizontal sections. (G,H) Reduced Gfap (red) staining indicating lower numbers of glia cells in mutant E18.5 forebrain (H). The wild-type brain (G) shows presence of those cells, which provide a scaffold for lateral neural cell migration from the ventricular zone. Scale bars: 50 μm. Horizontal sections. (I) Quantitation of apoptotic cells of the neopallial cortex (1, inset) and the ependymal layer (2) of the dorsal part of the third ventricle of three mutant and wild-type embryos. (J) Quantitation of BrdU positive nuclei in the posterior (3), anterior (4), lateral (5) and medial (6) telencephalic ventricular zones of three mutant and wild-type E17.5 embryos. Horizontal sections. A moderate reduction in proliferation could only be observed in the lateral area of Ndst1−/− embryos. Coronal sections also revealed similar proliferative rates in the medial and parietal cortex of wild-type and mutant embryos (not shown). Analysis of three E15.5 mutant and wild-type embryos showed no differences in proliferation (not shown).
Discussion
Ndst1-deficient mice were previously produced by conventional gene targeting, but craniofacial defects in those mice were not analyzed in detail (Fan et al., 2000; Ringvall et al., 2000). Mutant mice died perinatally because of hypoplastic lungs. We describe that Ndst1 mutant mice show striking developmental defects of the face, the forebrain and the eyes, regions that normally express Ndst1 at high levels (Fig. 1F,,HH,,JJ,,L).L). Abnormalities in mutant embryos are highly penetrant, but variable in expressivity.
Interestingly, eye defects have also been described in other HS-deficient mice. Mice that lack the uronyl 2-O-sulfotransferase (2-Ost) (Bullock et al., 1998) or uronyl C5-epimerase (Li et al., 2003) genes exhibit eye defects and also show a reduction in the thickness of the embryonic cerebral cortex (McLaughlin et al., 2003). Uronic acid 2-O-sulfation was strongly reduced in Ndst1 mutant embryos (Table 1), indicating some of the defects may result from altered 2-O-sulfation in addition to decreased N-sulfation. By contrast, 6-O-sulfation was not dramatically affected, which has also been shown for various organs in the Ndst1 E18.5 mutant embryos (Ledin et al., 2004) and murine ES cells deficient in Ndst1; Ndst2 function (Holmborn et al., 2004). The finding that the relative amount of GlcNAc in relation to GlcNS residues is elevated in the mutant indicates that Ndst1 plays a prominent role in HS modification, and that the other three isoforms do not fully compensate for the loss of Ndst1 function (Aikawa et al., 2001).
As an underlying reason, we assumed possible impairment of the two HS-binding factors Shh and Fgf based on a number of published observations. First, the phenotype of the most strongly affected Ndst1 mutant embryos strongly resembles chick embryos made deficient in Shh and Fgf8 signalling (Schneider et al., 2001). Oligodendrocyte lineage specification also depends on Fgf and Shh (Tekki-Kessaris et al., 2001) and oligodendrocyte precursors express highly sulfated, heparin-like HS (Stringer et al., 1999), which possibly explains the finding that the number of glia was found to be reduced in the telencephalon of Ndst1 mutant embryos. Moreover, the single Ndst ortholog in the fly, Sulfateless, is known to affect signalling mediated by Wingless, Hedgehog and Fgf (Lin et al., 1999; Lin and Perrimon, 1999; The et al., 1999). Thus, multiple pathways might be affected by altering Ndst1 expression throughout the brain and face.
Sonic hedgehog binds HS (Rubin et al., 2002), and mice lacking Shh function show defective axial patterning of the brain, eye, skeleton and limbs (Chiang et al., 1996a). Shh is also necessary for frontonasal, pituitary and brain development after initial patterning takes place (Ahlgren and Bronner-Fraser, 1999; Britto et al., 2002; Hu and Helms, 1999; Jeong et al., 2004; Treier et al., 2001). All of these developmental processes were found to be affected in Ndst1 mutant embryos, suggesting that the observed phenotypes could be explained in part by impaired Shh signalling. Facial and eye defects in 50% of Ndst1+/−; Shh+/− (n=4), like those found in 86% of Ndst1−/− (n=61) embryos, was consistent with this hypothesis. In addition, Ndst1−/−; Shh+/− embryos closely resemble reported phenotypes in which Shh signalling was inhibited after the initial patterning events occur, resulting in small head and brain size, craniofacial abnormalities, defects in all neural crest derived bones of the skull, lack of a tongue, and abnormal folding and collapse of the forebrain (Ahlgren and Bronner-Fraser, 1999; Britto et al., 2002; Hu and Helms, 1999; Jeong et al., 2004). Together, these results demonstrate a likely function of Ndst1 as a modifier of Shh signalling in the developing face. The simplest interpretation is that a decrease in sulfation of HS decreases the interaction of Shh with matrix and cell surface HS-proteoglycans expressed in the developing face, which in turn affects Shh gradient formation/signalling. Consistent with this idea, we found that AP-Shh binding to mutant HS was reduced by more than 50% in vitro. As the total amount of HS in E14.5 mutant embryos and wild-type littermates was found to be similar, a reduced number of Shh binding sites present on the undersulfated HS may be the most likely reason for this observation (Fig. 4H).
Loss of Shh signalling and mutations in other genes of the Shh signalling pathway, including patched, Gli2 and dispatched, cause holoprosencephaly (HPE) (Ma et al., 2002; Ming et al., 2002; Muenke and Beachy, 2000), the most common human forebrain defect with an incidence of 1:250 in embryos and 1:16,000 in newborn infants (Muenke and Beachy, 2000; Muenke and Cohen, 2000). Severe, alobar HPE is characterized by severe facial dysmorphism and the presence of a small monoventricular cerebrum, whereas the body is not strongly affected. In semilobar HPE, rudimentary cerebral lobes are present but the interhemispheric fissure is not complete, and olfactory tracts and bulbs are absent or hypoplastic. In mild lobar HPE, the brain may be cleaved and of normal size, but midline cleavage of the thalami and corpora striata may be incomplete, pituitary anomalies may be present and the eyes can be small. Olfactory tracts and bulbs may be absent as well. Various gradations of facial dysmorphism are commonly associated with HPE, including median cleft lip, midfacial hypoplasia and iris coloboma. (Muenke and Cohen, 2000). All mildly affected Ndst1 mutant mice (Fig. 2) show defects reminiscent of semilobar and lobar HPE, such as midfacial hypoplasia, reduced cerebral size, median cleft lip and palate, lack of olfactory bulbs, iris coloboma, a hypoplastic pituitary and commissural defects. In addition, the lack of neural crest cell derived skeletal structures commonly seen in the Ndst1 mutant has also been described in severe cases of HPE (Chiang et al., 1996b; Hu and Helms, 1999; Roessler et al., 1996; Schneider et al., 2001). This is supported by the finding that a partial loss of Ext function, another enzyme involved in HS synthesis, also leads to an HPE-like phenotype (Mitchell et al., 2001).
The following explanations may account for the finding that Ndst1−/− embryos do not completely phenocopy Shh mutant mice. First, only partial impairment of Shh signalling in these mice might occur based on the finding that sulfation of HS is not completely diminished (Table 1). Second, a variation in HS sulfation in the mutant might occur temporally and spatially, thus affecting developmental programs and tissues differentially. Modification of the Shh signalling pathway at different times of human embryonic development has been described to possibly account for the wide phenotypic spectrum of HPE (Cordero et al., 2004), and differences in Shh binding to HS derived from E14.5 and E17.5 mouse embryos have been observed in vitro (S.R.P. and K.G., unpublished), indicating that HS-coregulation of Shh signalling might depend on a specific chemical structure of HS. A third possibility explaining the variable phenotype of Ndst1 mutant embryos may be the dynamic expression of the other three Ndst isoforms (e.g. in the brain and developing face) that might account for a variable partial compensation of Ndst1 deficiency and thus the different degree in which specific tissues are affected.
Perhaps, the most intriguing explanation for the observed phenotypic variation is the impairment of different combinations of HS-binding growth factors, e.g. Wnt proteins, bone morphogenetic proteins (Bmps), Hh proteins and Fgf proteins. For example, Shh and Fgf8 act synergistically in cartilage outgrowth during cranial development in the chick (Abzhanov and Tabin, 2004). Simultaneous impairment of two soluble factors and their respective signal transduction pathways could result in a greater variation and severity of the resulting phenotypes that cannot be attributed to any of one of these factors acting independently. Wnt signalling might also contribute to some of the observed defects in Ndst1-mutant embryos, as developmental defects of the viscerocranium and neurocranium reminiscent of those found in the Ndst1−/− embryos (Fig. 3F) were described in β-catenin mutant embryos (Brault et al., 2001). Interestingly, these alterations are comparable with those found in severe HPE cases that include absence of most crest cell-derived skeletal structures (Chiang et al., 1996b; Hu and Helms, 1999; Roessler et al., 1996; Schneider et al., 2001). Last, HS is described to be required for axon guidance by interacting biochemically with known axon guidance molecules. Slit2, a repulsive guidance molecule, was purified in part by its binding to heparin (Wang et al., 1999) and mammalian Slits were isolated in a search for brain proteins that bind the HS proteoglycan glypican (Glp1) (Liang et al., 1999). Here, the HS chains of Glp1 mediate its binding to the Slits (Ronca et al., 2001). In Drosophila cell extracts, the HS proteoglycan Syndecan (Sdc) co-immunoprecipitates both with Slit and its receptor Robo (Johnson et al., 2004), and HS-Slit interaction was also shown to occur in mice (Inatani et al., 2003). In vitro, heparin was also shown to bind both netrin 1 and its receptor Dcc, which mediate attractive and repulsive responses (Bennett et al., 1997; Serafini et al., 1994). The axon guidance deficiencies found in Ndst1 mutant mice might thus have originated from affected molecules involved in attractive or repulsive axon guidance in the developing brain.
We also investigated Fgf activity in the Ndst1 mutant embryos. Fgf family members as well as their receptors are known to require HS for the formation of high affinity Fgf and Fgf receptor complexes and subsequent signalling (Rapraeger et al., 1991; Yayon et al., 1991). In addition, a crucial role of the Ndst homolog Sulfateless in the generation of Fgf-binding sites has been described in Drosophila (Lin et al., 1999) and in cell culture (Ishihara et al., 1993), which was confirmed by recent work showing that Fgf2 and Fgf8 signalling is affected in the developing mouse brain deficient in Ext1 function (Inatani et al., 2003). Thus, we assumed Fgfs to be affected in the Ndst1 mutant embryo as well, possibly contributing to some of the observed phenotypes. Relatively little expansion of either the mandibular or maxillary primordia, both being derived from the developing first branchial arch and resulting in agnathia, microglossia (small tongue) and forebrain defects were found in neural- and neural crest-specific Fgf8 mutant mouse embryos (Trumpp et al., 1999). Similar defects were also observed in 25% of Ndst1−/− embryos (Fig. 4I). Interestingly, agnathia alone occurs rarely but is often associated with forebrain defects such as holoprosencephaly, a combination also present in many severely affected Ndst1 mutant embryos. Additionally, the reduced cortex size and compression of the cortical layers seen in all Ndst1 mutant embryos has also been described in Fgf2 mutant mice (Dono et al., 1998; Raballo et al., 2000; Vaccarino et al., 1999). Fgf2 dependent activation of the Mapk pathway was strongly reduced in fibroblasts derived from facial mesenchyme of E14.5 mutant embryos (Fig. 4J), demonstrating that Ndst1 may modify Fgf signalling at least in facial primordia, but possibly also in the developing nervous system. Last, Fgfs play important roles in neurogenesis, axon growth, differentiation and neuronal survival (Reuss and von Bohlen und Halbach, 2003), all of which are affected in Ndst1-deficient embryos.
From this work, we conclude that Ndst1 is a crucial regulator of mammalian forebrain and facial development, that properly modified HS is necessary for neuronal, glia and neural crest development, and that impaired Shh and Fgf signalling (probably in combination with other soluble HS-binding factors) contributes to the observed phenotypes. Last, our findings imply that mutations in HS synthesizing genes, notably the Ext and Ndst genes, might contribute to developmental defects of the brain and face in humans, including defects of the mandible that are associated with more than 130 syndromes and thus are among the most common malformations.
Acknowledgments
The authors thank Dr Nissi Varki (University of California, San Diego, USA) for histological work and helpful discussions, Dr Bradley Hayes (Glycotechnology Core, University of California, San Diego, USA) for disaccharide analysis and Drs. L. Kjellén, M. Ringvall (Uppsala University, Sweden) and T. Fischer (Max-Planck Institute of Experimental Medicine, Göttingen, Germany) for helpful discussions. This work was supported by DFG (German Research Council) grants GR1748/1–1, GR1748/1–2 and SFB 492-B15 (to K.G.), National Institutes of Health grants GM33063 and HL57345 (to J.D.E.), and R01 NS41332 (to Y.Y.). The authors state that they have no competing interests.
References
- Abzhanov A and Tabin CJ (2004). Shh and Fgf8 act synergistically to drive cartilage outgrowth during cranial development. Dev. Biol 273, 134–148. [Abstract] [Google Scholar]
- Ahlgren SC and Bronner-Fraser M (1999). Inhibition of sonic hedgehog signaling in vivo results in craniofacial neural crest cell death. Curr. Biol 9, 1304–1314. [Abstract] [Google Scholar]
- Aikawa J, Grobe K, Tsujimoto M and Esko JD (2001). Multiple Isozymes of Heparan Sulfate/Heparin GlcNAc N-Deacetylase/N-Sulfotransferase: Structure and Activity of the Fourth Member, NDST4. J. Biol. Chem 276, 5876–5882. [Abstract] [Google Scholar]
- Allen BL and Rapraeger AC (2003). Spatial and temporal expression of heparan sulfate in mouse development regulates FGF and FGF receptor assembly. J. Cell Biol 163, 637–648. [Europe PMC free article] [Abstract] [Google Scholar]
- Bennett KL, Bradshaw J, Youngman T, Rodgers J, Greenfield B, Aruffo A and Linsley PS (1997). Deleted in colorectal carcinoma (DCC) binds heparin via its fifth fibronectin type III domain. J. Biol. Chem 272, 26940–26946. [Abstract] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O and Kemler R (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264. [Abstract] [Google Scholar]
- Britto J, Tannahill D and Keynes R (2002). A critical role for sonic hedgehog signaling in the early expansion of the developing brain. Nat. Neurosci 5, 103–110. [Abstract] [Google Scholar]
- Bullock SL, Fletcher JM, Beddington RSP and Wilson VA (1998). Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 12, 1894–1906. [Europe PMC free article] [Abstract] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H and Beachy PA (1996a). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413. [Abstract] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H and Beachy PA (1996b). Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383, 407–413. [Abstract] [Google Scholar]
- Cordero D, Marcucio R, Hu D, Gaffield W, Tapadia M and Helms JA (2004). Temporal perturbations in sonic hedgehog signaling elicit the spectrum of holoprosencephaly phenotypes. J. Clin. Invest 114, 485–494. [Europe PMC free article] [Abstract] [Google Scholar]
- Dono R, Texido G, Dussel R, Ehmke H and Zeller R (1998). Impaired cerebral cortex development and blood pressure regulation in FGF-2-deficient mice. EMBO J. 17, 4213–4225. [Europe PMC free article] [Abstract] [Google Scholar]
- Esko JD and Lindahl U (2001). Molecular diversity of heparan sulfate. J. Clin. Invest 108, 169–173. [Europe PMC free article] [Abstract] [Google Scholar]
- Esko JD and Selleck SB (2002). Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem 71, 435–471. [Abstract] [Google Scholar]
- Fan G, Xiao L, Cheng L, Wang X, Sun B and Hu G (2000). Targeted disruption of NDST-1 gene leads to pulmonary hypoplasia and neonatal respiratory distress in mice. FEBS Lett. 467, 7–11. [Abstract] [Google Scholar]
- Grobe K and Esko JD (2002). Regulated translation of heparan sulfate N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes by structured 5′-untranslated regions and internal ribosome entry sites. J. Biol. Chem 277, 30699–30706. [Abstract] [Google Scholar]
- Holmborn K, Ledin J, Smeds E, Eriksson I, Kusche-Gullberg M and Kjellen L (2004). Heparan sulfate synthesized by mouse embryonic stem cells deficient in NDST1 and NDST2 is 6-O-sulfated but contains no N-sulfate groups. J. Biol. Chem 279, 42355–42358. [Abstract] [Google Scholar]
- Hu D and Helms JA (1999). The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development 126, 4873–4884. [Abstract] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M and Yamaguchi Y (2003). Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science 302, 1044–1046. [Abstract] [Google Scholar]
- Ingham PW and McMahon AP (2001). Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087. [Abstract] [Google Scholar]
- Ishihara M, Guo Y, Wei Z, Yang Z, Swiedler SJ, Orellana A and Hirschberg CB (1993). Regulation of biosynthesis of the basic fibroblast growth factor binding domains of heparan sulfate by heparan sulfate-N-deacetylase/N-sulfotransferase expression. J. Biol. Chem 268, 20091–20095. [Abstract] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH and McMahon AP (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937–951. [Europe PMC free article] [Abstract] [Google Scholar]
- Johnson KG, Ghose A, Epstein E, Lincecum J, O’Connor MB and Van Vactor D (2004). Axonal heparan sulfate proteoglycans regulate the distribution and efficiency of the repellent slit during midline axon guidance. Curr. Biol 14, 499–504. [Abstract] [Google Scholar]
- Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L and Spillmann D (2004). Heparan sulfate structure in mice with genetically modified heparan sulfate production. J. Biol. Chem 279, 42732–42741. [Abstract] [Google Scholar]
- Li JP, Gong F, Hagner-McWhirter A, Forsberg E, Abrink M, Kisilevsky R, Zhang X and Lindahl U (2003). Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J. Biol. Chem 278, 28363–28366. [Abstract] [Google Scholar]
- Liang Y, Annan RS, Carr SK, Popp S, Mevissen M, Margolis RK and Margolis RU (1999). Mammalian homologues of the Drosophila slit protein are ligands of the heparan sulfate proteoglycan glypican-1 in brain. J. Biol. Chem 274, 17885–17892. [Abstract] [Google Scholar]
- Lin XH and Perrimon N (1999). Daily cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature 400, 281–284. [Abstract] [Google Scholar]
- Lin XH, Buff EM, Perrimon N and Michelson AM (1999). Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development 126, 3715–3723. [Abstract] [Google Scholar]
- Lindahl U, Kusche-Gullberg M and Kjellén L (1998). Regulated diversity of heparan sulfate. J. Biol. Chem 273, 24979–24982. [Abstract] [Google Scholar]
- Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K and Beachy P (2002). Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111, 63–75. [Abstract] [Google Scholar]
- McLaughlin D, Karlsson F, Tian N, Pratt T, Bullock SL, Wilson VA, Price DJ and Mason JO (2003). Specific modification of heparan sulphate is required for normal cerebral cortical development. Mech. Dev 120, 1481–1488. [Abstract] [Google Scholar]
- Ming JE, Kaupas ME, Roessler E, Brunner HG, Golabi M, Tekin M, Stratton RF, Sujansky E, Bale SJ and Muenke M (2002). Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum. Genet 110, 297–301. [Abstract] [Google Scholar]
- Mitchell KJ, Pinson KI, Kelly OG, Brennan J, Zupicich J, Scherz P, Leighton PA, Goodrich LV, Lu X, Avery BJ et al. (2001). Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet 28, 241–249. [Abstract] [Google Scholar]
- Muenke M and Beachy PA (2000). Genetics of ventral forebrain development and holoprosencephaly. Curr. Opin. Genet. Dev 10, 262–269. [Abstract] [Google Scholar]
- Muenke M and Cohen MM Jr (2000). Genetic approaches to understanding brain development: holoprosencephaly as a model. Ment. Retard. Dev. Disabil. Res. Rev 6, 15–21. [Abstract] [Google Scholar]
- Ornitz DM and Itoh N (2001). Fibroblast growth factors. Genome Biol. 2, 3005.1–3005.12. [Europe PMC free article] [Abstract] [Google Scholar]
- Perrimon N and Bernfield M (2000). Specificities of heparan sulphate proteoglycans in developmental processes. Nature 404, 725–728. [Abstract] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML and Vaccarino FM (2000). Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci 20, 5012–5023. [Europe PMC free article] [Abstract] [Google Scholar]
- Rapraeger AC, Krufka A and Olwin BB (1991). Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708. [Abstract] [Google Scholar]
- Reuss B and von Bohlen and Halbach O (2003). Fibroblast growth factors and their receptors in the central nervous system. Cell Tissue Res. 313, 139–157. [Abstract] [Google Scholar]
- Ringvall M, Ledin J, Holmborn K, Van Kuppevelt T, Ellin F, Eriksson I, Olofsson AM, Kjellén L and Forsberg E (2000). Defective heparan sulfate biosynthesis and neonatal lethality in mice lacking N-deacetylase/N-sulfotransferase-1. J. Biol. Chem 275, 25926–25930. [Abstract] [Google Scholar]
- Roelink H, Porter JA, Chiang C, Tanabe Y, Chang DT, Beachy PA and Jessell TM (1995). Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 81, 445–455. [Abstract] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC and Muenke M (1996). Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet 14, 357–360. [Abstract] [Google Scholar]
- Ronca F, Andersen JS, Paech V and Margolis RU (2001). Characterization of Slit protein interactions with glypican-1. J. Biol. Chem 276, 29141–29147. [Abstract] [Google Scholar]
- Rubin JB, Choi Y and Segal RA (2002). Cerebellar proteoglycans regulate sonic hedgehog responses during development. Development 129, 2223–2232. [Abstract] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ and Mohammadi M (2000). Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750. [Abstract] [Google Scholar]
- Schneider RA, Hu D, Rubenstein JL, Maden M and Helms JA (2001). Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development 128, 2755–2767. [Abstract] [Google Scholar]
- Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM and Tessier-Lavigne M (1994). The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell 78, 409–424. [Abstract] [Google Scholar]
- Stringer SE, Mayer-Proschel M, Kalyani A, Rao M and Gallagher JT (1999). Heparin is a unique marker of progenitors in the glial cell lineage. J. Biol. Chem 274, 25455–25460. [Abstract] [Google Scholar]
- Tekki-Kessaris N, Woodruff R, Hall AC, Gaffield W, Kimura S, Stiles CD, Rowitch DH and Richardson WD (2001). Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development 128, 2545–2554. [Abstract] [Google Scholar]
- The I, Bellaiche Y and Perrimon N (1999). Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell 4, 633–639. [Abstract] [Google Scholar]
- Treier M, O’Connell S, Gleiberman A, Price J, Szeto DP, Burgess R, Chuang PT, McMahon AP and Rosenfeld MG (2001). Hedgehog signaling is required for pituitary gland development. Development 128, 377–386. [Abstract] [Google Scholar]
- Trumpp A, Depew MJ, Rubenstein JL, Bishop JM and Martin GR (1999). Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 13, 3136–3148. [Europe PMC free article] [Abstract] [Google Scholar]
- Vaccarino FM, Schwartz ML, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin JD, Wyland JJ and Hung YT (1999). Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat. Neurosci 2, 848. [Abstract] [Google Scholar]
- Wang KH, Brose K, Arnott D, Kidd T, Goodman CS, Henzel W and Tessier-Lavigne M (1999). Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell 96, 771–784. [Abstract] [Google Scholar]
- Yayon A, Klagsbrun M, Esko JD, Leder P and Ornitz DM (1991). Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64, 841–848. [Abstract] [Google Scholar]
Full text links
Read article at publisher's site: https://doi.org/10.1242/dev.01935
Read article for free, from open access legal sources, via Unpaywall:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7851831
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1242/dev.01935
Article citations
Structural and mechanistic characterization of bifunctional heparan sulfate N-deacetylase-N-sulfotransferase 1.
Nat Commun, 15(1):1326, 13 Feb 2024
Cited by: 3 articles | PMID: 38351061 | PMCID: PMC10864358
In vivo activities of heparan sulfate differentially modified by NDSTs during development.
Proteoglycan Res, 2(1):e17, 01 Jan 2024
Cited by: 1 article | PMID: 38616954 | PMCID: PMC11011245
Neuronal Ndst1 depletion accelerates prion protein clearance and slows neurodegeneration in prion infection.
PLoS Pathog, 19(9):e1011487, 25 Sep 2023
Cited by: 0 articles | PMID: 37747931 | PMCID: PMC10586673
Ndst1, a heparan sulfate modification enzyme, regulates neuroectodermal patterning by enhancing Wnt signaling in Xenopus.
Dev Growth Differ, 65(3):153-160, 03 Mar 2023
Cited by: 2 articles | PMID: 36726238 | PMCID: PMC11520968
Establishing Hedgehog Gradients during Neural Development.
Cells, 12(2):225, 05 Jan 2023
Cited by: 7 articles | PMID: 36672161 | PMCID: PMC9856818
Review Free full text in Europe PMC
Go to all (136) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Heparan sulfate Ndst1 gene function variably regulates multiple signaling pathways during mouse development.
Dev Dyn, 236(2):556-563, 01 Feb 2007
Cited by: 42 articles | PMID: 17183530
Heparan sulfate expression in the neural crest is essential for mouse cardiogenesis.
Matrix Biol, 35:253-265, 05 Nov 2013
Cited by: 13 articles | PMID: 24200809 | PMCID: PMC4324438
Heparan sulfate biosynthetic gene Ndst1 is required for FGF signaling in early lens development.
Development, 133(24):4933-4944, 15 Nov 2006
Cited by: 78 articles | PMID: 17107998
Mice deficient in heparan sulfate N-deacetylase/N-sulfotransferase 1.
Prog Mol Biol Transl Sci, 93:35-58, 01 Jan 2010
Cited by: 22 articles | PMID: 20807640
Review
Funding
Funders who supported this work.
NHLBI NIH HHS (2)
Grant ID: HL57345
Grant ID: P01 HL057345
NIGMS NIH HHS (3)
Grant ID: R01 GM033063
Grant ID: R37 GM033063
Grant ID: GM33063
NINDS NIH HHS (2)
Grant ID: R01 NS41332
Grant ID: R01 NS041332




