Abstract
Free full text

Detection of Rhinoviruses by Tissue Culture and Two Independent Amplification Techniques, Nucleic Acid Sequence-Based Amplification and Reverse Transcription-PCR, in Children with Acute Respiratory Infections during a Winter Season
Abstract
Five hundred seventeen consecutive nasopharyngeal aspirates were collected between October 1998 and May 1999 for episodes of acute respiratory tract infections in children presenting at the University Hospital of Antwerp. Culture and nucleic acid amplification techniques—nucleic acid sequence-based amplification (NASBA) and reverse transcription-PCR (RT-PCR)—were applied to detect rhinoviruses (RVs). Other respiratory viruses were detected by immunofluorescence (IF) analysis of the specimens and IF analysis of shell vial cultures. Among the 517 specimens, 219 viral agents were identified. They were, in decreasing order, rhinoviruses (93 [18.0%]), respiratory syncytial virus (76 [14.7%]), adenoviruses (16 [3.1%]), influenza viruses (15 [2.9%]), enteroviruses (15 [2.9%]), and herpes simplex virus (4 [0.8%]). For the evaluation of rhinovirus detection, culture positivity and/or a positive reaction in the two independent amplification methods was used as an expanded “gold standard.” Based on this standard, the sensitivity, specificity, positive predictive value, and negative predictive value of culture were 44.7, 100, 100, and 99.8%, and those of NASBA and RT-PCR were 85.1, 98.3, 83.3, and 98.5% and 82.9, 93.4, 55.7, and 98.2%, respectively. NASBA and RT-PCR produced comparable results and were significantly more sensitive than virus culture. RVs showed the highest incidence in acute respiratory tract infections in children.
Rhinoviruses (RVs) are the most frequent cause of acute upper respiratory tract infections in humans and are usually associated with the common cold (7, 16, 29, 31). However, they may also be associated with more-severe lower respiratory tract infections in children, in the elderly (23, 24, 30, 33), and in immunocompromised patients (14, 39, 48, 49). RVs have been isolated from cases of cystic fibrosis (44), otitis media (3), sinusitis (37), and exacerbations of chronic bronchitis and asthma (12, 21, 22, 34).
Detection of rhinoviruses by tissue culture is slow and cumbersome due to the specific culture conditions required, limiting this diagnostic procedure to a few reference laboratories (35). Serologic diagnosis is virtually impossible because of the existence of more than 100 serotypes.
Increasing the sensitivity of the detection of rhinoviruses by nucleic acid amplification techniques (NAATs) would expand the epidemiological knowledge of these viruses and improve the management of patients by avoiding unnecessary use of antibiotics.
Two NAATs are presently available for the detection of RV: nucleic acid sequence-based amplification (NASBA) (26, 42) and reverse transcription-PCR (RT-PCR) (13). Both NASBA and RT-PCR have their advantages. NASBA is easier to perform and produces results more rapidly than RT-PCR.
Studies comparing two different molecular amplification techniques applied to a considerable number of clinical specimens are rare.
Here, the results obtained for the detection of RV in respiratory specimens by the standard tissue culture procedure and by NASBA and RT-PCR during one winter season are presented.
MATERIALS AND METHODS
Patients.
Between 1 October 1998 and 31 May 31 1999, 543 consecutive nasopharyngeal aspirates (NAs) were obtained from 427 children (range, 1 week to 18 years) seen at the outpatient clinic or hospitalized for acute respiratory symptoms at the University Hospital, Antwerp. Some patients were seen and specimens also sampled at a follow-up visit. Five hundred seventeen specimens were available for analysis by all methods.
Specimen collection and handling.
NAs were collected and processed as described earlier (36). In brief, aspirates were collected with a mucus extractor (Vygon, Ecouen, France) and, without addition of transport medium, delivered to the laboratory within 2 to 4 h. The specimens were processed immediately, except on Saturdays and Sundays, when they were kept at 4°C until the next Monday. One portion (100 μl) (517/543 NAs were available for NAAT) was put immediately in lysis buffer (pH 6.2) (26) and stored at −80°C until processed by the Boom nucleic acid extraction method (6). Molecules (1 × 104) of the RV internal control (IC) RNA were added to each extraction tube prior to nucleic acid extraction (27). The second portion was spotted on slides (ICN, Costa Mesa, CA) for detection by indirect immunofluorescence with monoclonal antibodies of respiratory syncytial virus (RSV), influenza A virus, and influenza B virus (Argene Biosoft Labconsult, Brussels, Belgium).
The third portion was mixed with an equal volume of minimum essential medium containing gentamicin, 50 μg/ml (Gibco, Paisley, United Kingdom), and vortexed with glass beads for 1 min; 200 μl was inoculated into respective roller tubes of MRC-5 cells in REGA medium (Gibco) maintained at 33°C for the detection of picornaviruses or into shell vial cultures of appropriate cell lines (MDCK and Vero cells) for the detection of influenza virus, parainfluenza virus, herpes simplex virus (HSV), adenovirus, and respiratory syncytial virus by immunofluorescence after 48 h of incubation at 36°C. The roller tubes were examined daily for the appearance of a cytopathic effect, and RV was identified by the nucleic acid being RNA, by using 5-iodo-2-deoxyuridine, and by ether resistance and sensitivity to pH 2.
Production of NASBA RV IC RNA.
The cDNA of the 5′ noncoding region (NCR) RNA of RV 15 (ATCC VR1125) was produced by an RT-PCR using adapted NASBA primers, containing an EcoRI site or a Csp45I site, and inserted into a pGEM plasmid. It contains a sequence hybridizing with the enhanced chemiluminescence (ECL) detection probe.
For the production of the IC, the segment of the cloned RV 15 5′ NCR sequence hybridizing with the ECL detection probe was modified by insertion of a nontarget sequence hybridizing with a specific ECL IC detection probe and inserted in a second pGEM plasmid. The plasmid for the production of the IC was transformed in Escherichia coli DH5α. Large-scale runoff transcripts after linearization with BamHI (Pharmacia Biotech, Roosendaal, The Netherlands) were prepared.
In vitro IC RNA was generated from the construct with T7 RNA polymerase (Pharmacia Biotech, Roosendaal, The Netherlands) as described by Sambrook etal. (41). Plasmid DNA was removed by treatment with DNase I (Pharmacia Biotech). The RNA was quantified spectrophotometrically. The material was stored in lysis buffer at −80°C.
NASBA primers and probes.
The primers and probes were described previously (26). One primer mix composed of EG 59 and pd 906 amplified group B RVs, and the amplicons were detected by ECL with the probe combination pd 910, pd 908, and pd 1147. The second primer mix consisted of pd 905, pd 907, pd 1274, and pd 1275 for the amplification of group A RVs to be detected by the probe combination pd 909, pd 911, pd 1276, and pd 1277. In negative control reactions, target nucleic acid was replaced by RNase-free/DNase-free H2O. Complete or partial NASBA inhibition was reflected by a total absence of or a decreased ECL detection signal of the IC.
RT-PCR.
Five microliters of the nucleic acid extracts were used in a 50-μl single-tube RT-PCR with the Access RT-PCR system (Promega, Belgium) (26) using primers OL-26 and OL-27 (13). After the RT reaction at 48°C for 45 min, the 5′ NCR was amplified by 1 cycle at 94°C for 2 min and 40 cycles at 94°C for 30 s, 60°C for 1 min, and 68°C for 2 min each and 1 cycle at 68°C for 7 min. Amplicons were detected by agarose electrophoresis.
Sequence analysis of RV isolates.
To classify RT-PCR-positive, NASBA-negative specimens as rhino- or enteroviruses, the 5′ NCRs of 38 strains displaying a band at 380 bp on agarose gel that were RT-PCR positive, NASBA negative and of 9 strains with a molecular size larger than 380 bp as shown on agarose gel were sequenced, as were those of the strains of 11 follow-up specimens taken from children during the same or a second hospital admission period and of 7 strains of culture-proven enteroviruses. Since RVs have an approximately 750-nucleotide 5′ noncoding region with a particular pattern of sequence variation of short stretches of generally 1 to 6 relatively well-conserved nucleotides separated by stretches (not always similar in size) of highly variable residues (1), it was decided to sequence the amplicons of the 9 strains with a molecular size larger than 380 bp. The sequencing using primer OL-26 was done by Eurogentec (Eurogentec, Herstal, Belgium). In cases where a positive result was obtained by NAAT, NASBA, and RT-PCR and if the RT-PCR amplicon had a molecular size of 380 bp, the result was considered to be true RV positive and no sequencing was done.
The sequence data were compared to those in GenBank or to data from “in-house” sequencing of RV amplicons. The entries of GenBank used were AF162711, S79276, S76768, L76398, and AJ133661 for the alignment of echovirus 30 sequences, U11707 and X89532 for echovirus 2 sequences, AF188359, X87328, X89535, AF188358, and AF083069 for echovirus 5 sequences, and X00595.1 for poliovirus 2, respectively.
Analysis of results.
In the absence of a reference method or “gold standard,” an expanded gold standard was applied: a specimen was considered positive if culture positive or if a positive result was obtained with both NASBA and RT-PCR.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences reported here are DQ316262 to DQ316315.
RESULTS
Sensitivity of the detection methods.
Among the 517 clinical specimens, 27 (5.2%) were completely inhibited in the NASBA and 40 (7.7%) were partially inhibited, as revealed by a total absence of or a decreased ECL detection signal of the IC, respectively. After a 1/5 dilution of the nucleic acid extracts, 10 (1.9%) samples were completely and 18 (3.5%) were partially inhibited. Since no IC was used in the RT-PCR, inhibition was not detected, and if the sample showed inhibition in the NASBA analysis, RT-PCR, if negative, was repeated on 1/5 and 1/10 dilutions.
A total of 517 NAs were studied. Culture detected 21 RV, and NASBA and RT-PCR detected 49 and 67, respectively. Based on the expanded gold standard, sensitivity, specificity, positive predictive value, and negative predictive value of culture were 44.7, 100, 100, and 99.8% and those of NASBA and RT-PCR were 85.1, 98.3, 83.3, and 98.5% and 82.9, 93.4, 55.7, and 98.2%, respectively (Table (Table1).1). All RVs found by NASBA belonged to group B as described by Andries (2).
TABLE 1.
Sensitivities, specificities, and positive and negative predictive values for three different rhinovirus detection proceduresa
| Detection method | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|
| Conventional culture | 44.7 | 100.0 | 100.0 | 94.8 |
| NASBA on NAb | 85.1 | 98.3 | 83.3 | 98.5 |
| RT-PCR on NA | 82.9 | 93.4 | 55.7 | 98.2 |
Sequence analysis.
Thirty-eight amplicon samples, displaying a band at 380 bp after RT-PCR only, were sequenced. Three showed a similarity with group A RVs, 24 with group B RVs, and 7 with echovirus 2, and 4 could not be aligned (Fig. (Fig.1).1). The sequences of these amplicons contained mutations causing mismatches between the wild-type 5′ NCR region and the primers and probes used (Fig. (Fig.1),1), affecting the efficiency of the NASBA amplification and detection assay.

RV 5′ NCR subregion sequences of NASBA-negative specimens. Superscript letters: a, group A RV; b, RV (MW > 380 bp). P1 and P2 are 5′ NCR subregions chosen for amplification by the NASBA primers. Biotin probe and ECL-probe sequences are 5′ NCR subregions chosen for amplicon hybridization with the NASBA ECL probes.
Additionally, the comparison of sequence data obtained with the amplicons of nine specimens with a molecular size larger than 380 bp revealed sequence similarity with RV group B in three cases (98108245, 99038325, and 99048153 [Fig. [Fig.1]),1]), and two were untypable RV. The sequences of four amplicons with a larger molecular size could not be determined.
Results during follow-up of patients.
Three of 10 patients whose first specimen was RV positive and who were seen 4 to 9 days later, still being ill, were RV positive. In five patients whose first specimen was RV negative, a second specimen collected 1 to 3 days later was RV positive.
The sequences of the amplicons of 11 follow-up specimens collected during the same or a second admission period were compared. No differences in nucleotide sequences were seen in amplicons from the same patient obtained during one admission period. Sequence similarities of >82% were noted in amplicons from the same patient obtained during different admission periods, while the amplicons of one patient, who was RV positive during 51 days, showed 100% sequence similarity of the isolates during two successive hospital admissions.
Epidemiology.
Between October 1998 and May 1999, a mean of 68 samples was collected monthly with a considerable peak in November and December (204 samples) caused by RSV (Fig. (Fig.22).
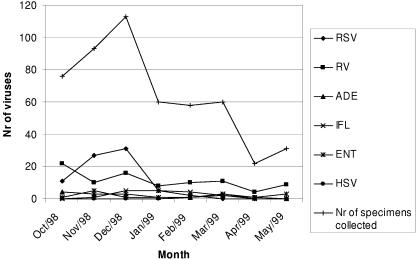
Temporal distribution of respiratory virus (RSV, influenza virus [IFL], adenovirus [ADE], HSV, enterovirus [ENT], and RV) cases by month between 1 October 1998 and 31 May 1999. Nr, number.
Among the 517 specimens studied, 219 viral agents were identified. They were, in decreasing order, RV, 93 specimens (18.0%); RSV, 76 specimens (14.7%); adenovirus, 16 specimens (3.1%); influenza virus, 15 specimens (2.9%); entero-virus, 15 specimens (2.9%); and HSV, 4 specimens (0.8%). There were no parainfluenza virus infections detected during this study period.
RSV was detected in 70 specimens using immunofluorescence on the specimen, and 6 additional specimens were positive after shell vial culture in combination with immunofluorescence. The detection of influenza virus yielded nine positive specimens by immunofluorescence and six additional positive specimens through the combination of shell vial culture and immunofluorescence.
Fifteen enteroviruses were detected, 7 by conventional viral culture and 8 by RT-PCR, including, as determined by sequence analysis, 7 echoviruses of serotype 2, 3 echoviruses of serotype 30, 1 echovirus of serotype 5, 1 echovirus of serotype 3, and 1 poliovirus 2 (strain Sabin), and 2 isolates could not be sequenced at all: one because of a mixed viral infection, and in the other case, the sequence data were not interpretable.
The distribution of the respiratory agents by age is presented in Table Table2.2. During the period studied, the global incidence of RV in the pediatric population did not differ considerably among the age groups, whereas RSV, enteroviruses, and adenoviruses were more prevalent in very young children and influenza viruses in children older than 10 years.
TABLE 2.
Etiologic agents in nasopharyngeal aspirates by age of patient
| Age (yr) of patient | No. (%) of respiratory virusesf
| Total no. of NAg | ||||||
|---|---|---|---|---|---|---|---|---|
| RSV | RV | ADE | IFL | ENT | HSV | Negative | ||
| <1a | 39 (21.2) | 34 (18.5) | 6 (3.3) | 3 (1.6) | 9 (4.9) | 1 (0.5) | 101 (54.9) | 184 |
| 1-2b | 15 (16.0) | 15 (16.0) | 6 (6.4) | 2 (2.1) | 3 (3.2) | 0 (0) | 56 (59.6) | 94 |
| 2-3c | 10 (14.9) | 12 (17.9) | 2 (3.0) | 3 (4.5) | 0 (0) | 0 (0) | 41 (61.2) | 67 |
| 3-4d | 7 (12.7) | 14 (25.5) | 2 (3.6) | 2 (3.6) | 1 (1.8) | 2 (3.6) | 28 (50.9) | 55 |
| 4-10e | 4 (4.8) | 13 (15.5) | 0 (0) | 3 (3.6) | 2 (2.4) | 0 (0) | 62 (73.8) | 84 |
| 10-15 | 1 (4.0) | 4 (16.0) | 0 (0) | 2 (8.0) | 0 (0) | 1 (4.0) | 17 (68.0) | 25 |
| 15-18 | 0 (0) | 1 (12.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (87.5) | 8 |
    Total Total | 76 (14.7) | 93 (18.0) | 16 (3.1) | 15 (2.9) | 15 (2.9) | 4 (0.8) | 329 (63.6) | 517 |
Mixed virus infections were detected in 12/517 (2.3%) NAs; 10 of these were revealed by NASBA and RT-PCR. Four different viral combinations were found, with the combination of an RV with RSV (8/12) being the most frequent, followed by RV and adenovirus (2/12), RV and HSV (1/12), and RSV and echovirus 30 (1/12). A mixed viral infection was found in 9/184 (4.9%) children younger than 12 months and in 1.1 to 1.8% of children between 13 and 48 months (Table (Table22).
DISCUSSION
Using NAAT, rhinoviruses were detected in 18.0% of the nasopharyngeal aspirates, twice as many as were obtained by the culture technique. During this study period, RVs (18.0%) were also the most prevalent, followed by RSV (14.7%), adenoviruses (3.1%), influenza viruses (2.9%), enteroviruses (2.9%), and HSV (0.8%). The figures are comparable to those found in other studies. In those studies, however, the role of RVs may have been underestimated as a result of the exclusive use of the insensitive tissue culture technique (20, 28). Furthermore, until the number of organisms present in clinical specimens of diseased individuals is known, it is impossible to state whether the RVs detected in this study are the true causes of the respiratory tract infections.
There are very few studies on the comparison of different NAAT protocols and/or comparison of NAAT and culture in respiratory specimens for the detection of RVs, and in some studies different extraction methods were applied in the two NAAT assays. In the present study, the same nucleic acid extracts were used for NASBA and RT-PCR amplification, eliminating one variable factor. NASBA and RT-PCR produced comparable results and were almost twice as sensitive as virus isolation. However, caution should be taken with the interpretation of the RT-PCR results. After sequencing of the NASBA-negative RT-PCR-positive amplification products, some of these turned out to be echoviruses.
Two culture-positive, RT-PCR-positive specimens were negative by NASBA. Similar observations have been reported by others (19). The presence of mismatches between the target RNA and the primers and probes causes an inefficient amplification and detection, as reported previously (8, 9, 15, 26, 27). Gobbers et al. (15) reported that single point mutations near the 3′ end of the primers, closely followed by a second mismatch, seem to hamper NASBA amplification. Christopherson et al. (8) and Coste et al. (9) mentioned that the presence of more than four point mutations in a primer decreases the amplification efficiency of an RT-PCR and that a number of mismatches in combination with one or two critical mismatches results in a complete absence of amplification.
The major difficulties in the interpretation of the NAAT data are the possible false-positive and false-negative results, the former due to contamination and the latter resulting from inhibitors present in the respiratory specimens. Internal controls are indispensable for the reliable detection of microorganisms in clinical specimens by NAATs. In the present study, false-negative results were observed due to complete inhibition in 5.1% of the specimens, and partial inhibition was observed in 7.8% of the specimens. After dilution of the nucleic acid extracts, the number of samples showing inhibition decreased considerably, as also shown previously by Ieven et al. (20). In this study, false-positive results due to contamination can be excluded by applying the expanded gold standard, considering a NAAT result positive only if confirmed in two independent amplification reactions. However, this approach my decrease the sensitivity of the most sensitive test.
Ninety RV isolates belonged to Andries group B and only three to Andries group A. In earlier studies, group A RVs were also detected less frequently than group B RVs (1, 2, 24, 26, 32). Although at present, a higher number of group B RVs is known than group A RVs, analysis of epidemiological data indicated that group B RVs produced more than twice as many clinical infections per serotype as group A RVs did.
All enteroviruses detected in this study were detected in specimens from children under 7 years of age. One isolate was poliovirus type 2 with a high level of sequence similarity to poliovirus 2 Sabin. This patient was 5 months old and in the middle of the oral polio vaccination scheme. The remaining strains were classified as echoviruses, with serotype 2 being the most prevalent.
In this study, no etiologic agent was found in 56.7% of the respiratory specimens from children younger than 4 years and in 76.4% of specimens from older children.
Until now, the precise incidence of RVs in acute respiratory tract infections was unknown because laboratory confirmation is usually not obtained. Still higher incidences (up to 55%) of rhinoviruses have been reported in several PCR-based studies (1, 4, 5, 19, 21, 22, 29, 34). Several factors could explain this discrepancy. Our patients were sampled not when the first clinical symptoms appeared but after presentation to the hospital because of persisting respiratory disease or because the general practitioner referred the patient to the hospital. The patients in our study were suffering from relatively severe disease. Therefore, RV infections typically associated with milder respiratory diseases, such as the common cold, were less likely to be observed in this study.
Since samples collected during the weekends were processed on Mondays, RNA degradation cannot be ruled out completely. But in our study, only a minority of the specimens (57/517 [11.0%]) were collected during the weekend. In 36/57 of these nasopharyngeal aspirates, 63.2%, no virus was detected, versus 63.3% when all nasopharyngeal aspirates are taken into account. In 8 of these 57 specimens (14.0%), an RV was detected, in 5/8 cases by NAAT and culture, versus 18.0% when all specimens are taken into account. Previously, experiments to monitor the stability of the RV RNA in fresh respiratory specimens showed that when spiking a pool of RV-negative NAs from children with respiratory tract infections caused by other respiratory viruses with a dilution series of RV-15, storage for 24 and 48 h at room temperature resulted in a loss of 2 and 3 logs, respectively. Storage at 4°C showed less RNA degradation (27).
The prevalence of mixed infections (10, 46) is influenced by multiple factors, including differences in patient populations (e.g., differences in age and comorbid conditions), time of study (e.g., winter versus summer or during epidemics of respiratory virus infections), and diagnostic methods used. Their prevalence increases with the range of diagnostic methods applied. Most reports of mixed viral infections involved children (11, 17, 18, 25, 38, 40, 43, 45, 47). In our study, all patients with mixed viral infections were <4 years old, with rhinoviruses and RSV being the most common because RSV was so prevalent during the study period.
It seems that mixed viral infections occur more frequently than is currently appreciated, and as viral diagnostic ability improves, their reported number may well increase. The clinical significance of mixed viral infections remains unknown and should be the subject for further studies.
A rhinovirus was detected in follow-up samples from patients, up to 51 days in the presence of persisting respiratory symptoms. The clinical significance of RV infections is not always straightforward, since asymptomatic infections are mentioned for 4 to 12% of healthy individuals (21, 22).
In conclusion, we evaluated NAAT for the detection of rhinoviruses in clinical specimens. Both NASBA and RT-PCR produced comparable results and were significantly more sensitive than virus isolation by cell culture technique. Using these techniques, we demonstrated that the acute respiratory tract disease of 18.0% of the children admitted to the hospital was associated with a rhinovirus infection. At the moment, no quantitative tests for the detection of RVs are available. Since it has not yet been reported that RVs may colonize the respiratory tract, the RVs detected with the qualitative NAATs are considered the true causes of the respiratory tract infections.
REFERENCES
Articles from Journal of Clinical Microbiology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jcm.44.1.166-171.2006
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1351952?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/jcm.44.1.166-171.2006
Article citations
Recent Advances in Molecular Diagnostics of Fungal Plant Pathogens: A Mini Review.
Front Cell Infect Microbiol, 10:600234, 11 Jan 2021
Cited by: 32 articles | PMID: 33505921 | PMCID: PMC7829251
Review Free full text in Europe PMC
Development of real-time fluorescent reverse transcription loop-mediated isothermal amplification assays for rhinovirus detection.
J Med Virol, 91(7):1232-1238, 22 Feb 2019
Cited by: 8 articles | PMID: 30735248 | PMCID: PMC7166982
Profile of the Alere i Influenza A & B assay: a pioneering molecular point-of-care test.
Expert Rev Mol Diagn, 18(5):403-409, 24 Apr 2018
Cited by: 9 articles | PMID: 29688086 | PMCID: PMC6153442
Genetic diversity of human rhinoviruses in Cambodia during a three-year period reveals novel genetic types.
Infect Genet Evol, 35:42-49, 29 Jul 2015
Cited by: 5 articles | PMID: 26231720 | PMCID: PMC7105975
Culture-Independent Diagnostics for Health Security.
Health Secur, 14(3):122-142, 01 May 2016
Cited by: 15 articles | PMID: 27314653
Review
Go to all (43) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Nucleotide Sequences (Showing 14 of 14)
- (1 citation) ENA - AF162711
- (1 citation) ENA - X89532
- (1 citation) ENA - X89535
- (1 citation) ENA - S76768
- (1 citation) ENA - AF188359
- (1 citation) ENA - AF188358
- (1 citation) ENA - AJ133661
- (1 citation) ENA - L76398
- (1 citation) ENA - DQ316315
- (1 citation) ENA - DQ316262
- (1 citation) ENA - AF083069
- (1 citation) ENA - U11707
- (1 citation) ENA - X00595
- (1 citation) ENA - S79276
Show less
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Detection of enterovirus RNA in cerebrospinal fluid (CSF) using NucliSens EasyQ Enterovirus assay.
J Clin Virol, 32(3):236-240, 01 Mar 2005
Cited by: 27 articles | PMID: 15722029
Early detection of acute rhinovirus infections by a rapid reverse transcription-PCR assay.
J Clin Microbiol, 39(1):129-133, 01 Jan 2001
Cited by: 37 articles | PMID: 11136760 | PMCID: PMC87691
Development and evaluation of nucleic acid sequence based amplification (NASBA) for diagnosis of enterovirus infections using the NucliSens Basic Kit.
J Clin Virol, 24(1-2):117-130, 01 Feb 2002
Cited by: 36 articles | PMID: 11744436
Molecular diagnosis of respiratory viruses.
Future Microbiol, 5(6):901-916, 01 Jun 2010
Cited by: 39 articles | PMID: 20521935
Review





