Abstract
Free full text

Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: Implications for tauopathies
Abstract
Frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) is caused by mutations in the gene encoding the microtubule-associated protein, tau. Some FTDP-17 mutations affect exon 10 splicing. To correct aberrant exon 10 splicing while retaining endogenous transcriptional control, we evaluated the feasibility of using spliceosome-mediated RNA trans-splicing (SMaRT) to reprogram tau mRNA. We designed a pre-trans-splicing molecule containing human tau exons 10 to 13 and a binding domain complementary to the 3′ end of tau intron 9. A minigene comprising tau exons 9, 10, and 11 and minimal flanking intronic sequences was used as a target. RT-PCR analysis of SH-SY5Y cells or COS cells cotransfected with a minigene and a pre-trans-splicing molecule using primers to opposite sides of the predicted splice junction generated products containing exons 9 to 13. Sequencing of the chimeric products showed that an exact exon 9–exon 10 junction had been created, thus demonstrating that tau RNA can be reprogrammed by trans-splicing. Furthermore, by using the same paradigm with a minigene containing full-length intronic sequences, we show that cis-splicing exclusion of exon 10 can be by-passed by trans-splicing and that conversion of exon 10– tau RNA into exon 10+ tau RNA could be achieved with ≈34% efficiency. Our results demonstrate that an alternatively spliced exon can be replaced by trans-splicing and open the way to novel therapeutic applications of SMaRT for tauopathies and other disorders linked to aberrant alternative splicing.
Tau is a microtubule-associated protein predominantly expressed in neurons and specifically localized in axons that promotes microtubule polymerization and stabilization (1, 2). Mutations in the MAPT gene, encoding tau, on chromosome 17, cause frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17) (3–5). FTDP-17 is characterized by intraneuronal aggregates of tau and, as such, belongs to the group of dementias that includes Alzheimer's disease and collectively referred to as tauopathies.
The human MAPT gene is a large gene of ≈134 kb comprising 16 exons (6). Alternative splicing of exons 2, 3, and 10 in the tau pre-mRNA results in the expression of six isoforms in the brain. Exon 10 encodes the second of four imperfect microtubulebinding repeats in the C-terminal half of the tau protein. Exclusion or inclusion of exon 10 gives rise to tau isoforms with three (tau3R, exon 10–) or four (tau4R, exon 10+) microtubulebinding repeats (7). Furthermore, inclusion or exclusion of exons 2 and 3 produces tau isoforms with zero, one, or two inserts near the N terminus. Only tau3R is expressed during embryogenesis whereas tau3R and tau4R are expressed in approximately equal amounts in adult human brain. Exon 10 splicing is regulated by multiple cis-acting elements comprising exonic and intronic silencers and enhancers (for reviews, see refs. 8 and 9). In particular, intron 10 contains a bipartite element comprising a silencer and a modulator downstream to the 5′ splice site (10). Furthermore, a potential stem-loop formed at the exon 10/intron 10 junction would have an inhibitory effect on exon 10 inclusion (11). Exon 10 inclusion is the default option (12), and exon 10 skipping requires the binding of specific trans-acting factors to silencer elements in exon 10 (13).
MAPT mutations in FTDP-17 are either missense or silent mutations or affect exon 10 splicing regulatory elements and alter exon 10 splicing (5, 11, 14). Exon 10 retention in FTDP-17 results in a 2- to 6-fold excess of tau4R over tau3R (3, 5, 15, 16). Attempts to correct isoform imbalance by conventional expression of tau cDNA or genomic fragments is complicated by the fact that overexpression of tau seems to be toxic per se, irrespective of the isoform (17–19). An attractive alternative would be to reprogram tau mRNA, which would maintain endogenous transcriptional control. Therefore, the modified transcript would not be overexpressed and would have the same tissue-specific expression pattern as endogenous tau.
A unique technology for RNA reprogramming is spliceosomemediated RNA trans-splicing, or SMaRT (20). RNA transsplicing refers to the splicing between the 5′ splice site of a pre-mRNA and the 3′ splice site of a separate pre-mRNA. SMaRT achieves a trans-splicing reaction mediated by the spliceosome between an endogenous target pre-mRNA and an exogenously delivered pre-trans-splicing RNA molecule (PTM). A typical PTM comprises a binding domain that binds to an intron of the targeted pre-mRNA, a trans-splicing domain, and a coding sequence corresponding to the new 3′ end of the reprogrammed mRNA. SMaRT has been used successfully in a number of cases (for reviews, see refs. 21 and 22). The most significant applications of SMaRT have been the functional repair of the CFTRΔF508 mutation of the cystic fibrosis transmembrane conductance regulator in human airway epithelial cells (23) and the phenotypic correction of hemophilia A in mice (24). SMaRT is also a promising technology for imaging gene expression in vivo (25).
Here, we show that tau RNA can be reprogrammed by trans-splicing and that exon 10– to exon 10+ tau RNA conversion can be achieved by SMaRT. Our results demonstrate that an alternatively spliced exon can be replaced by trans-splicing. Our data also suggest that SMaRT could be used to manipulate alternative splicing and could have therapeutic applications for tauopathies and other disorders linked to aberrant alternative splicing.
Materials and Methods
Pre-Trans-Splicing Molecules and Minigene Constructs. PTMs were constructed by using a combination of PCR, annealing of oligonucleotides, and subcloning into plasmid vectors. The coding sequence of the PTMs (525-bp) consisting of tau exons 10 to 13 was amplified from a human tau cDNA (a generous gift from M. Goedert, Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.) to which a sequence encoding the FLAG epitope was added: 5′-GACTACAAGGACGACGATGACAAG-3′. The different domains of the targeted PTM (TauP TM6) and the control P TMs (TauP TMnt and TauPTMΔTSD) are detailed in Results. PTMs were subcloned into the mammalian expression vector, pcDNA3.1 (Invitrogen), between the XhoI and AflII sites. All constructs were verified by sequencing. A tau minigene target, MI9/MI10, comprising exons 9, 10, and 11 and minimal flanking intronic sequences, was constructed by PCR amplification from genomic DNA and subcloned in pcDNA3.1, between the EcoRI and NotI sites (see Fig. 3A). The LI9/LI10 minigene in pCIneo comprising tau exons 9, 10, and 11 and full-length introns 9 and 10 has been described (26).
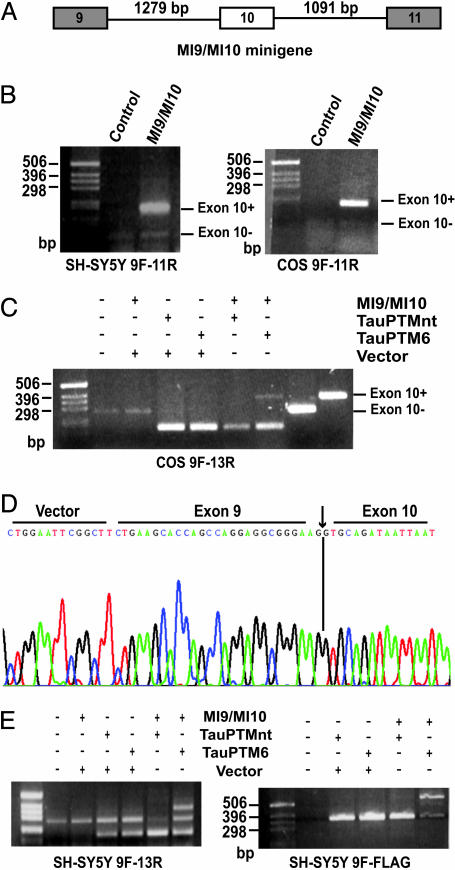
Trans-splicing between tau minigene-derived RNA and TauPTM6 in cultured cells. (A) The target minigene, MI9/MI10, comprises tau exons 9, 10, and 11 and the 3′ and 5′ ends of introns 9 and 10. (B) RT-PCR analysis using 9F and 11R primers of the splicing pattern of MI9/MI10 in SH-SY5Y cells and COS cells. The 189-bp product from minigene-transfected cells corresponds to exon 10+ tau RNA. The 96-bp product from untransfected SH-SY5Y cells arises from endogenous tau3R. RT-PCR products using RNA from untransfected COS cells suggests that they express low levels of tau, as reported for a number of nonneuronal cell lines (47–49). (C) RT-PCR analysis using primers 9F and 13R of COS cells cotransfected with MI9/MI10 and TauPTMs. A 367-bp trans-spliced product was generated by using RNA from TauPTM6-expressing cells, but not from cells transfected with TauPTMnt. (D) An exact exon 9–exon 10 junction was confirmed by sequencing of the 367-bp product. (E) RT-PCR analysis of SH-SY5Y cells cotransfected with MI9/MI10 and TauPTMs. The 274-bp product corresponds to endogenous tau3R. A 367-bp product was detected by using RNA from cells transfected with the minigene and TauPTM6, but not TauPTMnt. Trans-splicing was further confirmed by RT-PCR analysis by using 9F and FLAG primers. The products appearing in reactions using RNA from cells transfected with PTMs only (≈180-bp for 9F/13R and ≈360-bp for 9F-FLAG) most likely arise from the partial complementarity between primer 9F and the coding sequence of the PTM. This problem is inherent to the repeated nature of the tau sequence in the region being analyzed and has been observed previously (16, 40).
Cell Culture and Transfection. COS-7 cells and human neuroblastoma SH-SY5Y cells were maintained in DMEM or DMEM/F12, respectively, supplemented with 10% (vol/vol) heat-inactivated FBS (Invitrogen), 100 units/ml penicillin/100 μg/ml streptomycin and 2 mM l-glutamine, in a humidified atmosphere of 95% air/5% CO2 at 37°C. Cells were transiently transfected with Lipofectamine 2000 reagent (Invitrogen). Briefly, cells plated on 35-mm dishes were grown to 60–70% confluence and exposed to the DNA/liposome complex for 5 h in Optimem (Invitrogen) before being returned to normal culture medium. Cells were routinely analyzed 24 h after transfection. Cells were transfected with either tau minigene plus empty vector or PTM plus empty vector, or cotransfected with minigene and PTM. Typically, 2 μg of minigene and 2 μg of PTM vector DNA were used in each transfection.
RT-PCR Analysis of Cis- and Trans-Splicing. Total RNA was isolated from transfected cells by using TRIzol reagent (Invitrogen) and treated with DNaseI (Promega). Reverse transcription was performed by using the Multiscribe RT kit (Applied Biosystems) and random hexamers or oligo(dT). Each reaction contained 0.5–1 μg of RNA in a total volume of 10 μl. The reverse transcription conditions were 10 min at 25°C, 30 min at 48°C, and a final step of 5 min at 95°C. To detect cis- and trans-splicing events, reverse transcribed RNA was amplified by PCR by using primers spanning the splice junction and under the following conditions: 95°C for 5 min, 40 cycles of 30 s at 94°C, 30 s at 68°C, 45 s to 1 min at 72°C, and a final step of 10 min at 72°C. The sequences of the primers were as follows: forward primer (9F, exon 9), 5′-CTGAAGCACCAGCCAGGAGG-3′; reverse primers, exon 11 (11R), 5′-GGATGTTGCCTAATGAGCCAC-3′; exon 13 (13R), 5′-TGGTCTGTCTTGGCTTTGGC-3′; and FLAG, 5′-TTGTCATCGTCGTCCTTGTAG-3′. The predicted sizes for the corresponding PCR products are indicated in Table 1 and were confirmed by PCR analysis of tau3R or tau4R cDNA templates by using the same primers. RT-PCR products were separated by electrophoresis in 2% (wt/vol) agarose gels and stained with ethidium bromide.
Table 1.
Predicted sizes of the PCR amplification products generated from cis- and trans-acting products detected using the RT-PCR strategy illustrated in Fig. 2
| Predicted size, bp
| |||
|---|---|---|---|
| Strategy | 9F-11R | 9F-13R | 9F-FLAG |
| Minigene | |||
| Cis-splicing | 96/189 | — | — |
| Trans-splicing | 189 | 367 | 569 |
| Endogenous | |||
| Tau3R | 96 | 274 | — |
| Tau4R | 189 | 367 | — |
Levels of exon 10+ and exon 10– tau RNA were estimated by RT-PCR by using primers 9F and 11R and densitometric analysis of the gels by using the innotech software (Alpha Innotech, San Leandro, CA). Consistency between transfections was established through the total level of tau RNA. GAPDH mRNA (amplified by using the following primers: forward, 5′-CCATGGCACCGTCAAGGCTGA-3′; reverse, 5′-GCCAGTAGAGGCAGGGATGAT-3′) was used as a control for gel loadings.
Sequencing. For sequencing, RT-PCR products were excised from agarose gels and cloned into the pCR2.1 vector by using the TA cloning system (Invitrogen). Sequencing of both strands was performed by using M13 reverse and T7 primers.
Results
Design of Tau Pre-Trans-Splicing Molecules. To evaluate the possibility of reprogramming human tau mRNA at the level of exon 10, we engineered a PTM, TauPTM6, designed to bind to tau intron 9 (Fig. 1A). The coding sequence of TauPTM6 consists of exons 10–13 of human tau, followed by a 24-nt sequence encoding the FLAG epitope. The trans-splicing domain of TauPTM6 comprises a 125-nt binding domain corresponding to the 3′end of tau intron 9 in the antisense orientation, as well as elements that promote efficient trans-splicing, as defined (20, 27, 28). These elements include a strong conserved yeast branch point sequence, a 26-nt polypyrimidine tract, an AG acceptor site, and a spacer sequence separating the binding domain and the branch point. The predicted binding of TauPTM6 to tau pre-mRNA is illustrated in Fig. 1B. As controls, we constructed PTMs with the same coding sequence as TauPTM6 but in which the 5′end is not directly complementary to a tau intronic sequence (TauPTMnt), or lacking a trans-splicing domain, thus being trans-splicing incompetent (TauPTMΔTSD) (Fig. 1C). Cis- and trans-splicing patterns are illustrated in Fig. 2. To facilitate the analysis of tau splicing in cells, the targets used were minigenes comprising exons 9, 10, and 11 of human tau and minimal (MI9/MI10) or full-length (LI9/LI10) flanking intronic sequences. An RT-PCR strategy was designed to detect specifically RNA resulting from cis- and trans-splicing events by using a forward primer specific for exon 9 (9F) and reverse primers specific for exon 11 (11R), exon 13 (13R), or for the FLAG epitope sequence (FLAG) (Fig. 2). The predicted sizes of the various amplification products are indicated in Table 1.
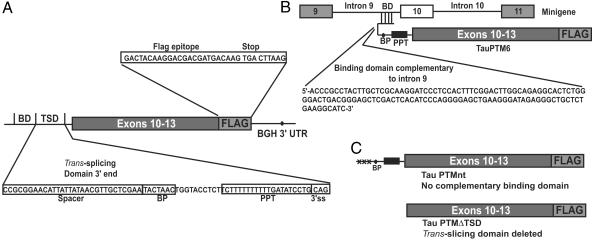
SMaRT strategy for tau reprogramming. (A) Schematic representation of TauPTM6 structure. The coding sequence of TauPTM6 consists of exons 10 to 13 of human tau followed by a 24-nt sequence encoding the FLAG epitope. The trans-splicing domain of TauPTM6 comprises a 125-nt binding domain (BD) complementary to the 3′ end of tau intron 9 as well as a strong conserved yeast branch point sequence (BP) and a 26-nt polypyrimidine tract (PPT), an AG acceptor site, and a spacer sequence separating the binding domain and the branch point. (B) Diagram showing the SMaRT strategy used to reprogram tau mRNA at the level of exon 10 splicing by using TauPTM6. (C) Control PTMs used included TauPTMnt, in which the 5′ end is not directly complementary to a tau intronic sequence, and TauPTMΔTSD, in which the trans-splicing domain has been deleted.
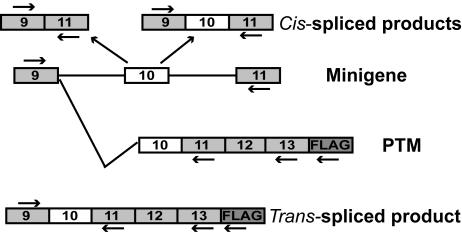
RT-PCR strategy to detect cis and trans-splicing. Shown is a schematic representation of cis- and trans-splicing patterns from a tau pre-mRNA minigene target. Arrows indicate the positions of the forward (9F) and reverse (11R, 13R, and FLAG) PCR primers in the target and in the PTMs. Primer 9F is specific for the target, and primers 13R and FLAG are specific for the PTMs. A list of the predicted sizes of the corresponding RT-PCR products can be found in Table 1.
Trans-Splicing Replacement of Tau Exon 10. The MI9/MI10 tau minigene comprises tau exons 9, 10, and 11 and the 3′ and 5′ ends of the introns flanking exon 10 (Fig. 3A). RT-PCR analysis of MI9/MI10-transfected SH-SY5Y cells with primers 9F and 11R revealed a 189-bp product, corresponding to exon 10+ tau RNA, in addition to the 96-bp product arising from endogenous tau3R RNA (29) (Fig. 3B). An identical 189-bp RT-PCR product was detected by using RNA from MI9/MI10-transfected COS cells. Thus, the pre-mRNA transcribed from MI9/MI10 is spliced to give rise to an RNA including exon 10.
We next tested whether exons 10 and 11 from minigenederived RNA could be replaced by exons 10 to 13 derived from TauPTM6 in transfected cells. The 3′ end of intron 9 present in the minigene contains the sequence complementary to the binding domain of TauPTM6. COS cells were cotransfected with MI9/MI10 and TauPTM6 or a nontargeted PTM, TauPTMnt. Trans-splicing was analyzed by RT-PCR by using primer 9F (minigene-specific) and 13R (PTM-specific). A 367-bp product was obtained by using RNA from cells cotransfected with MI9/MI10 and TauPTM6, the size predicted for a product generated from tau RNA containing exons 9 to 13 and matching the size of the product obtained with the same primers by using a tau4R cDNA template (Fig. 3C). No trans-spliced product was observed in RNA from cells cotransfected with the minigene and TauPTMnt. This finding excludes the possibility of jumping PCR or of recombination between the PTMs and the minigene at the DNA level. Recombination events were also excluded by the analysis of genomic DNA from cotransfected cells with the same set of primers (data not shown). Sequencing of the 367-bp RT-PCR product confirmed that an exact exon 9–exon 10 junction had been generated (Fig. 3D). This result demonstrates that trans-splicing had occurred between RNA derived from the MI9/MI10 tau minigene and the PTM at the predicted splice site.
Tau RNA Reprogramming in Neuroblastoma Cells. To evaluate whether tau RNA reprogramming could occur in a neuronal environment, trans-splicing was analyzed in SH-SY5Y cells cotransfected with MI9/MI10 and TauPTM6 or TauPTMnt. RT-PCR analysis of cells transfected with the minigene and TauPTM6 with primers 9F and 13R demonstrated a 367-bp product, the predicted size of a product generated by a transspliced product, in addition to a 274-bp product, corresponding to endogenous tau3R (Fig. 3E). The 367-bp product was not observed in reactions using RNA from cells transfected with TauPTM6 only. Trans-splicing between RNA derived from the minigene and TauPTM6 in SH-SY5Y cells was further confirmed by RT-PCR analysis by using primer 9F and a reverse primer corresponding to the FLAG epitope sequence, specific for the PTM. By using this primer combination, a 569-bp product was detected by using RNA from cells transfected with the minigene and TauPTM6, the predicted size of a product originating from tau RNA containing exon 9 to exon 13 with a FLAG epitope sequence at the 3′ end (Fig. 3E).
Trans-Splicing Inclusion of Exon 10. The above experiments show that tau exon 10 derived from MI9/MI10 can be substituted for PTM-derived exon 10 by trans-splicing. To determine whether cis-splicing of exon 10 could be by-passed by trans-splicing under normal alternative splicing regulation we used the LI9/LI10 minigene, which contains full-length introns 9 and 10 (Fig. 4A). RT-PCR analysis of LI9/LI10-transfected SH-SH5Y with primers 9F and 11R revealed a major 96-bp product corresponding to both endogenous tau3R and ectopic tau transcript. A faint band at 189 bp was also observed, indicating that some exon 10+ RNA was also produced. (Fig. 4B). By contrast, both exon 10– and exon 10+ products were detected in LI9/LI10-transfected COS cells (Fig. 4B).
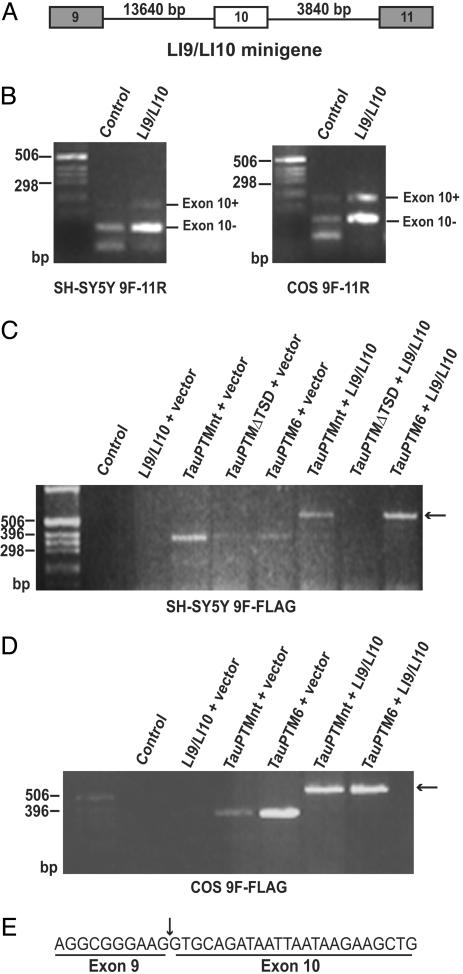
Trans-splicing inclusion of exon 10 in tau transcripts. (A) Schematic structure of the minigene, LI9/LI10, comprising tau exons 9, 10, and 11 and full-length introns 9 and 10. (B) RT-PCR analysis of the cis-splicing pattern of LI9/LI10 minigene in SH-SY5Y cells and COS cells using 9F and 11R primers. A major 96-bp product was detected by using RNA from LI9/LI10-transfected SH-SY5Y cells, corresponding to both endogenous tau3R and ectopic tau transcript. Some exon 10+ tau RNA was also observed. In COS cells, both exon 10+ and exon 10– forms were produced. SH-SY5Y cells (C) and COS cells (D) were cotransfected with LI9/LI10 and TauPTMs and analyzed by RT-PCR by using 9F and FLAG primers. RT-PCR products of 569 bp, the size predicted for a product generated from a trans-spliced product, were detected in cells transfected with LI9/LI10 and either TauPTM6 or TauPTMnt (arrows). No such product was observed in cells cotransfected with the minigene and the trans-splicing incompetent TauPTMΔTSD. (E) The generation of an accurate exon 9–exon 10 junction was confirmed by sequencing of the 569-bp RT-PCR product.
Trans-splicing was analyzed in SH-SY5Y cells cotransfected with LI9/LI10 and TauPTM6 or PTMnt by using primers 9F and FLAG. A 569-bp product, the size predicted for a trans-spliced product, was detected in reactions using RNA from cells cotransfected with LI9/LI10 and TauPTM6 or TauPTMnt (Fig. 4C). Trans-splicing observed by using TauPTMnt suggests some degree of complementarity between the 5′ end of TauPTMnt and sequences in intron 9 and is most likely the consequence of high levels of target and PTM expression, as observed previously in similar experimental paradigms (30, 31). No such products were observed using RNA from cells cotransfected with the minigene and the trans-splicing incompetent TauPTMΔTSD (Fig. 4C). This result demonstrates that the chimeric products obtained in cells cotransfected with LI9/LI10 and TauPTM6 or TauPTMnt are the result of trans-splicing between minigeneand PTM-derived RNA. Similar results were obtained in COS cells (Fig. 4D). Sequencing of the 569-bp RT-PCR product confirmed that an accurate exon 9–exon 10 junction had been generated (Fig. 4E).
We next analyzed the change in exon 10+/exon 10– tau RNA ratio in transfected cells to determine whether trans-splicing could achieve exon 10– to exon 10+ conversion and not be limited to transcripts that failed to recruit factors required for exon 10 skipping. The exon 10+/exon 10– ratio was estimated by RT-PCR with primers 9F and 11R by using RNA from COS cells cotransfected with LI9/LI10 and TauPTM6. An increase in exon 10+ RNA and a concomitant decrease in exon 10– RNA was evident in cells cotransfected with the minigene and TauPTM6 as compared with cells transfected with the minigene only or cells cotransfected with the minigene and TauPTMΔTSD (Fig. 5A). The exon 10+/exon 10– ratio was ≈0.5 in cells transfected with the minigene only or cotransfected with the minigene and TauPTMΔTSD and raised to 1.5 in cells cotransfected with the minigene and TauPTM6 (Fig. 5B). Assuming that a high proportion of transfected cells have incorporated both constructs, a trans-splicing efficiency of ≈34% was deduced from the change in the ratio.
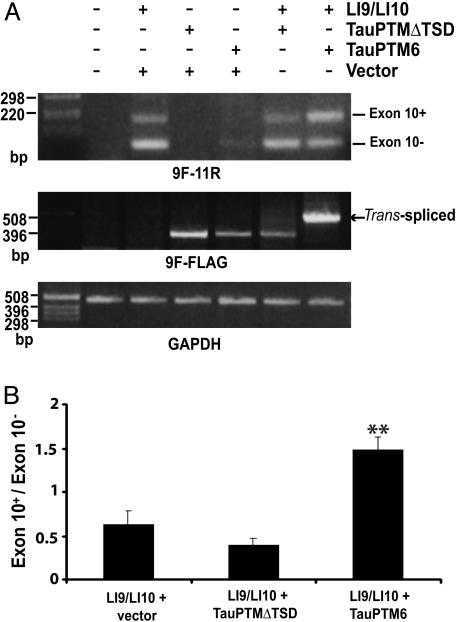
Exon 10– to exon 10+ tau RNA conversion by trans-splicing. (A) RT-PCR analysis with primers 9F and 11R of COS cells transfected with the LI9/LI10 minigene and tau PTMs. Cotransfection of TauPTM6 with LI9/LI10 resulted in a clear increase in exon 10+ tau RNA and a concomitant decrease in exon 10– RNA. Trans-splicing was confirmed by RT-PCR analysis with primers 9F and FLAG. Loadings were normalized by comparison with GAPDH mRNA. (B) Analysis of the change in exon 10+/exon 10– tau RNA ratio. The exon 10+/exon 10– ratios are expressed as mean ± SEM (**, P < 0.01, Student's t test, n = 4).
Discussion
We have demonstrated that trans-splicing could compete effectively with tau exon 10 cis-splicing in a neuronal environment. This finding has important implications for expanding the range of applications of the SMaRT technology. Specifically, this study (i) adds tau to the growing number of targets that can be modified by SMaRT; (ii) is an example of trans-splicing reprogramming of a neuronal transcript; (iii) shows that the complement of splicing factors expressed in neurons is capable of supporting trans-splicing; and (iv) demonstrate that an alternatively spliced exon can be replaced by trans-splicing.
Until now, the successful application of SMaRT has been limited to the replacement of constitutively spliced exons. As compared with constitutive splicing, the selection of specific 5′ and 3′ splice sites in alternative splicing strongly depends on nonspliceosomal RNA-binding proteins (32). Significantly, we have shown that exon 10– tau RNA could be converted into exon 10+ RNA by trans-splicing under normal alternative splicing regulation. Tau exon 10 splicing is regulated by a complex mechanism involving the binding of specific trans-acting factors to exonic and intronic enhancer and silencer elements (8, 9). Exon 10 inclusion is the default option, and its inhibition requires specific splicing factors, such as SRp30c or SRp55, that bind to silencer elements at the 5′ end of exon 10 (12, 33). Exon 10 incorporation through trans-splicing shows that splicing factors inhibiting exon 10 inclusion do not interfere with trans-splicing. This finding has also important consequences for the construction of PTMs designed to replace alternatively spliced exons because it shows that such PTMs do not require the inclusion of specific intronic enhancer or silencer elements in their transsplicing domain to compete efficiently with cis-splicing.
The efficiency of exon 10– to exon 10+ tau RNA conversion was ≈34%. This value is consistent with trans-splicing efficiency measured in other studies. For instance, 24% and 12% transsplicing efficiencies were reported with a collagen XVII minigene target (28) and a CFTR minigene (27), respectively. Trans-splicing represented 12–27% of cis-splicing in cells cotransfected with a minigene comprising lacZ exons separated by a truncated intron from the CFTR gene and a targeted PTM (30).
To date, the only attempt to modulate tau exon 10 splicing has been by using blocking oligonucleotides binding to exon 10 splice junctions in PC12 cells, which express predominantly tau4R (34). Other, very elegant strategies to interfere with splicing events are based on the design of bifunctional oligoribonucleotides with a first domain complementary to an exonic enhancer or silencer element in the target pre-mRNA, and a second domain recruiting specific splicing factors near the splice site (35–37). Although effective, these technologies require the precise knowledge of cis-acting elements regulating splicing of the targeted exon. Another way of modulating tau isoform ratio would be to down-regulate trans-acting factors, such as Tra2β, by RNA interference (38) or to overexpress such factors (12, 13). Again, this strategy requires precise knowledge of the mechanisms regulating exon 10 splicing and is likely to result in abnormal splicing of transcripts other than tau. Thus, due to its versatility and selectivity, SMaRT would have a distinctive advantage over these methods.
Therapeutic applications of SMaRT will require further optimization, in particular to increase efficiency. Possible transsplicing on targets other than the intended target can be minimized by increasing the length of the binding domain (30) and by incorporating inverted repeats in the trans-splicing domain that form an intramolecular stem designed to mask the 3′ splice site until the binding domain has interacted with the target (20, 27). However, nontargeted trans-splicing is unlikely to represent a major problem because many such products will be lost through nonsense-mediated decay, and this issue has been discussed elsewhere (39). In the context for tauopathies, SMaRT could be used to convert excess tau4R into tau3R. Some forms of FTDP-17 are caused by mutations in the MAPT gene that affect exon 10 splicing, resulting in a 2- to 6-fold excess of tau4R (3, 5, 15, 16). Elevated 4R/3R tau mRNA ratio in the sporadic tauopathy, progressive supranuclear palsy, is at least, in part, the result of aberrant splicing of tau pre-mRNA (40). Excess tau4R is likely to affect microtubule properties because tau4R and tau3R have different qualitative and quantitative effects on microtubule dynamics (41–43). In vivo conversion of tau4R into tau3R by using SMaRT could be achieved with a PTM containing exons 11 to 13 only in their coding sequence. In vivo applications of SMaRT for tau will require optimization of PTM design, including choice of the binding domain and incorporation of 3′UTR elements regulating RNA stability and localization.
The demonstration that alternatively spliced exons can be replaced by trans-splicing opens the way to novel applications of SMaRT. SMaRT could be used to mimic differential usage of specific exons and help establish the biological significance of maintaining a specific balance of isoform of tau or other proteins. Moreover, a growing number of neurological disorders are being associated with dysregulation of alternative splicing of specific transcripts (44–46), and SMaRT would be a method of choice to restore a normal isoform balance from the transcripts involved.
Acknowledgments
We thank Dr. Michel Goedert for tau cDNA constructs. This work was supported by the Wellcome Trust and the Medical Research Council.
Notes
Author contributions: T.R.-M. and S.G.M. performed research; T.R.-M. and J.-M.G. analyzed data; M.A.G.-B., B.H.A., and J.-M.G. designed research; A.C.G., M.H., Q.Y., and J.Z. contributed new reagents/analytic tools; and J.-M.G. wrote the paper.
Conflict of interest statement: M.A.G.-B. is a founder of and consultant for Intronn, which owns and commercializes the SMaRT technology. S.G.M. is an employee of Intronn.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FTDP-17, frontotemporal dementia with parkinsonism linked to chromosome 17; SMaRT, spliceosome-mediated RNA trans-splicing; PTM, pre-trans-splicing molecule; Tau4R, tau isoform with four microtubule-binding repeats; Tau3R, tau isoform with three microtubule-binding repeats.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0503150102
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1266082?pdf=render
Citations & impact
Impact metrics
Article citations
RNA exon editing: Splicing the way to treat human diseases.
Mol Ther Nucleic Acids, 35(3):102311, 16 Aug 2024
Cited by: 0 articles | PMID: 39281698 | PMCID: PMC11401238
Review Free full text in Europe PMC
Therapeutic potential of alternative splicing in cardiovascular diseases.
EBioMedicine, 101:104995, 12 Feb 2024
Cited by: 1 article | PMID: 38350330 | PMCID: PMC10874720
Review Free full text in Europe PMC
Alternative splicing in neurodegenerative disease and the promise of RNA therapies.
Nat Rev Neurosci, 24(8):457-473, 19 Jun 2023
Cited by: 27 articles | PMID: 37336982
Review
SMaRT modulation of tau isoforms rescues cognitive and motor impairments in a preclinical model of tauopathy.
Front Bioeng Biotechnol, 10:951384, 05 Oct 2022
Cited by: 3 articles | PMID: 36277399 | PMCID: PMC9581281
RNA-based therapeutics for neurological diseases.
RNA Biol, 19(1):176-190, 31 Dec 2021
Cited by: 25 articles | PMID: 35067193 | PMCID: PMC8786337
Review Free full text in Europe PMC
Go to all (38) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Correction of tau mis-splicing caused by FTDP-17 MAPT mutations by spliceosome-mediated RNA trans-splicing.
Hum Mol Genet, 18(17):3266-3273, 04 Jun 2009
Cited by: 36 articles | PMID: 19498037 | PMCID: PMC2722988
An SRp75/hnRNPG complex interacting with hnRNPE2 regulates the 5' splice site of tau exon 10, whose misregulation causes frontotemporal dementia.
Gene, 485(2):130-138, 30 Jun 2011
Cited by: 27 articles | PMID: 21723381 | PMCID: PMC3163755
Regulation of alternative splicing of tau exon 10.
Neurosci Bull, 30(2):367-377, 14 Mar 2014
Cited by: 53 articles | PMID: 24627328 | PMCID: PMC5562657
Review Free full text in Europe PMC
A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10.
J Neurochem, 90(1):164-172, 01 Jul 2004
Cited by: 42 articles | PMID: 15198676
Funding
Funders who supported this work.






