Abstract
Free full text

The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair
Abstract
The molecular mechanism by which foreign DNA integrates into the human genome is poorly understood yet critical to many disease processes, including retroviral infection and carcinogenesis, and to gene therapy. We hypothesized that the mechanism of genomic integration may be similar to transposition in lower organisms. We identified a protein, termed Metnase, that has a SET domain and a transposase/nuclease domain. Metnase methylates histone H3 lysines 4 and 36, which are associated with open chromatin. Metnase increases resistance to ionizing radiation and increases nonhomologous end-joining repair of DNA doublestrand breaks. Most significantly, Metnase promotes integration of exogenous DNA into the genomes of host cells. Therefore, Metnase is a nonhomologous end-joining repair protein that regulates genomic integration of exogenous DNA and establishes a relationship among histone modification, DNA repair, and integration. The data suggest a model wherein Metnase promotes integration of exogenous DNA by opening chromatin and facilitating joining of DNA ends. This study demonstrates that eukaryotic transposase domains can have important cell functions beyond transposition of genetic elements.
The ability of a eukaryotic cell to integrate foreign DNA into its genome and faithfully pass that DNA on to progeny is a critical biological process. It is a key step in retroviral infection and subsequent persistence of the retroviral clone during replication of the host cell (1, 2). Genomic integration of foreign DNA also has more benign functions. It is occasionally used advantageously in gene therapy of human disease, such as in retroviral correction of inherited immunologic deficiencies (3, 4). Of much wider utility, it has been a foundation of molecular laboratory investigation for decades, allowing for stable modification of a cell's genomic content for construction of in vivo and in vitro laboratory models (5, 6).
Although less appreciated, the integration of exogenous DNA into a host cell's genome can also occur in many normal cell functions. For example, the integration of DNA from apoptotic bodies can horizontally pass oncogenes from one dying malignant cell to another viable cell (7). This phenomenon can contribute to temporal alterations in a tumor phenotype, perhaps even mediating changes in metastatic capability or drug resistance. In addition, extrachromosomal DNA can contribute to repair of chromosomal breaks. For example, damaged chromosomes have been found to repair by insertion of mitochondrial DNA (8). Finally, in lower organisms, internal retrotransposition is a common natural occurrence to produce genomic alteration and is widely used in the laboratory study of such organisms (9).
Exogenous DNA is usually integrated into the genome by two mechanisms: homology-dependent integration and illegitimate integration (10). Homology-dependent integration depends on homologous recombination (HR) repair proteins and the extent of homology between foreign and endogenous DNA (10-12). Illegitimate integration inserts exogenous DNA into nonhomologous genomic sequences and is far more efficient, probably in part because of the much larger number of potential illegitimate integration sites. Illegitimate integration sites are not random but probably depend on open chromatin associated with transcription or repair, allowing access of foreign DNA and integration factors to the genomic DNA (10-12). Illegitimate integration resembles nonhomologous end-joining (NHEJ) repair, because it is enhanced by DNA damage, especially double-strand breaks (DSBs); it is homology-independent and involves end joining (12-14). In addition, there are several reports of reduced integration of retroviral DNA and Arabidopsis T elements in NHEJ-defective cells (15-18). However, it is also clear that NHEJ proteins are not essential for illegitimate integration, because many reports describe reasonable levels of integration in cells lacking NHEJ proteins (19-23). A potential explanation for reduced integration in NHEJ-defective cells is that foreign DNA is degraded more rapidly (24), consistent with NHEJ components protecting exogenous DNA from cellular nucleases. These results suggest that integration may involve factors distinct from known NHEJ proteins (25). Despite the critical importance of foreign DNA integration into host genomes for so many biological and laboratory processes, relatively little is understood about the molecular mechanisms by which integration occurs. In this study, we isolated and characterized a histone methylase, termed Metnase, that stimulates foreign DNA integration and enhances NHEJ repair. Thus, Metnase appears to be an important link among chromatin modification, DSB repair, and foreign DNA integration.
Materials and Methods
Expression Analysis. cDNAs from multiple tissue sources were obtained from Clontech. These were used as templates for Metnase PCR amplification for 30 cycles in the logarithmic phase by using forward primer 5′-GACGACACGGCCCTTGTGGGG and reverse primer 5′-TCTGAACTTCAGAGAATCC at 94°C for 30 seconds, 50°C for 30 seconds, and 68°C for 40 seconds. 18S rRNA served as an amplification control with forward primer 5′-aaacggctaccacatccaag and reverse primer 5′-cctccaatggatcctcgtta.
Antibody Production. An anti-Metnase antiserum (polyclonal) was generated from rabbits by using two peptides representing hydrophilic amino acids 483-495 (DEKWILYDNRRRS) and 659-671 (WQKCVDCNGSYFD), not present in any other protein species.
Histone Methylation. Recombinant Metnase was produced in TOP 10 bacteria by using the pGEX 4T3 expression vector and isolated on a glutathione-Sepharose column after induction with isopropyl β-d-thiogalactoside (IPTG). Methyl transfer was tested by using high specific activity 3H-S-adenosyl methionine (SAM; Amersham Pharmacia) and recombinant human histone H3 (Upstate Biotechnology, Lake Placid, NY), as described (26). Each 40-μl reaction contained 7.5 mM Hepes (pH 7.9), 50 mM KCl, 2.5 mM MgCl2, 0.25 mM DTT, 0.5 mM EDTA, 10% glycerol, 1 mM PMSF (Roche, Penzberg, Germany), 5 μCi (1 Ci = 37 GBq) 3H-SAM, and varying concentrations of Metnase and was incubated at 30°C for 2 h. The reaction was spotted on phosphocellulose paper, dried, washed five times in 0.1 M NaCO3, pH 9.0, dried, and analyzed with a scintillation counter. Histone methyl transfer was validated by Western analysis of the reaction product, substituting unlabeled SAM at a final concentration of 2 mM for the 3H-SAM, using different anti-H3 lysine antibodies (Abcam, Cambridge, U.K.).
Illegitimate Integration Analysis. Illegitimate integration was analyzed by the ability of cells to pass foreign nonhomologous DNA containing a selectable marker onto progeny. Metnase was cloned into the pcDNA3.1-TOPO expression vector, which contains a neo resistance marker, and into the pJ6 expression vector, which does not contain a selectable marker, as described (27, 28). Calcium phosphate transfection was performed as described (27) and transfection efficiencies were tested by using β-galactosidase. Forty-eight hours after transfection, varying numbers of cells were seeded into 100-mm dishes, and 24 h later, selective agents were added (0.8 mg/ml G418/0.15 mg/ml hygromycin/0.0008 mg/ml puromycin). Cells were incubated for 14 (puromycin or hygromycin) or 21 days (G418), washed twice with PBS, stained with 0.17% methylene blue in methanol, and colonies defined as >100 cells were counted.
Metnase small interfering RNA (siRNA) (5′-GGATCCCGGTCTACAGTTCCACTTCCTTCAAGAGAGGAAGTGGAACTGTAGACCTTTTTTCCAAAAGCTT) was stably transduced by using the pRNA/U6.Hygro plasmid into 293 cells. Western analysis was performed as described (27) to assess Metnase expression. There was no difference in unmanipulated colony formation ability between Metnase underexpressors and vector controls. For all transfections, DNA concentrations were normalized with empty vectors. All experiments were performed three or more times in triplicate.
DSB Repair by HR or NHEJ. HR repair was assayed as described by using HT256 cells (29, 30) and the pJ6-Metnase expression vector. To assess the effect of Metnase on total and precise NHEJ repair, a modification of the plasmid-rejoining assay was used (31, 32). pBluescript plasmid DNA (Stratagene), which carries ampicillin resistance, was digested with EcoRI within the β-galactosidase gene and purified from an agarose gel. Five micrograms of linearized pBluescript was transfected into 106 recently thawed 293 cells per plate by using calcium phosphate, as described (27). The 293 cells were stably transduced with pRNA/U6-Metnase siRNA vector for underexpression, pcDNA3.1-Metnase for overexpression, or empty vectors for controls. Twenty-four hours posttransfection, cells were washed in PBS, and plasmid DNA was isolated from cell pellets by using the Qiagen (Valencia, CA) miniprep spin kit according to the manufacturer's instructions. Rescued plasmid DNA concentrations were normalized by using nanospectroscopy, and equivalent amounts were transformed into TOP10 bacteria, the bacteria plated on ampicillin LB plates in the presence of IPTG and X-GAL, and blue versus white colonies counted under oblique white light the next day. Mutations in Metnase were constructed by using overlap extension PCR, sequenced, and subcloned into either pcDNA3.1 or pCAGGS, an expression vector with the chicken β actin promoter and the cytomegalovirus enhancer (gift from M. Jasin, Memorial Sloan-Kettering Cancer Center, New York). Each mutant was tagged with the V5E sequence for discrimination of exogenous protein production by Western blot.
Radiation Survival. Human 293 cells stably transduced with pcDNA3.1-Metnase (for overexpression), pRNA/U6-Metnase (for underexpression) or empty vectors were seeded in varying numbers to 100-mm tissue culture dishes. After incubation for 4 h to allow for adherence, cells were irradiated with a 137Cs source (Gammacell 40, Atomic Energy, Ottawa) at a dose rate of 1.03 Gy per min. Cells were then incubated for 14 days and colonies counted as above.
Results
Expression of a Transposase/SET Protein. We hypothesized that integration might rely on uncharacterized cellular proteins structurally related to transposases, a family of DNA integrases/nucleases that mediate intragenomic movement of their own sequence (25). Although almost all human transposase-like sequences are untranslated pseudogenes, one EST had been identified (U80776, ref. 33) that contained a transposase domain. It also contained a SET domain, associated with histone lysine methyltransferase activity. We used RACE RT-PCR to clone the entire coding sequence of this gene and termed the gene product Metnase (AY952295). The Metnase gene has three exons spread over 13.8 kB located at 3p26, a region of frequent abnormalities in cancer, especially non-Hodgkin's lymphoma, acute and chronic lymphocytic leukemia, myeloma, myelodysplasia, hereditary prostate cancer, and breast cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman). By sequencing the RT-PCR product, we found that this EST indeed encoded a potential translated protein, which was confirmed by Western analysis. The Metnase transposase domain contains the DDE acidic motif that is conserved among retroviral integrase and transposase families and is essential for strand cleavage and end joining. However, Metnase does not have any internal tandem repeats at the ends of the gene common to transposases (25, 32). The Metnase pre-SET and SET domains are most closely related to SuvHar91, with 31% and 36% homology for each domain, respectively (Fig. 1 A and B; ref. 34). The Metnase SET domain contains homologies to the two conserved amino acid sequences in the SET domain of SUV39H1 thought to be responsible for the histone methyltransferase activity (NHSCXPN and ELXFNY in SUV39H1, and NHSCXPN and ELXYDY in Metnase, respectively; ref. 35). There is also a perfect consensus post-SET domain (CXCX4C) in Metnase that is in SUV39H1. The Metnase protein sequence was most closely related to Planarian Mariner-9 in the transposase domain, where it had 50% identity (Fig. 1C). Also, the Metnase transposase domain sequence was 18% identical to the HIV-1 Integrase core domain (Fig. 1D; ref. 36). Metnase is expressed in all tissues tested to various extents (Fig. 1E), with the highest expression in placenta and ovary and the lowest expression in skeletal muscle. This is reminiscent of expression patterns of other DNA repair proteins (37).

Metnase has pre-SET, SET, and transposase domains. (A) Metnase pre-SET domain aligned with the consensus pre-SET sequence and with human SUV39H1. (B) Metnase SET domain aligned with the consensus SET sequence and with SUV39H1. (C) Metnase transposase domain aligned with the transposase domain consensus and with planarian Mariner-9 transposase. (D) Metnase integrase domain aligned with the integrase domain consensus and with the HIV-1 integrase core domain. (E) Metnase expression in normal human tissues as measured by RT-PCR.
Metnase Is a Histone Methyltransferase. Because it has a canonical SET domain, Metnase was tested for histone methyltransferase activity by using a SAM donor, as described (27, 28). Metnase transferred a radiolabeled (3H)-methyl group from SAM to recombinant human histone H3 as assayed by scintillation counting and autoradiography (Fig. 2 A and B). By using Western analysis, Metnase was found to reproducibly stimulate the specific dimethylation of histone H3 lysine 36, and to a lesser extent histone H3 lysine 4, in a concentration-dependent manner (Fig. 2C). This activity was significantly less than recombinant Set9 methylation of H3 lysine 4, which may indicate either that Metnase is intrinsically less active or that it requires an unknown cofactor for maximal activity, as described for SMYD9 (38). Western analysis revealed little methylation by Metnase of histone H3 K9, K27, and K79 residues or histone H4 K20 and K79 (Fig. 2C and data not shown). The histone H3 K36 methylase activity of Metnase depended on the presence of the SAM methyl donor and histone H3 (Fig. 2D). However, despite also having an integrase domain, Metnase by itself had no retroviral integrase activity as measured by in vitro 3′ CA dinucleotide strand cleavage and subsequent U3 or U5 internal repeat integration (36) (data not shown).
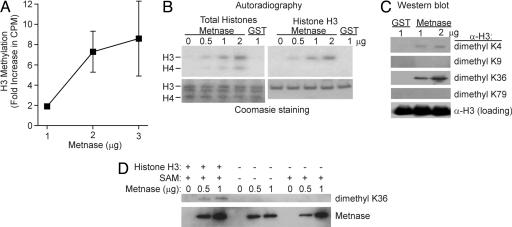
Histone methylation by Metnase. (A) Recombinant GST-Metnase transferred 3H-methyl groups from SAM to recombinant human histone H3 and total human histones as analyzed by scintillation counting. 3H levels (cpm) were subtracted from GST alone as a baseline control. (B) Recombinant GST-Metnase transferred 3H-methyl groups from SAM to recombinant human histone H3 and total human histones as analyzed by autoradiography. (C) Recombinant GST-Metnase in vitro transferred unlabeled methyl groups from SAM to pure recombinant human histone H3 lysines as detected by Western blot. (D) The transfer of unlabeled methyl groups from SAM to H3 K36 requires both SAM and histone H3.
Metnase Stimulates Plasmid and Viral DNA Integration. We next determined whether Metnase stimulates genomic integration of exogenous DNA, measured by the assimilation and passage to progeny of a selectable marker. Stable overexpression of Metnase in human 293 cells did not affect plating efficiency, but it increased integration of a Metnase expression vector carrying neo by 6.6-fold (Fig. 3B), and it increased integration of a cotransfected vector (pBOS) carrying a puromycin resistance cassette by up to 2.6-fold (Fig. 3C). Thus, Metnase promotes genomic integration in cis (when it is expressed from the same segment of DNA as the selectable marker) and in trans (when it is expressed on a separate segment of DNA).
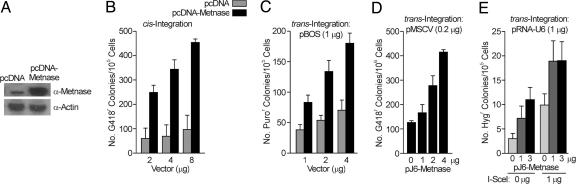
Metnase promotes foreign DNA integration. (A) Western blot analysis of the protein expression of transfected Metnase. (B) Metnase promotes cis integration of foreign DNA. Human 293 cells were transfected with the pCDNA3.1-Metnase expression vector, which also carries neo. The number of G418-resistant colonies is a measure of integration. (C) Metnase promotes trans integration of foreign DNA. Human 293 cells were cotransfected with the pcDNA-Metnase expression vector and the plasmid, pBOS, which expresses a puromycin resistance cassette. The number of puromycin-resistant colonies is a measure of pBOS integration. (D) Metnase promotes integration of Moloney leukemia virus (MLV) DNA. pJ6-Metnase was cotransfected into human 293 cells with the MLV vector MSCV2.1, which carries neo, and MLV integration was scored as the number of G418-resistant colonies. (E) A single DSB does not enhance the trans integration of foreign DNA induced by Metnase. Inducing a DSB by using the restriction enzyme I-SceI did not increase the ability of Metnase to integrate foreign DNA. Metnase was expressed from the pcDNA vector and the hygromycin selectable marker was carried on a separate pRNA-U6 vector.
To determine whether Metnase promotes integration of retroviral DNA, we cotransfected a plasmid containing Moloney leukemia virus DNA and neo with a Metnase expression vector that did not contain a selectable marker. Metnase enhanced retroviral DNA integration by up to 4.1-fold (Fig. 3D). Metnase promotion of the integration of widely varied exogenous DNA sequences indicates that its activity is DNA sequence-independent. This distinguishes Metnase from transposases, which act only on transposon-specific sequences.
Because integration is stimulated by DNA DSB damage (10-12), we next asked whether Metnase promotion of trans integration is further enhanced by a defined DSB. The pJ6-Metnase expression vector and a linearized plasmid that confers resistance to hygromycin were cotransfected into HT256 cells, which contain a single I-SceI site in a neo direct repeat HR substrate (30). These transfections were carried out with or without an I-SceI expression vector. I-SceI may repeatedly cleave a precisely rejoined I-SceI site until it is destroyed by HR, NHEJ, or targeted integration of the plasmid into the I-SceI site. This plasmid may also integrate elsewhere, and both targeted and illegitimate integration events yield hygromycin-resistant transfectants. As expected, I-SceI-induced DSBs or Metnase overexpression alone enhanced integration (Fig. 3E). Metnase-enhanced integration was further stimulated by DSBs, although the degree to which Metnase enhanced integration was the same with or without DSB induction. Interestingly, PCR analysis revealed that with Metnase overexpression, 6 of 33 colonies tested had the hygromycin resistance plasmid inserted into the I-SceI site; without Metnase overexpression, 0 of 11 colonies tested had inserted the resistance plasmid into the I-SceI site. Sequencing the I-SceI sites in which the hygromycin plasmid had been inserted revealed that in five of those six clones, the plasmid was inserted into the sequence 5′-CTGTAT/CCCTA, demonstrating that the I-SceI site remained intact, without exonuclease end-processing, when Metnase was overexpressed. This indicates that Metnase promotes precise end joining of exogenous DNA during integration into a chromosomal DSB.
The results above indicate that overexpression of Metnase promotes integration but do not address whether Metnase is required for integration. We tested this by decreasing the expression of Metnase with siRNA. When Metnase expression was reduced by stable expression of siRNA from the pRNA-U6 vector, integration of a plasmid conferring puromycin-resistance (pBOS) or Moloney leukemia virus DNA (MSCV2.1) decreased by up to 4- and 5.7-fold, respectively (Fig. 4). Thus, Metnase plays an important role in exogenous DNA integration.
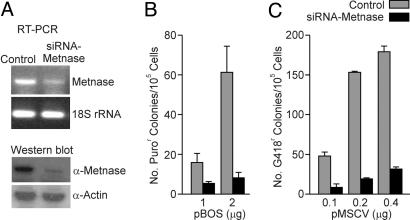
Metnase mediates foreign DNA integration. (A) siRNA was used to reduce the RNA and protein expression of Metnase. (B and C) Integration of plasmid (pBOS) and Moloney Leukemia Virus DNA (pMSCV) in control and Metnase knockdown cells.
It was possible that the increase in integration stimulated by Metnase was due to an increase in transfection efficiencies stimulated by Metnase overexpression. However, transfecting a β-galactosidase expression vector into control and cell lines over- or underexpressing Metnase did not result in statistically different β-galactosidase expression when normalized for protein concentration (not shown).
Metnase Enhances NHEJ and Resistance to Ionizing Radiation. Because Metnase promoted integration of DNA into a DSB that could be repaired by HR or NHEJ, we examined the effect of Metnase expression on DSB repair by these pathways. We transfected HT256 cells with the I-SceI expression vector and with a Metnase expression vector or empty vector control and selected G418-resistant HR products (30). Metnase overexpression did not produce any significant changes in HR repair in HT256 cells (data not shown). We then tested whether Metnase promotes precise and/or imprecise NHEJ of plasmid DNA. NHEJ was examined by testing the ability of Metnase to precisely or imprecisely rejoin a plasmid DNA that was linearized within the β-galactosidase gene and transfected into human 293 cells (31, 32). Overexpressing Metnase increased precise NHEJ by 2-fold and imprecise NHEJ by 2.7-fold (Fig. 5A). In addition, PCR assessment of the presence of rejoined plasmid in the control versus Metnase overexpressing cells revealed a significantly higher amount of rejoined plasmid with increased Metnase expression (Fig. 5A). To determine whether NHEJ requires Metnase, we repeated the plasmid NHEJ assay in cells in which Metnase levels were reduced by siRNA and found that imprecise NHEJ was reduced by 12-fold and precise NHEJ by 20-fold (Fig. 5A). Together, these results are consistent with the precise insertion of the selectable marker into the I-SceI site with Metnase expression.
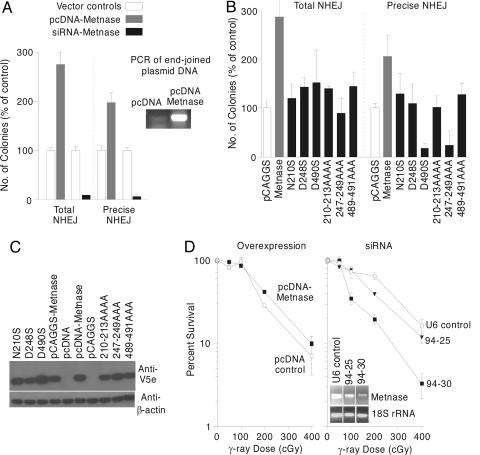
Metnase enhances NHEJ. (A) NHEJ was measured by the ability of a linearized vector to have its free ends rejoined and transform bacteria to ampicillin resistance. Precise end joining was measured by the ability of the pBluescript plasmid linearized within a β-galactosidase gene to make functional β-galactosidase protein when rescued from human 293 cells with stably increased or decreased expression of Metnase. PCR analysis by using T7/SP6 primers of standardized concentrations of rescued plasmid showed an increased amount of end-joined plasmid when Metnase is overexpressed. (B) Mutations of residues known to be required for histone methylase or transposase function in Metnase reduced the ability of Metnase to stimulate NHEJ. (C) Western analysis shows that the Metnase mutants are all expressed well at the protein level. (D) Cell survival after γ-radiation in cells over- or underexpressing Metnase.
We then assessed whether mutating required amino acids in the SET domain or the transposase domain, which would abrogate histone methylase or transposase function, would decrease NHEJ. Three point mutants were constructed, N210S and D248S, which alters known conserved essential SET domain amino acids, and D490S, which alters a known essential conserved amino acid in the Transposase domain (32-36). Three mutants were also generated that altered three or four amino acids in these same regions that harbored these essential residues: the NHSC at 210-213 was altered to AAAA, the YDY at 247-249 to AAA, and the YDN at 489-491 to AAA. Each of these six mutations showed no increase in precise and total NHEJ as compared with vector controls. Indeed, the D490S and YDY247AAA mutants have dominant negative activity, repressing end-joining repair below the level of endogenous Metnase. Thus, mutations in amino acids known to be required for either histone methylase function or transposase function significantly reduced the activity of Metnase in DSB repair.
Because NHEJ is the major determinant of radiation resistance in mammalian cells (39-41), we tested the effects of Metnase overexpression on cell survival after exposure to ionizing radiation. Metnase overexpression resulted in a 2-fold survival advantage compared with vector controls (Fig. 5B), whereas Metnase underexpression using siRNA resulted in up to an 8-fold decrease in survival compared with vector controls, providing further evidence of a linkage between Metnase and NHEJ.
Discussion
Metnase has detectable histone methyltransferase activity and therefore may regulate chromatin configuration (34, 35). Metnase transfers methyl groups to recombinant human histone H3, mainly to K36 but also to K4. Such dimethylation of H3 K36 is associated with open chromatin (35, 38, 42, 43). This would be important for integration of foreign DNA into the host genome by enhancing access of the foreign DNA to host DNA free ends. Although there are conflicting reports, methylation of H3 K4 is also associated with transcription, which implies an open chromatin configuration (44). Thus, one mechanism by which Metnase enhances integration may be to improve access of foreign DNA to host DNA by opening chromatin.
Metnase plays a critical role in illegitimate integration of DNA into host genomic DNA. Overexpression of Metnase enhances foreign DNA integration, and reduced expression of Metnase inhibits integration. Metnase also enhances DSB end joining and improves survival after radiation exposure, implying that it may be a component of the NHEJ repair system (39-41). It is likely that Metnase stimulation of NHEJ accounts for its ability to promote survival after radiation. The mechanism by which Metnase enhances NHEJ is not clear. The ability of Metnase to enhance precise NHEJ may reflect a direct role in repair through an interaction with one or more NHEJ proteins. This may be limited, for example to an interaction with DNA ligase IV, which would account for why cells lacking other NHEJ components such as DNA PKcs or Ku can still integrate DNA (19-23). Alternatively, Metnase may promote precise NHEJ indirectly. The promotion of integration by Metnase and its histone methylase activity suggests that Metnase may open chromatin and enhance accessibility of repair factors to integration sites, speeding foreign DNA integration and thereby reducing the time for end degradation of foreign DNA before integration. Supporting this hypothesis is the finding that mutating essential amino acid residues in the histone methylase domain of Metnase reduced the ability of Metnase to stimulate NHEJ repair.
It was recently shown that hepatitis B virus integration is enhanced at sites of chromosomal DSBs induced by I-SceI, and that these events are mediated by NHEJ (45). We found that Metnase enhanced precise integration of exogenous DNA into a defined chromosomal DSB and significantly enhanced precise NHEJ of a plasmid substrate. Based on these observations, we propose that Metnase functions by opening chromatin and facilitating conservative end joining between foreign and genomic DNA. The transposase domain in Metnase suggests additional roles, specifically implying a nuclease activity. However, the enhancement of precise end joining by Metnase makes it unlikely that it has exonuclease activity but does not exclude double-strand endonuclease activity. Thus, Metnase may have intrinsic DNA modifying functions in addition to its chromatin-modifying ability.
It is imperative to develop a better understanding of foreign DNA integration. One of the most compelling reasons is the tragic occurrence of T cell leukemias in children who have had severe combined immunodeficiency successfully corrected by retroviral gene therapy (46). Two of these leukemias occurred because of vector insertion into the LMO2 oncogene site, stimulating host cell proliferation. Although gene therapy for inherited diseases is a rare and expensive undertaking, a far more common problem is the HIV pandemic, where genomic insertion in quiescent node lymphocytes underlies not just the initial infection but also viral escape from multidrug antiretroviral therapy (47). If the integration of HIV could be disrupted, perhaps by targeting Metnase, it would be far more difficult for HIV to propagate. In addition, there are several significant oncologic implications of our findings. It is possible that the frequent neoplastic chromosomal alterations seen at 3p26 may alter Metnase's expression or function. For example, defects in Metnase could be an underlying first step toward aberrant chromosomal events that lead to neoplasia; if Metnase enhances precise end joining, then its malfunction may contribute to nonhomologous chromosomal ligation. Alternatively, its overexpression in malignancy could result in resistance to therapeutics that induce DSBs. Metnase overexpression could be selected for in tumors treated with radiation or chemotherapy, providing increased resistance to these agents and allowing these cells to overgrow and repopulate the tumor.
Recently, a plant transposase termed DAYSLEEPER was found to be essential for proper growth, although the mechanism of its action was not elucidated (48). This study demonstrated that a eukaryotic transposase is essential for a normal physiologic cellular function outside of transposition of genetic elements. Metnase is the second example of such a eukaryotic transposase that has a normal cellular function. Finally, Metnase could also be mutated in one of the uncharacterized combined immunodeficiency syndromes. An example of a recently identified NHEJ component mutated in severe combined immunodeficiency is Artemis (49). Artemis appears to be an essential component of Ig and probably T cell receptor rearrangements. One could also envision similar roles for Metnase in physiologic DNA rearrangements.
In conclusion, the ability of Metnase to methylate histone H3, improve NHEJ, and enhance foreign DNA integration places it at a key juncture in several critical aspects of chromosome dynamics. Metnase may ultimately prove to link these processes temporally and spatially in a coordinated fashion to enhance the normal repair and integration of DNA segments within a cell.
Acknowledgments
This research was supported by National Institutes of Health Grants CA102283 and HL075783 (to R.H.), CA100862 (to J.A.N.), and NIH CA92111 (to S.-H.L.)
Notes
Author contributions: J.A.N. and R.H. designed research; S.-H.L., M.O., S.T.D., K.K.R., E.A.W., H.R., L.K., J.A.N., and R.H. performed research; S.-H.L., M.O., and J.A.N. contributed new reagents/analytic tools; J.A.N., S.-H.L., and R.H. analyzed data; and J.A.N. and R.H. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY952295).
Abbreviations: HR, homologous recombination; NHEJ, nonhomologous end joining; DSB, double-strand break; SAM, 3H-S-adenosyl methionine; siRNA, small interfering RNA.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0503676102
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1312370?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Article citations
Epistasis between mutator alleles contributes to germline mutation spectrum variability in laboratory mice.
Elife, 12:RP89096, 21 Feb 2024
Cited by: 3 articles | PMID: 38381482 | PMCID: PMC10942616
DNA Damage Responses, the Trump Card of Stem Cells in the Survival Game.
Adv Exp Med Biol, 1470:165-188, 01 Jan 2024
Cited by: 1 article | PMID: 37923882
Review
Leveraging dominant-negative histone H3 K-to-M mutations to study chromatin during differentiation and development.
Development, 150(21):dev202169, 17 Oct 2023
Cited by: 2 articles | PMID: 37846748 | PMCID: PMC10617616
Review Free full text in Europe PMC
Chromosome-Wide Distribution and Characterization of H3K36me3 and H3K27Ac in the Marine Model Diatom Phaeodactylum tricornutum.
Plants (Basel), 12(15):2852, 02 Aug 2023
Cited by: 1 article | PMID: 37571007 | PMCID: PMC10421102
Role of Transposable Elements in Genome Stability: Implications for Health and Disease.
Int J Mol Sci, 23(14):7802, 15 Jul 2022
Cited by: 10 articles | PMID: 35887150 | PMCID: PMC9319628
Review Free full text in Europe PMC
Go to all (102) article citations
Other citations
Wikipedia
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (2)
- (2 citations) ENA - AY952295
- (1 citation) ENA - U80776
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Human Pso4 is a metnase (SETMAR)-binding partner that regulates metnase function in DNA repair.
J Biol Chem, 283(14):9023-9030, 08 Feb 2008
Cited by: 54 articles | PMID: 18263876 | PMCID: PMC2431028
Metnase/SETMAR: a domesticated primate transposase that enhances DNA repair, replication, and decatenation.
Genetica, 138(5):559-566, 23 Mar 2010
Cited by: 34 articles | PMID: 20309721 | PMCID: PMC2847698
Review Free full text in Europe PMC
The human set and transposase domain protein Metnase interacts with DNA Ligase IV and enhances the efficiency and accuracy of non-homologous end-joining.
DNA Repair (Amst), 7(12):1927-1937, 18 Sep 2008
Cited by: 35 articles | PMID: 18773976 | PMCID: PMC2644637
Expression levels of the human DNA repair protein metnase influence lentiviral genomic integration.
Biochimie, 90(9):1422-1426, 23 May 2008
Cited by: 14 articles | PMID: 18549821 | PMCID: PMC2556226
Funding
Funders who supported this work.
NCI NIH HHS (6)
Grant ID: CA92111
Grant ID: R01 CA100862
Grant ID: R01 CA102283
Grant ID: CA102283
Grant ID: CA100862
Grant ID: R01 CA092111
NHLBI NIH HHS (2)
Grant ID: R01 HL075783
Grant ID: HL075783





