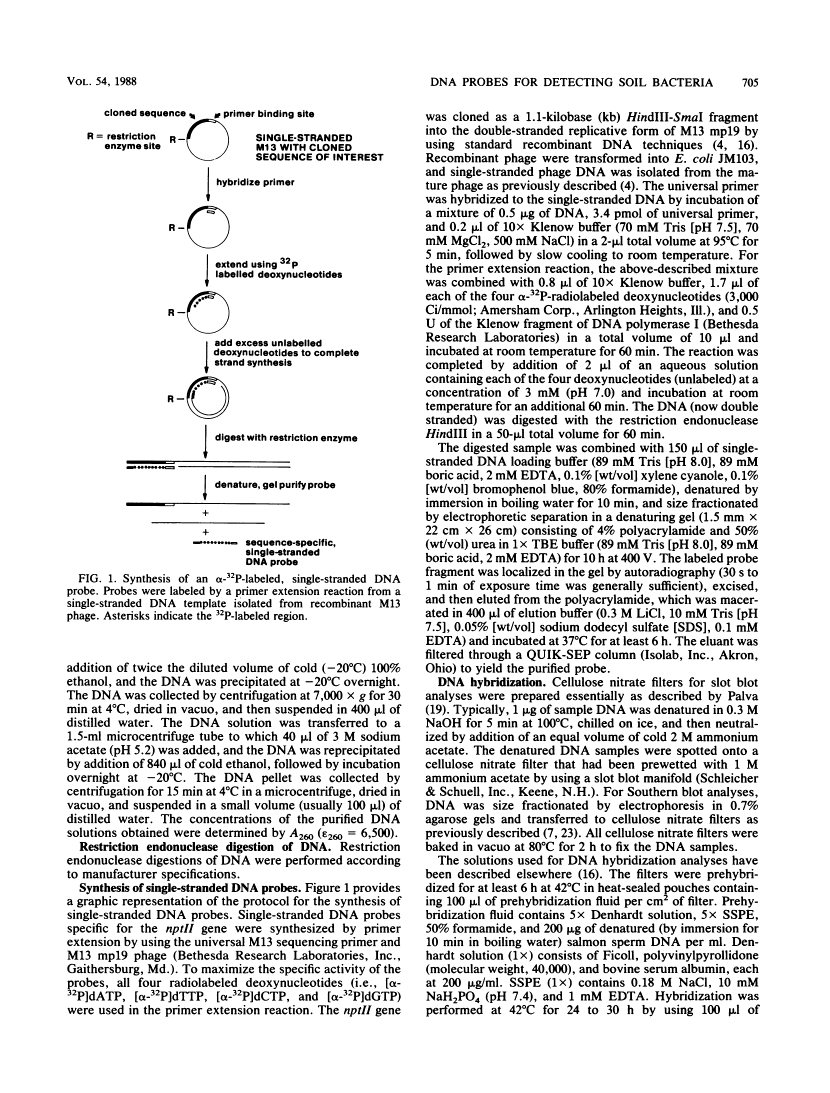
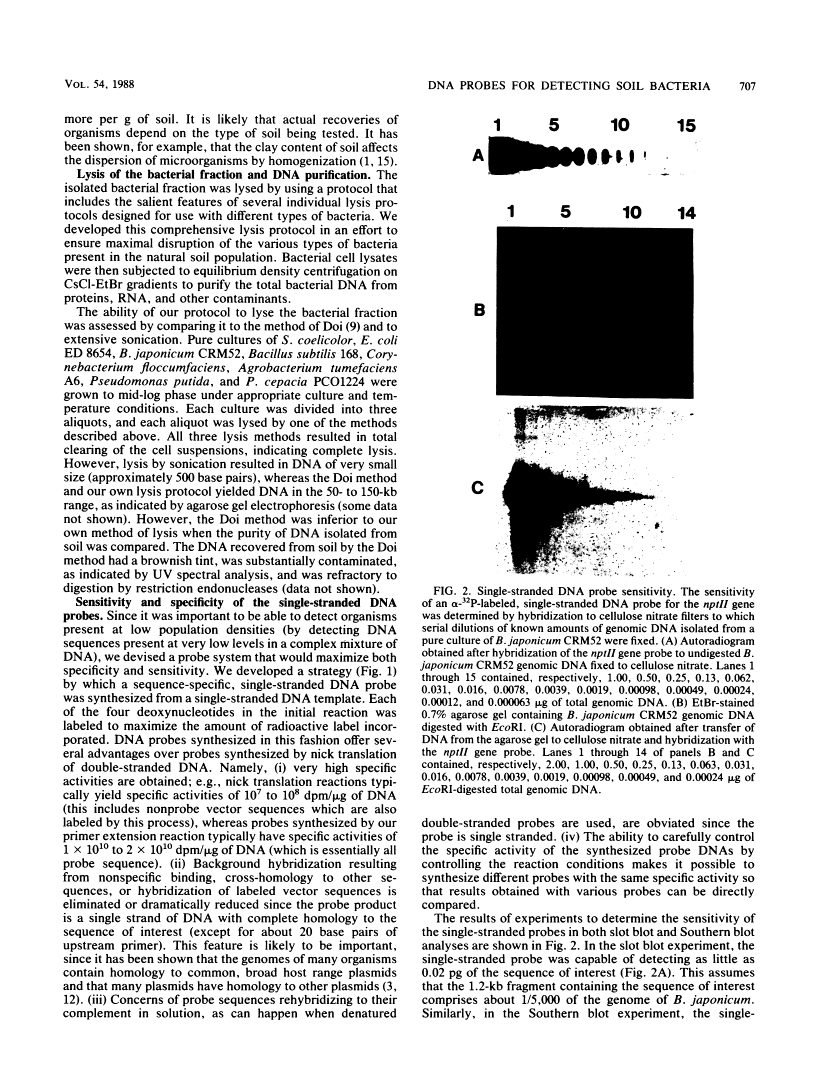
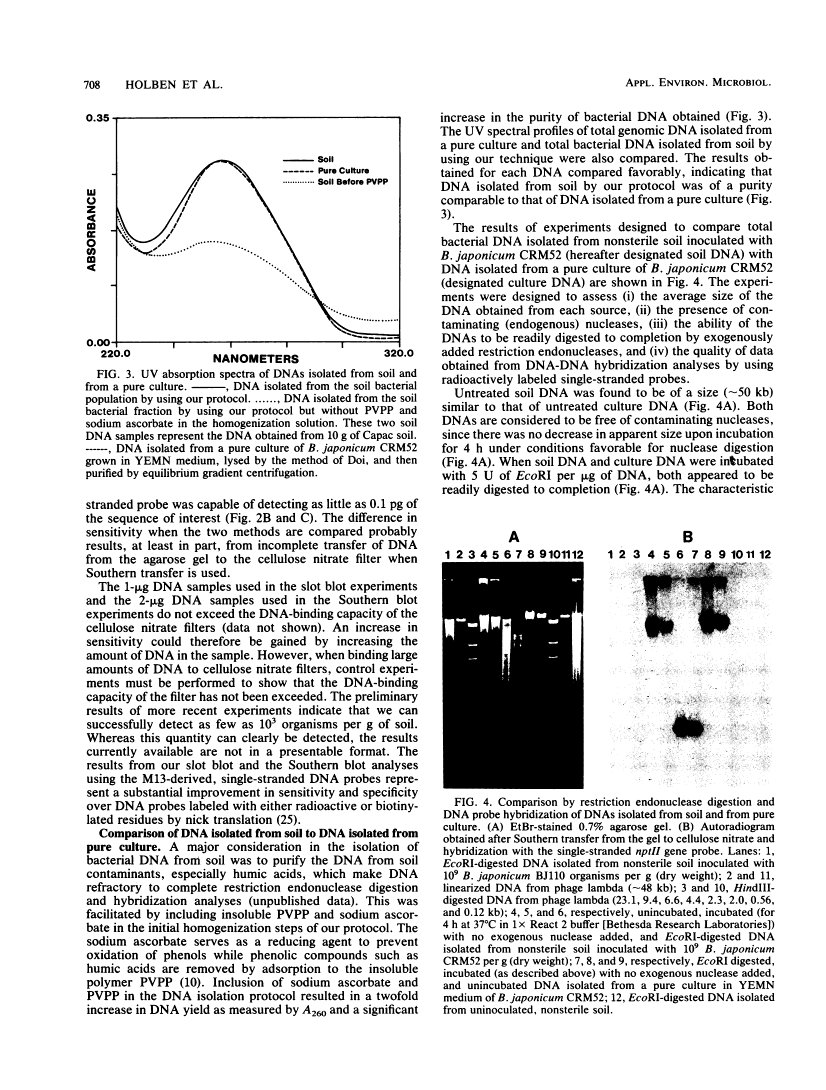
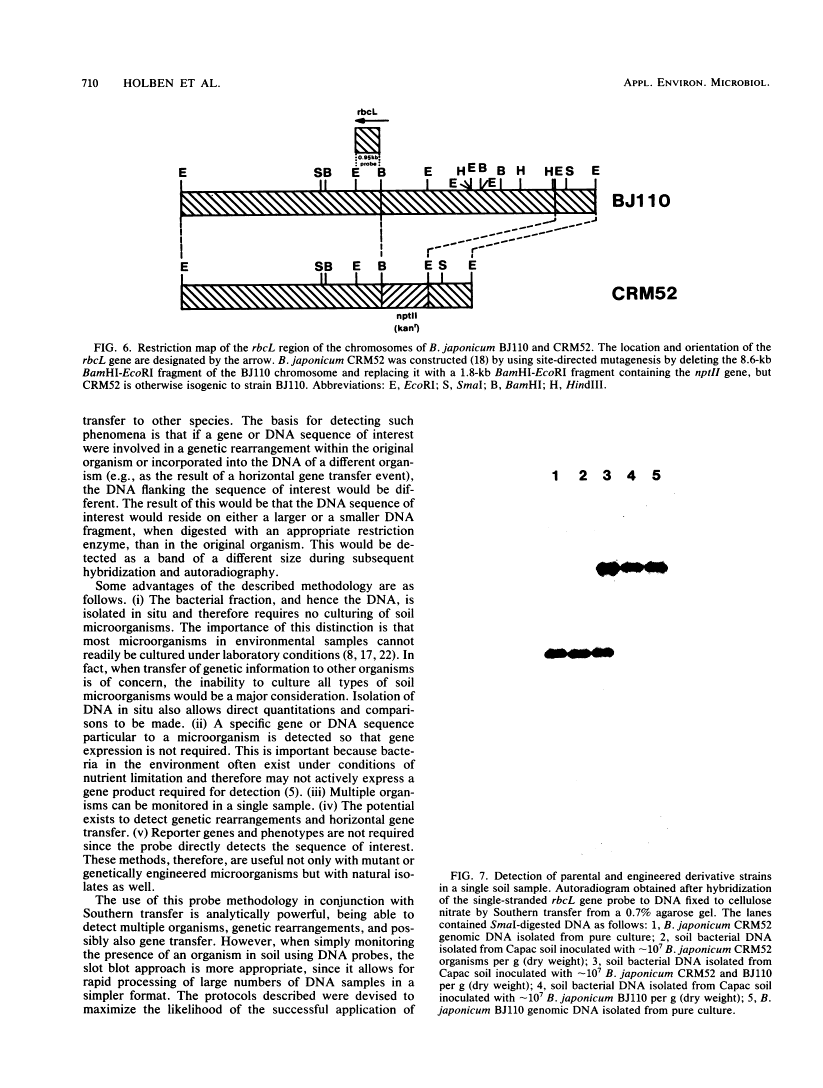
Abstract
Free full text

DNA Probe Method for the Detection of Specific Microorganisms in the Soil Bacterial Community †
Abstract
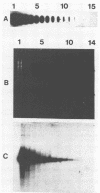
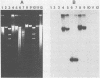
We developed a protocol which yields purified bacterial DNA from the soil bacterial community. The bacteria were first dispersed and separated from soil particles in the presence of polyvinylpolypyrrolidone, which removes humic acid contaminants by adsorption to this insoluble polymer. The soil bacteria were then collected by centrifugation and lysed by using a comprehensive protocol designed to maximize disruption of the various types of bacteria present. Total bacterial DNA was purified from the cell lysate and remaining soil contaminants by using equilibrium density gradients. The isolated DNA was essentially pure as determined by UV spectral analysis, was at least 48 kilobases long, and was not subject to degradation, which indicated that there was no contaminating nuclease activity. The isolated DNA was readily digested by exogenously added restriction endonucleases and successfully analyzed by slot blot and Southern blot hybridizations. Using single-stranded, 32P-labeled DNA probes, we could detect and quantitate the presence of a specific microbial population in the natural soil community on the basis of the presence of a DNA sequence unique to that organism. The sensitivity of our methodology was sufficient to detect Bradyrhizobium japonicum at densities as low as 4.3 × 104 cells per g (dry weight) of soil, which corresponds to about 0.2 pg of hybridizable DNA in a 1-μg DNA sample.
Full text
Full text is available as a scanned copy of the original print version. Get a printable copy (PDF file) of the complete article (2.0M), or click on a page image below to browse page by page. Links to PubMed are also available for Selected References.
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakken LR. Separation and purification of bacteria from soil. Appl Environ Microbiol. 1985 Jun;49(6):1482–1487. [Europe PMC free article] [Abstract] [Google Scholar]
- Balkwill DL, Labeda DP, Casida LE., Jr Simplified procedures for releasing and concentrating microorganisms from soil for transmission electron microscopy viewing as thin-sectioned and frozen-etched preparations. Can J Microbiol. 1975 Mar;21(3):252–262. [Abstract] [Google Scholar]
- Bender CL, Cooksey DA. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J Bacteriol. 1986 Feb;165(2):534–541. [Europe PMC free article] [Abstract] [Google Scholar]
- Bissonnette GK, Jezeski JJ, McFeters GA, Stuart DG. Influence of environmental stress on enumeration of indicator bacteria from natural waters. Appl Microbiol. 1975 Feb;29(2):186–194. [Europe PMC free article] [Abstract] [Google Scholar]
- Bone TL, Balkwill DL. Improved flotation technique for microscopy of in situ soil and sediment microorganisms. Appl Environ Microbiol. 1986 Mar;51(3):462–468. [Europe PMC free article] [Abstract] [Google Scholar]
- Botchan M, Topp W, Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. [Abstract] [Google Scholar]
- Evans HJ, Koch B, Klucas R. Preparation of nitrogenase from nodules and separation into components. Methods Enzymol. 1972;24:470–476. [Abstract] [Google Scholar]
- Golub EI, Low KB. Unrelated conjugative plasmids have sequences which are homologous to the leading region of the F factor. J Bacteriol. 1986 May;166(2):670–672. [Europe PMC free article] [Abstract] [Google Scholar]
- Hanson CW, Martin WJ. Microwave oven for melting laboratory media. J Clin Microbiol. 1978 Apr;7(4):401–402. [Europe PMC free article] [Abstract] [Google Scholar]
- McClung CR, Chelm BK. A genetic locus essential for formate-dependent growth of Bradyrhizobium japonicum. J Bacteriol. 1987 Jul;169(7):3260–3267. [Europe PMC free article] [Abstract] [Google Scholar]
- Roszak DB, Grimes DJ, Colwell RR. Viable but nonrecoverable stage of Salmonella enteritidis in aquatic systems. Can J Microbiol. 1984 Mar;30(3):334–338. [Abstract] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. [Abstract] [Google Scholar]
Associated Data
Articles from Applied and Environmental Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/aem.54.3.703-711.1988
Read article for free, from open access legal sources, via Unpaywall:
https://aem.asm.org/content/aem/54/3/703.full.pdf
Free to read at aem.asm.org
http://aem.asm.org/cgi/content/abstract/54/3/703
Free after 4 months at aem.asm.org
http://aem.asm.org/cgi/reprint/54/3/703
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/aem.54.3.703-711.1988
Article citations
Maximizing efficiency in sedimentary ancient DNA analysis: a novel extract pooling approach.
Sci Rep, 14(1):19388, 20 Aug 2024
Cited by: 0 articles | PMID: 39169089 | PMCID: PMC11339378
Estimating the bias related to DNA recovery from hemp stems for retting microbial community investigation.
Appl Microbiol Biotechnol, 107(14):4665-4681, 25 May 2023
Cited by: 2 articles | PMID: 37227475
Comparing sediment DNA extraction methods for assessing organic enrichment associated with marine aquaculture.
PeerJ, 8:e10231, 27 Oct 2020
Cited by: 11 articles | PMID: 33194417 | PMCID: PMC7597629
Extracellular DNA (eDNA): Neglected and Potential Sources of Antibiotic Resistant Genes (ARGs) in the Aquatic Environments.
Pathogens, 9(11):E874, 23 Oct 2020
Cited by: 9 articles | PMID: 33114079 | PMCID: PMC7690795
Review Free full text in Europe PMC
Inter-laboratory testing of the effect of DNA blocking reagent G2 on DNA extraction from low-biomass clay samples.
Sci Rep, 8(1):5711, 09 Apr 2018
Cited by: 5 articles | PMID: 29632323 | PMCID: PMC5890260
Go to all (225) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Development and application of a new method to extract bacterial DNA from soil based on separation of bacteria from soil with cation-exchange resin.
Appl Environ Microbiol, 58(8):2458-2462, 01 Aug 1992
Cited by: 73 articles | PMID: 16348750 | PMCID: PMC195803
Analysis of broad-scale differences in microbial community composition of two pristine forest soils.
Syst Appl Microbiol, 21(4):579-587, 01 Dec 1998
Cited by: 33 articles | PMID: 9924826
Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast.
Appl Environ Microbiol, 59(8):2657-2665, 01 Aug 1993
Cited by: 289 articles | PMID: 7690221 | PMCID: PMC182335
Construction and applications of DNA probes for detection of polychlorinated biphenyl-degrading genotypes in toxic organic-contaminated soil environments.
Appl Environ Microbiol, 56(1):254-259, 01 Jan 1990
Cited by: 27 articles | PMID: 2106826 | PMCID: PMC183298