Abstract
Free full text

Living with two extremes: Conclusions from the genome sequence of Natronomonas pharaonis
Associated Data
Abstract
Natronomonas pharaonis is an extremely haloalkaliphilic archaeon that was isolated from salt-saturated lakes of pH 11. We sequenced its 2.6-Mb GC-rich chromosome and two plasmids (131 and 23 kb). Genome analysis suggests that it is adapted to cope with severe ammonia and heavy metal deficiencies that arise at high pH values. A high degree of nutritional self-sufficiency was predicted and confirmed by growth in a minimal medium containing leucine but no other amino acids or vitamins. Genes for a complex III analog of the respiratory chain could not be identified in the N. pharaonis genome, but respiration and oxidative phosphorylation were experimentally proven. These studies identified protons as coupling ion between respiratory chain and ATP synthase, in contrast to other alkaliphiles using sodium instead. Secretome analysis predicts many extracellular proteins with alkaline-resistant lipid anchors, which are predominantly exported through the twin-arginine pathway. In addition, a variety of glycosylated cell surface proteins probably form a protective complex cell envelope. N. pharaonis is fully equipped with archaeal signal transduction and motility genes. Several receptors/transducers signaling to the flagellar motor display novel domain architectures. Clusters of signal transduction genes are rearranged in haloarchaeal genomes, whereas those involved in information processing or energy metabolism show a highly conserved gene order.
Strains of Natronomonas pharaonis were first isolated from highly saline soda lakes in Egypt (Soliman and Truper 1982) and Kenya (Tindall et al. 1984), which show pH values around 11. Such alkaline brines are enriched with carbonate and chloride, resulting in a scarcity of magnesium and calcium. The aerobic haloalkaliphilic euryarchaeon N. pharaonis thrives optimally in 3.5 M NaCl and at a pH of 8.5, but is sensitive to high magnesium concentrations.
Since plasma membranes and protoplasts lose their stability at high pH, it has been suggested that one of the key features of alkaliphilic specialization is associated with the cell envelope protecting the cell from alkaline conditions (Horikoshi 1999). Bacillus spp. contain acidic polymers that may support the adsorption of sodium and protons but repulse hydroxide ions. Haloalkaliphilic archaea have also been reported to possess unique cell walls consisting of glutaminylglycan polymers (Kandler and Konig 1998) as well as characteristic membranes containing C20–C25 in addition to C20–C20 diether core lipids (Tindall et al. 1984). Low extracellular proton concentrations further affect membrane-linked energetics because of immediate neutralization of extruded protons. In order to deal with alkaline conditions, some bacteria replace protons by sodium ions as the coupling ion rather than increasing ΔΨ (Skulachev et al. 1999).
A wide range of extracellular enzymes such as alkaline proteases, amylases, and cellulases, has been isolated from alkaliphiles, and were used for industrial production of laundry detergent additives and cyclodextrins (Horikoshi 1999). Most alkaline enzymes have been described in Bacillus spp., but the haloalkaliphilic archaeon Natronococcus occultus was also found to produce a haloarchael α-amylase and to exhibit extracellular proteolytic activity (Horikoshi 1999).
N. pharaonis has been phylogenetically classified within the order Halobacteriales, which includes the intensively studied Halobacterium salinarum. Natronomonas cells are motile (Soliman and Truper 1982) and actively search for optimal growth conditions with the help of retinal proteins responsible for light-dependent ion transport and sensory functions. For N. pharaonis (strain SP1), the chloride pump halorhodopsin (Lanyi et al. 1990) and sensory rhodopsin II (Seidel et al. 1995) with its transducer HtrII (Klare et al. 2004) have been described in detail.
Here we report the complete N. pharaonis genome and complementing experimental results. This study identified novel adaptation strategies of alkaliphiles regarding its respiratory chain, nitrogen metabolism, and its cell envelope.
Results and Discussion
Genome and gene statistics
The genome of N. pharaonis type strain Gabara (DSM 2160) consists of three circular replicons, the 2.6-Mb chromosome, a typical haloarchaeal 131-kb plasmid (PL131) and a unique multicopy 23-kb plasmid (PL23) (Table 1). The GC-rich chromosome (63.4% GC) contains an integrated copy of PL23, and features four regions of reduced GC content (GC-poor regions I–IV) as well as several transposases (illustrated in Supplemental Fig. S5). (Additional information on transposases, plasmids, and GC-poor regions is provided as Supplemental material.) The replication origin of N. pharaonis is delineated by a 30-bp inverted repeat (302280–302311, 302712–302681) and an adjacent Cdc6 homolog (cdc6_1, NP0596A), which are very similar to the recently identified inverted repeat and the orc7 gene of the Halobacterium NRC1, respectively (Berquist and DasSarma 2003). Interestingly, haloarchaeal origins are found around the maximum, not the minimum, of the cumulative GC skew plot.
Table 1.
Basic data for the N. pharaonis replicons
| CHR | PL131 | PL23 | |
|---|---|---|---|
| Length (bp) | 2,595,221 | 130,989 | 23,486 |
| GC content | 63.4% | 57.2% | 60.6% |
| Sequence coverage (normalized values) | 5.8 (1.0) | 3.9 (0.67) | 100.5 (17.3) |
| % coding | 90.8% | 82.3% | 83.9% |
| Encoded proteins | 2675 | 132 | 36 |
| Average protein length (amino acids) | 293 | 271 | 183 |
| Encoded stable RNAs | 51 | - | - |
By rigorous evaluation of the automatic gene finder data, 2843 protein-coding and 51 RNA genes were predicted for the GC-rich N. pharaonis genome (Supplemental Table S2 lists all proteins from N. pharaonis and Table S1 lists those mentioned in the paper). This process was greatly facilitated by the close relationship between Natronomonas and other halophilic archaea with completely sequenced genomes, Halobacterium salinarum (of which two strains have been sequenced with minimal sequence deviation: strain R1 [F. Pfeiffer, S. Schuster, M. Falb, P. Palm, K. Rodewald, A. Ruepp, J. Soppa, J. Tittor, and D. Oesterhelt, unpubl.; http://www.halolex.mpg.de] and strain NRC-1 (published as Halobacterium sp. NRC-1 [Ng et al. 2000])) as well as Haloarcula marismortui (Baliga et al. 2004). The accuracy of our gene selection is further enhanced by the availability of genome-scale proteomic data for H. salinarum strain R1 (Klein et al. 2004; Tebbe et al. 2004; http://www.halolex.mpg.de) and confirmed by genome-wide proteomic data for N. pharaonis (F. Siedler, unpubl.; a single false negative was uncovered in a set of several hundred identified proteins).
The subsequent assessment of the applied gene prediction tools by comparison with the validated N. pharaonis gene set revealed significant improvement of gene selection by the REGANOR approach as compared to that of the underlying programs CRITICA and adequately trained GLIMMER (see Methods). However, ~400 of the start codons predicted by REGANOR and CRITICA (14%–15%), and even a third of the GLIMMER starts were reassigned, mostly because predicted genes were too long.
A total of 43% of the N. pharaonis genes are likely to be cotranscribed, and 322 gene pairs revealed typical transcription unit overlaps of one or four bases resulting from a shift of open reading frames by -1. The gene order of transcription units coding for ribosomal proteins, RNA polymerase, and membrane complexes involved in energy metabolism (atp, nuo clusters) and pH adaptation (pha cluster) is very well conserved between Halobacterium and Natronomonas. In contrast, gene clusters involved in signal transduction and motility are extensively rearranged between the genomes of halophiles (see Supplemental Fig. S6).
Cytoplasmic N. pharaonis proteins contain a high proportion of acidic amino acids (average 19.3%) resulting in low isoelectric points (average pI 4.6). These are typical adaptative features of haloarchaea, which are known to apply the salt-in strategy (high internal salt concentrations) in order to cope with their hypersaline environment. In addition, N. pharaonis was reported to produce the compatible osmolyte 2-sulfotrehalose (Desmarais et al. 1997), and its genome codes for a typical compatible solute transporter (tp58, NP3588A). However, homologs of genes for the de novo synthesis of common compatible solutes such as glycine betaine (betAB) and trehalose (otsAB) or an osmolyte-binding protein (cosB) could not be detected.
Function analysis
Of the 2843 proteins, 65% were grouped to an orthologous cluster (COG), and specific and general functions could be assigned for 45% and 12% of the predicted proteome, respectively. Proteins with specific function belong to the functional categories metabolism (17%), transport and cellular processes (9%), genetic information processing (7%), environmental information processing (5%), and miscellaneous (7%) (see also Supplemental Table S2). For the remaining proteins, only general functions (12%), partly derived through gene-context analyses, or no functions (43%) were assigned. One-fifth of the N. pharaonis proteins have no homologs in other species (singletons). Among them are two probable Natronomonas-specific membrane complexes, each encoded by nine adjacent genes (NP2336A–NP2352A, NP5354A–NP5338A), which are highly similar and arranged in the same gene order. All 15 previously published N. pharaonis genes could be identified within the genome, although minor sequence variations were detected usually owing to strain differences. However, a partially sequenced protease from N. pharaonis, which is extremely similar to vertebrate chymotrypsin (Stan-Lotter et al. 1999), could not be found.
Central metabolism and transport
Metabolic enzymes comprise a large number of fatty-acid degradation genes, and the complete set of enzymes involved in biosynthetic pathways leading to amino acids and coenzymes. Thus, the chemo-organotrophic N. pharaonis, which is usually grown on media with amino acids as the carbon source, has a high degree of nutritional self-sufficiency. In agreement with this, we were able to simplify the synthetic medium for this species omitting all amino acids except leucine. Requirement of this amino acid might be caused by disruption of the isopropylmalate synthase gene (leuA_1, NP2206A) in the 5′-region. As H. salinarum, N. pharaonis is likely not capable of sugar utilization because of the lack of genes encoding key enzymes of glycolytic pathways. A gene cluster (NP4962A–NP4944A) encoding a set of probable anaerobic dehydrogenase subunits might indicate growth of N. pharaonis cells under anaerobic or micro-aerophilic conditions. Genes encoding mevalonate pathway enzymes are present, as are other genes required for the synthesis of membrane lipids and other components such as menaquinones (Soliman and Truper 1982) and retinal pigments (Seidel et al. 1995), which are derived from prenyl-precursors. A large set of predicted transporter genes was found, including homologs of transporters for metal ions such as iron, manganese, copper, and cobalt, which are scarce in a highly alkaline environment. A more extensive description of the predicted metabolism and transport of N. pharaonis is given as Supplemental material.
Nitrogen metabolism
Natronomonas grows under highly alkaline conditions in brines of pH around 11 (Soliman and Truper 1982; Tindall et al. 1984). These extreme pH conditions cause not only low availability of metal ions, but also reduced levels of ammonium ions. For the uptake of ammonium, which can be assimilated in the central metabolite glutamate, N. pharaonis possesses transporters for several exogenous nitrogen sources, ammonium (AmtB, NP0922A), nitrate/nitrite (NarK, NP4228A), and urea (ABC transporter Urt-ABCDE, NP1996A–NP2004A) (Fig. 1). Genes involved in nitrate reduction (narB_1, NP4226A; nirA_1, NP4224A) and urea conversion (ure cluster, NP2008A–NP2020A) to ammonium were found to be clustered with their respective transporter genes. The observed nitrate assimilation pathway has also been described for cyanobacteria (Hirasawa et al. 2004), and the enzymes involved show 34%–51% sequence identity to those present in N. pharaonis. It is likely that Natronomonas uses ferredoxin and not NADH as the electron donor for all three reductive conversions. This view is supported by the occurrence of conserved ferredoxin-binding residues within the N. pharaonis NirA protein (Hirasawa et al. 1998) and ferredoxin dependence of nitrate and nitrite reductases in the halophile Haloferax mediterranei (Martinez-Espinosa et al. 2001). Nine ferredoxins of four orthologous groups (COG0633, COG1141, COG1146, COG3411) are present in N. pharaonis, and ferredoxin appears to be the common proteinaceous electron carrier for functional N-assimilation as well as the conversion of 2-oxoacids and aldehydes.
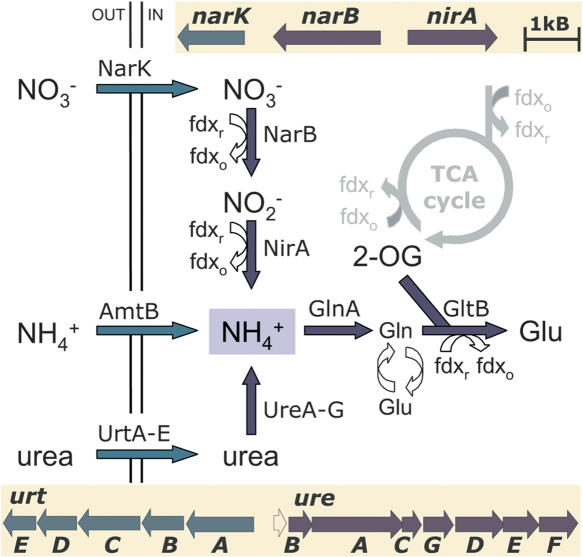
Schematic representation of the nitrogen metabolism of N. pharaonis. The figure illustrates proposed transport (green arrows) and metabolic processes (blue arrows) for nitrogen compounds and the associated gene clusters (nark–narB–nirA, urtABCDE, ureBACGDEF). Ammonia can be supplied by (1) direct uptake, (2) uptake and reduction of nitrate (

While H. marismortui also possesses all necessary genes for urea conversion and nitrate assimilation but also for nitrate respiration, H. salinarum lacks these genes. Urea-cycle enzymes for the conversion of ornithine to arginine are encoded in all three halophilic strains. With respect to arginine degradation, halophiles adopted different strategies. H. marismortui splits arginine into ornithine and urea by arginase (EC 3.5.3.1; rrnAC0383, rrnAC0453), while H. salinarum uses plasmid-encoded enzymes of the arginine deiminase pathway for the fermentation of arginine. In this pathway, arginine is converted to ornithine and carbamoylphosphate, which is further degraded to CO2 and NH3 with concomitant ATP generation (Ruepp and Soppa 1996). N. pharaonis lacks both pathways for arginine utilization.
Respiratory chain
The available biochemical data on electron transport chain components for N. pharaonis (Scharf et al. 1997) and other respiratory archaea (Schafer et al. 1996), as well as the genomic data from Halolex (http://www.halolex.mpg.de) and STRING databases (von Mering et al. 2003), were used to generate an archaeal profile of the electron transport chain (Fig. 2A). The profile revealed a high degree of plasticity in the composition of the respiratory chain, often differing greatly from the “classical” five complex systems found in mitochondria. Notably, type, number, and composition of terminal oxidases vary widely in the archaeal domain of life. Furthermore, NADH is not oxidized by proton-pumping type I NADH dehydrogenase complexes because the NADH acceptor module (nuoEFG) is absent. In N. pharaonis, NADH dehydrogenation is likely to occur via the non-proton-pumping type II NADH dehydrogenase encoded by a homolog (NP3508A) of the ndh gene characterized in Acidianus ambivalens (Gomes et al. 2001). All subunits of the succinate dehydrogenase (sdhCDBA, NP4264A-NP4270A) are present, and menaquinone biosynthesis genes were found, supporting its proposed function as a mobile carrier (Scharf et al. 1997). However, no complex III subunits could be identified in the genome in spite of searching with the respective H. salinarum genes (petABC). The crenarchaeote A. ambivalens is able to channel electrons from complex I/II directly into a terminal quinole oxidase using a mobile quinol carrier (Gomes et al. 2001). None of the three terminal oxidases in N. pharaonis, though, shows sufficient sequence similarity to support the idea that one of them is a quinole oxidase. The N. pharaonis cytochrome ba3-type oxidase (cba cluster, NP2960A–NP2968A) (Mattar and Engelhard 1997) interacts with the blue copper protein halocyanin (hcp_1, NP3954A), and the cbaD subunit of the orthologous complex in H. salinarum occurs as a fusion protein with halocyanin. The probable electron transport from halocyanin to the terminal oxidase complex in haloarchaea indicates a novel yet unknown type of complex III, which mediates the electron transfer step between menaquinone and halocyanin.
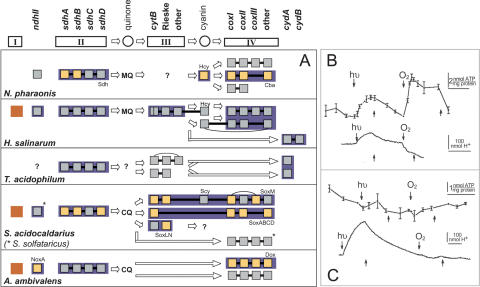
Genomic and experimental studies of the N. pharaonis electron transport chain. (A) The electron transport chain profile for several respiratory archaea displays subunits (squares) of respiratory complexes (boxes I–IV). These subunits are often encoded adjacently in the archaeal genomes (straight connections) and can be fused to each other (curved connections). The proposed electron flow between respiratory complexes is indicated by arrows. Complexes, which have been characterized experimentally, are indicated in blue, and protein-sequenced subunits of isolated complexes are yellow. The profile for Sulfolobus acidocaldarius (Sa) was established by complementation with genetic data from the completely sequenced Sulfolobus solfataricus (Ss) (asterisks). Menaquinone (MQ), caldariellaquinone (CQ), halocyanin (Hcy), and sulfocyanin (Scy) are shown or predicted to function as mobile carriers in archaeal respiratory chains. Homologs to complex I subunits have been found in archaeal genomes, but genes encoding the NADH acceptor module are missing. A functional NADH-dehydrogenating complex I has been experimentally excluded for three species (red). Instead, it is replaced by NADH dehydrogenase type II, which is not capable of proton translocation. (B,C) Oxidative and photo-phosphorylation processes in N. pharaonis cells were investigated through measurements of ATP levels (upper curves, mean values with error bars from triplicates) and extracellular pH (lower curves, continuous recording at a rate of 10/sec) in the (B) absence or (C) presence of the protonophore CCCP (0.2 mM). The effects (ON: above curve arrow, OFF: below curve arrow) of light (hν, λ > 515 nm, 32 mW/cm2) and aeration (O2) were determined. All experiments were performed at pH 8.1. Vertical scaling bars indicate ATP level and amount of proton uptake, whereas horizontal scaling bars indicate a 10-min time interval.
Through physiological experiments, we showed that N. pharaonis is able to eject protons during respiration (oxygen-induced acidification) with a subsequent increase in ATP levels (oxidative phosphorylation) (Fig. 2B,C). Since both effects are sensitive to the protonophore CCCP, N. pharaonis needs to possess a functional respiratory chain including a component responsible for proton pumping upon electron transfer (either one of the oxidases or the postulated complex III analog). Furthermore, CCCP did not prevent light-induced alkalinization2 by the light-driven chloride pump halorhodopsin, but did prevent the subsequent increase in ATP levels. Thus, the N. pharaonis ATP synthase also operates with protons, and completes a full proton circuit. This finding shows that N. pharaonis does not replace protons with sodium ions as the coupling ion between respiratory chain and ATP synthase. In contrast, the alkaliphilic bacterium Bacillus halodurans FTU switches from proton to sodium energetics in case of alkaline conditions or the addition of a protonophore (Skulachev 1992). Induction of respiratory complexes and ATPases with altered ion-specificity permits alkaliphiles to cope with inverted proton gradients requiring no increase of ΔΨ and avoiding reduced ATP yield as well as the risk of membrane electric breakdown (Skulachev et al. 1999). However, dependent on the external pH, varying internal pH values were observed for alkaliphilic Bacillus strains, and the internal pH reaches values greater than pH 9 (Horikoshi 1999). For N. pharaonis we measured internal pH values up to 9.3. By accepting high pH values in the cytoplasm, the difference between intra- and extracellular pH remains moderate, and consequently protons are permissible as the coupling ion. It should be noted, that our data are not in agreement with the previous suggestion that chloride is used as an alternative coupling ion by N. pharaonis (Avetisyan et al. 1998).
Secretion and membrane anchoring
N. pharaonis encodes the same probable components of the Sec and Tat (twin-arginine) protein translocation pathways that were found in the Halobacterium genome (Pohlschroder et al. 2004). For the transport of archaeal flagellins, involvement of flaI and flaJ genes was proposed (Thomas et al. 2001), since those show similarity to components of type II/IV secretion systems and were described to influence flagella formation (Patenge et al. 2001). To estimate the contribution of different systems for protein secretion in N. pharaonis, the complete secretome was predicted as described in Methods (Fig. 3A; see also Supplemental Table S3). This and previous genome surveys for H. salinarum strain NRC-1 (Bolhuis 2002; Rose et al. 2002) found that the Tat system, which secretes folded proteins, might be extensively used in haloarchaea compared to nonhalophilic archaea. Haloarchaea probably use the Tat pathway not only for coenzyme-containing redox components like halocyanins (e.g., hcp_4, NP0050A) and thioredoxins (e.g., trx_1, NP3914A), but also for the export of non-redox proteins such as substrate-binding proteins of ABC transport systems (e.g., dppA_2, NP3578A) and chemotactic signal transduction (e.g., H. salinarum cosB, OE3476R). Thus, an adaptation to the high-salt and alkaline environment may involve the avoidance of protein folding in the extracellular space where chaperones are absent (Rose et al. 2002). However, there are also several proteins in Natronomonas that are likely cotranslationally delivered to and exported through the Sec system, among them three extracellular subtilisin-like proteases (NP1682A, NP2654A, NP4628A).
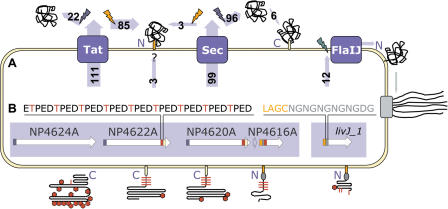
Schematic representation of protein secretion, anchoring, and glycosylation in N. pharaonis. (A) Substrates of the Tat, Sec and flagellin-specific protein translocation systems (blue boxes) are cleaved by signal peptidases (flash signs), and partly remain C- or N-terminally anchored to the cell membrane. Secreted proteins are cleaved by signal peptidase type I (blue), whereas lipobox-containing proteins are cleaved by signal peptidase type II (orange) and N-terminally attached to a lipid anchor (orange box). Lipoproteins are frequently transported via the Tat pathway (substrate numbers indicated in light-blue arrows). For three lipobox-containing proteins, the export pathway remains as yet unassigned. Furthermore, six proteins are likely to be modified by a C-terminally attached lipid anchor (yellow box). After cleavage by membrane-bound preflagellin peptidase (green), the substrates of the flagellin-specific export pathway reveal an N-terminal hydrophobic stretch possibly involved in membrane retention. (B) Signal sequence and peptide repeat modules (indicated by colored boxes) for a representative gene cluster (white arrows) are presented diagrammatically in models of the cell surface proteins. A Thr-rich tetrapeptide repeat (red box in genes, red line in proteins), likely to be O-glycosylated, occurs in several cell surface proteins adjacent to the C-terminal or N-terminal lipid anchor. An Asn-Gln dipeptide repeat (gray box, oval) follows directly after the lipobox-containing Tat-related signal sequence (orange box) of several membrane components. Other indicated features are Sec-related signal sequences (blue box in genes) and N-glycosylation sites (red hexagons in proteins).
For the N. pharaonis halocyanin (hcp_1, NP3954A), attachment of a diphytanyl chain to an N-terminal cysteine residue has been shown previously (Mattar et al. 1994). Prokaryotic lipoproteins are cleaved by signal peptidase II upstream of a cysteine residue that is part of a conserved motif (lipobox), and subsequently modified by lipid-anchor attachment to the cysteine residue of the processed N-terminus (Hayashi and Wu 1990). N. pharaonis halocyanin exhibits a probable lipobox motif (LAGC), indicating signal peptidase II-like processing before the observed lipid attachment. However, a signal peptidase II homolog is absent as in other archaeal genomes, and an alternative signal peptidase II-like protease remains to be identified. The N. pharaonis genome encodes a large number of proteins (91) containing lipobox motifs as adapted from PROSITE (PS00013) (see Methods). Probable N-terminal-anchored lipoproteins comprise a third of the predicted secretome, and seem to be commonly translocated via the Tat pathway (85 proteins) (Fig. 3A). More than 20 transporter subunits, six of the halocyanin homologs, and many Natronomonas-specific proteins of unknown function are found among predicted lipoproteins. Halophilic archaea have a higher fraction of lipobox-containing proteins than nonhalophilic archaea, the highest fraction being found in N. pharaonis (N. pharaonis: 91 [3.20% of all proteins], H. marismortui: 116 [2.74%], H. salinarum: 49 [1.74%] vs. Archaeoglobus fulgidus: 16 [0.66%], Methanosarcina mazei: 23 [0.68%], Pyrococcus furiosus: 9 [0.42%]). Lipoproteins seem to be more common in bacteria (Bacillus subtilis: 57 [1.39%], Escherichia coli: 90 [2.12%], Corynebacterium glutamicum: 77 [2.57%]). The retention of 34% of secreted N. pharaonis proteins by lipid anchors may reflect a protection mechanism against alkaline extraction of proteins from the cell membrane, an effect commonly exploited to deplete membrane preparations of peripheral membrane proteins (Klein et al. 2004).
Interestingly, eight of the putative lipoproteins have Asn-Gly dipeptide repeats directly following the lipobox (Fig. 3B), among them two halocyanins (hcp_1, NP3954A; hcp_2, NP4744AA) and three substrate-binding proteins (dppA_1, NP0758A; livJ_1, NP4140A; sfuA, NP5000A). These proteins are likely involved in interactions with membrane protein complexes (respiratory complexes, ABC transporters); thus, the repeat regions (length up to 24 amino acids) might function as flexible hinges promoting protein interactions.
Apart from the putative lipobox-containing proteins, 12 proteins with probable cleavage sites (RGQ) for pre-flagellin-peptidase (flaK, NP1276A), as previously proposed for halobacterial flagellins (Thomas et al. 2001), were identified. These proteins include not only the three flagellins (NP2086A–NP2090A), but also proteins with unknown functions, most of them encoded adjacent to each other and to genes similar to components of type II/IV secretion systems.
Cell envelope
Thr-rich tetrapeptide repeats varying in length and amino acid composition were detected in nine Natronomonas-specific proteins, three of whose genes (NP4616A, NP4620A, NP4622A) clustered within one genomic region (Fig. 3B). Four of the repeat-containing proteins revealed high regional similarity to the N- and C-termini of halophilic cell surface glycoproteins (csg), which form regular S-layer cell envelopes. Within these similar regions, three topological features have been described for the H. salinarum Csg; an N-terminal signal sequence, a C-terminal lipid-anchor region (Kikuchi et al. 1999), and—directly in front of this membrane anchor—a pattern of O-glycosylated threonines (Lechner and Sumper 1987). In all N. pharaonis proteins with Thr-rich tetrapeptide repeats, secretion, lipid-retention, and glycosylation signals were present (Fig. 3B). Interestingly, the Thr-rich repeating units are located not only next to the C-terminal lipid anchor regions as in H. salinarum, but also adjacent to N-terminal lipid anchors. This finding suggests that the likely function of observed Thr-rich tetrapeptide repeat patterns is as a glycosylated spacing region that forms a periplasma-like reaction space between cell envelope and membrane. Because N. pharaonis encodes several proteins with Csg-like features, it might not possess a typical S-layer, but instead form a more complex cell envelope consisting of various glycoprotein species with distinct saccharides.
Motility and signal transduction
The proposed signal transduction cascade of Natronomonas and Haloarcula is very similar to that described for Halobacterium (Rudolph and Oesterhelt 1996), and consists of signal receptors/transducers, the two-component regulatory system (cheA/Y, NP2172A, NP2102A), an adaptation module (cheR/B, NP2170A, NP2174A), and the flagellum with its motor. However, there are many differences in the organization of the genes in the genome and the domain architecture of individual components (further detailed as Supplemental material).
Major domain shuffling is also evident when comparing the 19 Natronomonas and the 18 Halobacterium transducers (also known as methyl-accepting chemotactic proteins) that participate in the signal transduction cascade. An example are the transducers mediating the phototactic response. Natronomonas has a single blue-light photoreceptor (sensory rhodopsin II; SRII, NP4834A) that is photochemically very similar to the blue-light photoreceptor SRII from Halobacterium but rather different from the orange/UV light photoreceptor SRI (Lutz et al. 2001). Although the transducers HtrII from these two archaea form a complex with their respective blue-light photoreceptors SRII (thus mediating the same photophobic response) and genes for receptor and transducer are cotranscribed (Seidel et al. 1995; Zhang et al. 1996), their domain architecture differs. Natronomonas HtrII (NP4832A) is characterized by a short loop between its two transmembrane domains (TM2SL) and shares its domain architecture with that of Halobacterium HtrI and three paralogs from Natronomonas (NP1756A, NP3122A, NP3134A) (Fig. 4). Four of the five TM2SL-type transducers also show similarity in their gene context. Their genes are shown (Yao and Spudich 1992; Seidel et al. 1995; Zhang et al. 1996) or are predicted to be cotranscribed by forming a transcription unit with a retinal protein or a distant homolog thereof (NP1758A, NP3132A). In contrast, HtrII of Halobacterium shares its domain architecture, characterized by a long extracellular domain between its two transmembrane domains (TM2ED), with six paralogs, several of which are involved in chemotaxis as determined experimentally (Kokoeva et al. 2002) or indicated by gene context. Whereas Halobacterium and Haloarcula have multiple TM2ED-type transducers and share several orthologous gene pairs, Natromononas has mainly TM2SL-type transducers, which lack an extracellular domain. As a result, either Natronomonas may have reduced chemotactic capabilities or chemotaxis may be mediated by other transducers with different domain architectures. A likely candidate for a chemotactic transducer is Htr32 (NP6128A), which has a substrate-binding domain directly fused to its signaling domain. Further transducers with unusual domain architectures are Htr34 (NP1642A) and Htr35 (NP1486A), which have an N-terminal signaling domain with a long C-terminal extension.
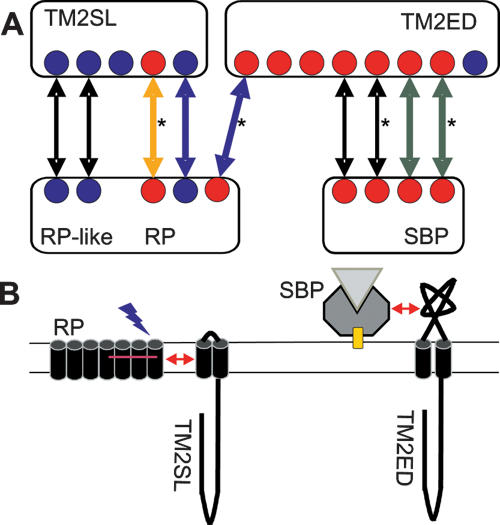
Domain architecture and gene context of several transducers from Natronomas and Halobacterium. (A) Two groups of transducers (upper boxes) with distinct domain architectures and their adjacent genes (arrows and lower boxes) from Natronomonas (blue dots) and Halobacterium (red dots) are schematically represented. Transducers with a long extracellular domain between their two transmembrane domains (box TM2ED) are frequently involved in chemotaxis, and are cotranscribed with the genes for periplasmic substrate-binding proteins (box SBP). Several of the transducers with a short loop between their two transmembrane domains (short-loop transducers, box TM2SL) are cotranscribed with retinal-containing photoreceptors (box RP) or distant homologs thereof. Experimental environmental response data are indicated by colored arrows (green: chemotaxis [Kokoeva et al. 2002]; blue: blue-light phototaxis [Seidel et al. 1995; Zhang et al. 1996]; orange: orange-light phototaxis [Yao and Spudich 1992]). The existence of orthologous gene pairs in Haloarcula is indicated by asterisks. (B) The interaction between a short-loop transducer (TM2SL) and a retinal protein (RP) occurs within the membrane. The interaction between a chemotactic extracellular-domain transducer (TM2ED) with a lipid-anchored (yellow box) periplasmic substrate-binding protein (SBP) may occur outside of the membrane.
In conclusion, we describe several features that permit N. pharaonis to cope with its extremely salty and alkaline environment. As protection, N. pharaonis may form a complex cell envelope consisting of different types of glycoproteins, probably glycosylated at Thr-rich tetrapeptide repeats. A large fraction of the extracellular proteins, being directly exposed to these extreme conditions, is predicted to be retained by the cell membrane by N- or C-terminal lipid anchors to prevent their alkaline extraction. As in other halophilic archaea, these are mainly secreted in the folded state through the Tat pathway as to avoid folding in a hostile and chaperone-free environment. N. pharaonis further encodes three potential alkaline proteases, which are probably secreted into the extracellular space by the Sec system.
Several key components of the energy metabolism are present in the cell membrane, in particular the ATP synthase and the respiratory chain complexes, of which a complex III analog remains to be detected in the genome. However, a functional respiratory chain was experimentally validated for N. pharaonis. Its determined ion specificity differs from that of other alkaliphiles using protons rather than sodium as the coupling ion.
A versatile nitrogen metabolism and a large number of transport systems for nitrogenous compounds and heavy metals, which are scarce in the habitat, reflect metabolic adaptation to the alkaline environment.
Genome annotation showed that N. pharaonis possesses high biosynthetic capabilities, which were exploited to develop a simple synthetic growth medium. Halophilic archaea have a highly similar signal transduction cascade for chemo- and phototaxis, but signal transduction and motility genes differ with respect to gene organization and domain architecture. Thus, high plasticity of environmental responses can be predicted reflecting variable halophilic environments.
Methods
Genome sequencing and assembly
N. pharaonis type strain Gabara (DSM 2160) was sequenced with 5.8-fold sequence coverage using a shotgun clone library (average insert size of 2 kb), and assembled with the PHRED-PHRAP-CONSED package (Gordon et al. 1998). The coverage for the two plasmids differed from that of the chromosome (100.5 for PL23 and 3.9 for PL131). The sequence is of high quality (0.01 errors/10 kb for the chromosome, 0.00 for PL23, and 0.04 for PL131). A copy of PL23 is integrated into the chromosome, and this copy was found to carry itself a 13-kb GC-poor insert (confirmed by clones bridging the 2.8 kb between the integration points and by a coverage typical for the chromosome). Apart from this integration, no other polymorphism between integrated and free plasmid could be detected.
Gene prediction and annotation
For gene prediction, REGANOR (McHardy et al. 2004) from the annotation package GENDB (Meyer et al. 2003) was used, which integrates results from CRITICA (Badger and Olsen 1999) and GLIMMER (Delcher et al. 1999). In addition, sixframe translation (>100 codons) was performed. From the resulting raw set of 11,874 distinct ORFs, a set of 2843 validated genes was selected by the following procedure: Using BLAST (Altschul et al. 1997), predicted amino acid sequences were bidirectionally compared to the H. salinarum strain R1 ORF set (http://www.halolex.mpg.de) containing 1060 genes experimentally verified by proteomic analysis (Klein et al. 2004; Tebbe et al. 2004). This allowed identification of undetected small genes, the discrimination between real proteins and spurious ORFs, the improvement of start codon selection, and the initial assignment of protein function. The ORF set was also analyzed by BLAST against itself, and against the NR database. Overlapping ORFs were adjusted based on gene context as well as characteristic halophilic pI and amino acid distribution patterns. tRNAs and other RNAs were predicted using tRNAscan (Lowe and Eddy 1997) and BLAST against H. salinarum, respectively. In the final validated gene set, 90.9% of the start codons were ATG, 7.9% GTG, and the residual 1.2% are pseudogenes, for example, due to interruption by ISH elements.
The generated validated gene set was used to assess performance of the three gene predictors, REGANOR, CRITICA, and GLIMMER (trained by CRITICA ORF set for optimal results) (McHardy et al. 2004). For the chromosome, GLIMMER predicted 12.9% false positives (FP) and 2.4% false negatives (FN), while CRITICA predicted no FP and 9.3% FN ORFs. The REGANOR gene finder performed best with 1.2% FP and 4.2% FN. One-third of the GLIMMER starts were reassigned (31.3% of the genes were shortened, 6.5% extended), while 13.2% (13.9%) of the genes predicted by CRITICA (REGANOR) were shortened and 0.9% (1.1%) were extended.
Each N. pharaonis protein was assigned to a cluster of orthologous groups (COG) and to a functional category (Tatusov et al. 1997), with a minimal BLAST e-value of e-05. BLAST results against H. salinarum and other databases were carefully evaluated for annotation of gene functions or descriptions. Intergene distances and configurations between gene pairs were analyzed, and genes <35 bases apart were considered to be cotranscribed. Regions were defined as GC-poor when the GC content of 30 adjacent coding regions was 5% below the replicon average.
Motif searches
The PROSITE pattern PS00013 (Hulo et al. 2004) was used to search for lipid attachment sites (lipobox) within the first 50 amino acids of predicted proteins of N. pharaonis and other prokaryotes. The lipobox is highly variable, resulting in a rather unspecific sequence motif, but with organism-specific preferences (e.g., LAGC in E. coli and N. pharaonis). We counted only those lipobox variations that occur frequently in a given species (i.e., at least three times) to reduce the number of false positives. Proteins with a predicted lipobox that contain an additional twin-arginine motif and proteins identified through predictions from TATFIND2.2 (Rose et al. 2002) were used to determine the total number of twin-arginine translocation pathway (Tat) substrates. The number of general secretion pathway (Sec) substrates results from SIGNALP3.0 (Nielsen et al. 1997), excluding specific Tat substrates and proteins with TMHMM-based transmembrane helix predictions past the first 50 residues (Krogh et al. 2001). Proteins with signal peptide predictions were searched for the proposed pre-flagellin peptidase cleavage site in haloalkaliphilic archaea (RGQ) (Thomas et al. 2001). N-/O-Glycosylation sites were predicted using NetNGlyc and NetOGlyc tools (http://www.cbs.dtu.dk/services/). NG-dipeptide repeats/T-rich tetrapeptide repeats ([VP][TE][ED]T) with 4/2 repeating units were detected by pattern searches within a sequence window of 30/50 amino acids.
ATP and pH measurements
N. pharaonis cells grown to late logarithmic phase in DSM medium 205 were harvested and resuspended in basal salt (medium without casamino acids) to result in an OD600 of 4. Cells were kept under a continuous nitrogen flow in a thermostated (20°C) glass vessel. For illumination, a 100-W mercury lamp (HBO 100 W-2; Oriel) was used, fitted with a heat protection filter (Calflex 3000; Balzers), and a yellow cutoff filter (OG 515; Schott) resulting in an irradiance of 32 mW/cm2. Air was flushed through the medium for oxygenation. The pH traces were recorded with a standard glass electrode. For ATP determination by a luciferin/luciferase assay, 0.1 mL of cells were lysed in 5 mL of ice-cold buffer (10 mM MgCl2, 0.02% NaN3, 0.1 mM EDTA, 25 mM HEPES at pH 7.5). Luminescence was measured (Lumac Biocounter) in triplicate by adding 0.1 mL of a 1:2 (w/w) mixture of D-Luciferin (0.1 mg/mL) and Photinus pyralis luciferase (0.2 mg/mL; Sigma-Aldrich) to 0.5 mL of lysed cells. The protonophore CCCP (carbonyl cyanide m-chloro-phenylhydrazone) was added to a final concentration of 0.2 mM.
For measurement of the internal pH, freshly harvested cells were disrupted by sonification after washing twice with unbuffered basal salt without citrate (Koch and Oesterhelt 2005). When grown between pH 9.0 and pH 9.5, the internal pH was similar to that of the medium (external/internal: 9.0/9.3 and 9.5/9.2).
Synthetic medium
A synthetic medium for N. pharaonis (M. Engelhard, pers. comm.) was further simplified by omitting amino acids and vitamins. The minimal medium for N. pharaonis consists of 20 mM sodium acetate, 10 mM sodium pyruvate, 12 mM NH4Cl, 5 mM leucine, 3.4 M NaCl, 27 mM KCl, 175 mM Na2CO3, 1 mM MgSO4, 2 mM Na2HPO4, 2 mM NaH2PO4, 5 μm FeSO4, 4 μM CuSO4, 4 μM MnCl2, 3 μM ZnSO4, and 3 μM CaCl2 (pH 9.2).
Acknowledgments
We acknowledge Hermann Lederer, Markus Rampp, and Reinhard Tisma for their help with the MIGENAS system (http://www.migenas.mpg.de); Burkhard Linke and Folker Meyer for successful cooperation regarding GenDB (Meyer et al. 2003); Günter Raddatz and Stephan Schuster for their expert advise on using genome assembly and annotation tools; Douglas Griffith and Martin Engelhard for careful manuscript reading and critical discussions; and Jan Wolfertz for continuous maintenance and improvements of the HaloLex system. We further thank the reviewers for valuable advice on the manuscript.
Notes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3952905. Article published online before print in September 2005.
Footnotes
[Supplemental material is available online at www.genome.org. The Natronomonas genome is accessible through HaloLex (http://www.halolex.mpg.de). The sequence data from this study have been submitted to EMBL under the accession numbers CR936257 (chromosome), CR936258 (plasmid PL131), and CR936259 (plasmid PL23).]
2Pumping of chloride ions into the cell causes a passive cation flow. The relative contribution of the different cations depends on their membrane permeability. The protonophore CCCP selectively increased membrane permeability for protons and accordingly results in increased alkalinization.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [Europe PMC free article] [Abstract] [Google Scholar]
- Avetisyan, A.V., Kaulen, A.D., Skulachev, V.P., and Feniouk, B.A. 1998. Photophosphorylation in alkalophilic halobacterial cells containing halorhodopsin: Chloride-ion cycle? Biochemistry (Mosc.) 63: 625-628. [Abstract] [Google Scholar]
- Badger, J.H. and Olsen, G.J. 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16: 512-524. [Abstract] [Google Scholar]
- Baliga, N.S., Bonneau, R., Facciotti, M.T., Pan, M., Glusman, G., Deutsch, E.W., Shannon, P., Chiu, Y.L., Gan, R.R., Hung, P.L., et al. 2004. Genome sequence of Haloarcula marismortui: A halophilic archaeon from the Dead Sea. Genome Res. 14: 2221-2234. [Europe PMC free article] [Abstract] [Google Scholar]
- Berquist, B.R. and DasSarma, S. 2003. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 185: 5959-5966. [Europe PMC free article] [Abstract] [Google Scholar]
- Bolhuis, A. 2002. Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: A major role for the twin-arginine translocation pathway? Microbiology 148: 3335-3346. [Abstract] [Google Scholar]
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with Glimmer. Nucleic Acids Res. 27: 4636-4641. [Europe PMC free article] [Abstract] [Google Scholar]
- Desmarais, D., Jablonski, P.E., Fedarko, N.S., and Roberts, M.F. 1997. 2-Sulfotrehalose, a novel osmolyte in haloalkaliphilic archaea. J. Bacteriol. 179: 3146-3153. [Europe PMC free article] [Abstract] [Google Scholar]
- Gomes, C.M., Bandeiras, T.M., and Teixeira, M. 2001. A new type-II NADH dehydrogenase from the archaeon Acidianus ambivalens: Characterization and in vitro reconstitution of the respiratory chain. J. Bioenerg. Biomembr. 33: 1-8. [Abstract] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. CONSED: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [Abstract] [Google Scholar]
- Hayashi, S. and Wu, H.C. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22: 451-471. [Abstract] [Google Scholar]
- Hirasawa, M., Dose, M.M., Kleis-Sanfrancisco, S., Hurley, J.K., Tollin, G., and Knaff, D.B. 1998. A conserved tryptophan at the ferredoxin-binding site of ferredoxin:nitrite oxidoreductase. Arch. Biochem. Biophys. 354: 95-101. [Abstract] [Google Scholar]
- Hirasawa, M., Rubio, L.M., Griffin, J.L., Flores, E., Herrero, A., Li, J., Kim, S.-K., Hurley, J.K., Tollin, G., and Knaff, D.B. 2004. Complex formation between ferredoxin and Synechococcus ferredoxin: Nitrate oxidoreductase. Biochim. Biophys. Acta 1608: 155-162. [Abstract] [Google Scholar]
- Horikoshi, K. 1999. Alkaliphiles: Some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 63: 735-750. [Europe PMC free article] [Abstract] [Google Scholar]
- Hulo, N., Sigrist, C.J.A., Le Saux, V., Langendijk-Genevaux, P.S., Bordoli, L., Gattiker, A., De Castro, E., Bucher, P., and Bairoch, A. 2004. Recent improvements to the PROSITE database. Nucleic Acids Res. 32: D134-D137. [Europe PMC free article] [Abstract] [Google Scholar]
- Kandler, O. and Konig, H. 1998. Cell wall polymers in Archaea (Archaebacteria). Cell. Mol. Life Sci. 54: 305-308. [Europe PMC free article] [Abstract] [Google Scholar]
- Kikuchi, A., Sagami, H., and Ogura, K. 1999. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J. Biol. Chem. 274: 18011-18016. [Abstract] [Google Scholar]
- Klare, J.P., Gordeliy, V.I., Labahn, J., Buldt, G., Steinhoff, H.J., and Engelhard, M. 2004. The archaeal sensory rhodopsin II/transducer complex: A model for transmembrane signal transfer. FEBS Lett. 564: 219-224. [Abstract] [Google Scholar]
- Klein, C., Garcia-Rizo, C., Bisle, B., Scheffer, B., Zischka, H., Pfeiffer, F., Siedler, F., and Oesterhelt, D. 2004. The membrane proteome of Halobacterium salinarum. Proteomics 5: 180-197. [Abstract] [Google Scholar]
- Koch, M.K. and Oesterhelt, D. 2005. MpcT is the transducer for membrane potential changes in Halobacterium salinarum. Mol. Microbiol. 55: 1681-1694. [Abstract] [Google Scholar]
- Kokoeva, M.V., Storch, K.-F., Klein, C., and Oesterhelt, D. 2002. A novel mode of sensory transduction in archaea: Binding protein-mediated chemotaxis towards osmoprotectants and amino acids. EMBO J. 21: 2312-2322. [Europe PMC free article] [Abstract] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567-580. [Abstract] [Google Scholar]
- Lanyi, J.K., Duschl, A., Hatfield, G.W., May, K., and Oesterhelt, D. 1990. The primary structure of a halorhodopsin from Natronobacterium pharaonis. Structural, functional and evolutionary implications for bacterial rhodopsins and halorhodopsins. J. Biol. Chem. 265: 1253-1260. [Abstract] [Google Scholar]
- Lechner, J. and Sumper, M. 1987. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 262: 9724-9729. [Abstract] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [Europe PMC free article] [Abstract] [Google Scholar]
- Lutz, I., Sieg, A., Wegener, A.A., Engelhard, M., Boche, I., Otsuka, M., Oesterhelt, D., Wachtveitl, J., and Zinth, W. 2001. Primary reactions of sensory rhodopsins. Proc. Natl. Acad. Sci. 98: 962-967. [Europe PMC free article] [Abstract] [Google Scholar]
- Martinez-Espinosa, R.M., Marhuenda-Egea, F.C., and Bonete, M.J. 2001. Assimilatory nitrate reductase from the haloarchaeon Haloferax mediterranei: Purification and characterisation. FEMS Microbiol. Lett. 204: 381-385. [Abstract] [Google Scholar]
- Mattar, S. and Engelhard, M. 1997. Cytochrome ba(3) from Natronobacterium pharaonis—An archaeal four-subunit cytochrome-c-type oxidase. Eur. J. Biochem. 250: 332-341. [Abstract] [Google Scholar]
- Mattar, S., Scharf, B., Kent, S.B.H., Rodewald, K., Oesterhelt, D., and Engelhard, M. 1994. The primary structure of halocyanin, and archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J. Biol. Chem. 269: 14939-14945. [Abstract] [Google Scholar]
- McHardy, A.C., Goesmann, A., Puehler, A., and Meyer, F. 2004. Development of joint application strategies for two microbial gene finders. Bioinformatics 20: 1622-1631. [Abstract] [Google Scholar]
- Meyer, F., Goesmann, A., McHardy, A.C., Bartels, D., Bekel, T., Clausen, J., Kalinowski, J., Linke, B., Rupp, O., Giegerich, R., et al. 2003. GenDB: An open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31: 2187-2195. [Europe PMC free article] [Abstract] [Google Scholar]
- Ng, W.V., Kennedy, S.P., Mahairas, G.G., Berquist, B., Pan, M., Shukla, H.D., Lasky, S.R., Baliga, N.S., Thorsson, V., Sbrogna, J., et al. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. 97: 12176-12181. [Europe PMC free article] [Abstract] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1-6. [Abstract] [Google Scholar]
- Patenge, N., Berendes, A., Engelhardt, H., Schuster, S.C., and Oesterhelt, D. 2001. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol. Microbiol. 41: 653-663. [Abstract] [Google Scholar]
- Pohlschroder, M., Dilks, K., Hand, N.J., and Rose, R.W. 2004. Translocation of proteins across archaeal cytoplasmic membranes. FEMS Microbiol. Rev. 28: 3-24. [Abstract] [Google Scholar]
- Rose, R.W., Bruser, T., Kissinger, J.C., and Pohlschroder, M. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45: 943-950. [Abstract] [Google Scholar]
- Rudolph, J. and Oesterhelt, D. 1996. Deletion analysis of the che operon in the archaeon Halobacterium salinarium. J. Mol. Biol. 258: 548-554. [Abstract] [Google Scholar]
- Ruepp, A. and Soppa, J. 1996. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): Genes, gene products, and transcripts of the arcRACB gene cluster. J. Bacteriol. 178: 4942-4947. [Europe PMC free article] [Abstract] [Google Scholar]
- Schafer, G., Purschke, W.G., Gleissner, M., and Schmidt, C.L. 1996. Respiratory chains of archaea and extremophiles. Biochim. Biophys. Acta 1275: 16-20. [Abstract] [Google Scholar]
- Scharf, B., Wittenberg, R., and Engelhard, M. 1997. Electron transfer proteins from the haloalkaliphilic archaeon Natronobacterium pharaonis: Possible components of the respiratory chain include cytochrome bc and a terminal oxidase cytochrome ba(3). Biochemistry 36: 4471-4479. [Abstract] [Google Scholar]
- Seidel, R., Scharf, B., Gautel, M., Kleine, K., Oesterhelt, D., and Engelhard, M. 1995. The primary structure of sensory rhodopsin II: A member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc. Natl. Acad. Sci. 92: 3036-3040. [Europe PMC free article] [Abstract] [Google Scholar]
- Skulachev, V.P. 1992. The laws of cell energetics. Eur. J. Biochem. 208: 203-209. [Abstract] [Google Scholar]
- Skulachev, V.P., Kobayashi, H., Krulwich, T.A., Schafer, G., Fillingame, R.H., Poole, R.K., Cook, G.M., Dimroth, M.J., Konings, W.N., Stock, J.B., et al. 1999. Bacterial energetics at high pH: What happens to the H+ cycle when the extracellular H+ concentration decreases? Bacterial Response to pH—Novartis Foundation Symposium 221: 200-217. [Abstract] [Google Scholar]
- Soliman, G.S.H. and Truper, H.G. 1982. Halobacterium pharaonis sp. nov., a new, extremely haloalkaliphilic archaebacterium with low magnesium requirement. Zbl. Bakt. Hyg. I Abt. Orig. C 3: 318-329. [Google Scholar]
- Stan-Lotter, H., Doppler, E., Jarosch, M., Radax, C., Gruber, C., and Inatomi, K.-I. 1999. Isolation of a chymotrypsinogen B-like enzyme from the archaeon Natronomonas pharaonis and other halobacteria. Extremophiles 3: 153-161. [Abstract] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631-637. [Abstract] [Google Scholar]
- Tebbe, A., Klein, C., Bisle, B., Siedler, F., Scheffer, B., Garcia-Rizo, C., Wolfertz, J., Hickmann, V., Pfeiffer, F., and Oesterhelt, D. 2004. Analysis of the cytosolic proteome of Halobacterium salinarum and its implication for genome annotation. Proteomics 5: 168-179. [Abstract] [Google Scholar]
- Thomas, N.A., Bardy, S.L., and Jarrell, K.F. 2001. The archaeal flagellum: A different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25: 147-174. [Abstract] [Google Scholar]
- Tindall, B.J., Ross, H.N.M., and Grant, W.D. 1984. Natronobacterium gen. nov. and Natronococcus gen. nov., two new genera of haloalkaliphilic archaebacteria. System. Appl. Microbiol. 5: 41-57. [Google Scholar]
- von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. 2003. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 31: 258-261. [Europe PMC free article] [Abstract] [Google Scholar]
- Yao, V.J. and Spudich, J.L. 1992. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc. Natl. Acad. Sci. 89: 11915-11919. [Europe PMC free article] [Abstract] [Google Scholar]
- Zhang, W.S., Brooun, A., Mueller, M.M., and Alam, M. 1996. The primary structures of the archaeon Halobacterium salinarium blue light receptor sensory rhodopsin II and its transducer, a methyl-accepting protein. Proc. Natl. Acad. Sci. 93: 8230-8235. [Europe PMC free article] [Abstract] [Google Scholar]
WEB SITE REFERENCES
- http://www.cbs.dtu.dk/services; Center for Biological Sequence Analysis Prediction Server providing the NetNGlyc and NetOGlyc tools.
- http://www.halolex.mpg.de; Information System for Halophilic Archaea.
- http://www.migenas.mpg.de; Microbial Genome Analysis System of the Max-Planck Society.
Articles from Genome Research are provided here courtesy of Cold Spring Harbor Laboratory Press
Full text links
Read article at publisher's site: https://doi.org/10.1101/gr.3952905
Read article for free, from open access legal sources, via Unpaywall:
https://genome.cshlp.org/content/15/10/1336.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Haloarchaea as Promising Chassis to Green Chemistry.
Microorganisms, 12(8):1738, 22 Aug 2024
Cited by: 0 articles | PMID: 39203580 | PMCID: PMC11357113
Review Free full text in Europe PMC
Ion-combination specific effects driving the enzymatic activity of halophilic alcohol dehydrogenase 2 from Haloferax volcanii in aqueous ionic liquid solvent mixtures.
RSC Sustain, 2(9):2559-2580, 08 Jul 2024
Cited by: 0 articles | PMID: 39211508 | PMCID: PMC11353702
The biosynthesis mechanism of bacterioruberin in halophilic archaea revealed by genome and transcriptome analysis.
Appl Environ Microbiol, 90(7):e0054024, 03 Jun 2024
Cited by: 0 articles | PMID: 38829054
Protein length distribution is remarkably uniform across the tree of life.
Genome Biol, 24(1):135, 08 Jun 2023
Cited by: 5 articles | PMID: 37291671 | PMCID: PMC10251718
Sec-Dependent Secretion of Subtilase SptE in Haloarchaea Facilitates Its Proper Folding and Heterocatalytic Processing by Halolysin SptA Extracellularly.
Appl Environ Microbiol, 88(8):e0024622, 29 Mar 2022
Cited by: 6 articles | PMID: 35348390 | PMCID: PMC9040580
Go to all (115) article citations
Other citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (3)
- (1 citation) ENA - CR936259
- (1 citation) ENA - CR936258
- (1 citation) ENA - CR936257
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
A halorhodopsin-overproducing mutant isolated from an extremely haloalkaliphilic archaeon Natronomonas pharaonis.
FEBS Lett, 582(19):2931-2936, 29 Jul 2008
Cited by: 12 articles | PMID: 18671971
Characterization of growth and metabolism of the haloalkaliphile Natronomonas pharaonis.
PLoS Comput Biol, 6(6):e1000799, 03 Jun 2010
Cited by: 16 articles | PMID: 20543878 | PMCID: PMC2881530
Bacterial-like nitric oxide synthase in the haloalkaliphilic archaeon Natronomonas pharaonis.
Microbiologyopen, 9(11):e1124, 14 Oct 2020
Cited by: 6 articles | PMID: 33306280 | PMCID: PMC7658456
From genomes to function: haloarchaea as model organisms.
Microbiology (Reading), 152(pt 3):585-590, 01 Mar 2006
Cited by: 77 articles | PMID: 16514139
Review





