Abstract
Background
Thrombospondin-1 (TSP-1) has been implicated in many different processes based in part on inhibitory activities of anti-TSP-1 monoclonal antibodies (mAbs).Objective
To map epitopes of 13 anti-TSP-1 mAbs to individual modules or groups of modules spanning TSP-1 and the closely related TSP-2 homolog.Results
The mapping has led to assignment or reassignment of the epitopes of four mAbs, refinement of the epitopes of six mAbs, and confirmation of the epitopes of the remaining three mAbs. ESTs10, P12, and MA-II map to the N-terminal domain; 5G11, TSP127.6, and ESTs12 to the third properdin module; C6.7, HB8432, and P10 to epidermal growth factor (EGF)-like modules 1 and/or 2; and A6.1, mAb133, MA-I, and D4.6 to the calcium-binding wire module. A6.1, which recognizes a region of the wire that is identical in mouse and human TSP-1, reacts with TSP-1 from both species, and also reacts weakly with human TSP-2. Two other mouse antihuman TSP-1 mAbs, A4.1 and D4.6, also react with mouse TSP-1.Conclusions
Consideration of previous literature and mapping of epitopes of inhibitory mAbs suggest that biological activities are present throughout TSP-1, including the EGF-like modules that have not been implicated in the past. Because the epitopes for 10 of the antibodies likely are within 18 nm of one another in calcium-replete TSP-1, some of the inhibitory effects may result from steric hindrance. Such seems to be the case for mAb133, which binds the calcium-binding wire but is still able to interfere with the activation of latent TGF-beta by the properdin modules.Free full text

Function-blocking antithrombospondin-1 monoclonal antibodies
Summary
Background
Thrombospondin-1 (TSP-1) has been implicated in many different processes based in part on inhibitory activities of anti-TSP-1 monoclonal antibodies (mAbs).
Objective
To map epitopes of 13 anti-TSP-1 mAbs to individual modules or groups of modules spanning TSP-1 and the closely related TSP-2 homolog.
Results
The mapping has led to assignment or reassignment of the epitopes of four mAbs, refinement of the epitopes of six mAbs, and confirmation of the epitopes of the remaining three mAbs. ESTs10, P12, and MA-II map to the N-terminal domain; 5G11, TSP127.6, and ESTs12 to the third properdin module; C6.7, HB8432, and P10 to epidermal growth factor (EGF)-like modules 1 and/or 2; and A6.1, mAb133, MA-I, and D4.6 to the calcium-binding wire module. A6.1, which recognizes a region of the wire that is identical in mouse and human TSP-1, reacts with TSP-1 from both species, and also reacts weakly with human TSP-2. Two other mouse antihuman TSP-1 mAbs, A4.1 and D4.6, also react with mouse TSP-1.
Conclusions
Consideration of previous literature and mapping of epitopes of inhibitory mAbs suggest that biological activities are present throughout TSP-1, including the EGF-like modules that have not been implicated in the past. Because the epitopes for 10 of the antibodies likely are within 18 nm of one another in calcium-replete TSP-1, some of the inhibitory effects may result from steric hindrance. Such seems to be the case for mAb133, which binds the calcium-binding wire but is still able to interfere with the activation of latent TGF-β by the properdin modules.
Introduction
Thrombospondins (TSP) are calcium-binding extracellular matrix glycoproteins. The vertebrate TSP family consists of the homotrimeric TSP-1 and TSP-2, and the homopentameric TSP-3, TSP-4 and TSP-5/COMP (cartilage oligomeric matrix protein) [1]. TSPs are modular in design and thus have logical boundaries for dividing the proteins into smaller units. The TSP-1 and -2 monomers consist of a N-terminal module (N), an oligomerization domain (o), a procollagen module (C), three properdin or type I modules (P123), three epidermal growth factor (EGF)-like or type II modules (E123), several type III repeats or calcium-wire module (Ca), and a lectin-like globular module or G domain (G) (Fig. 1A).
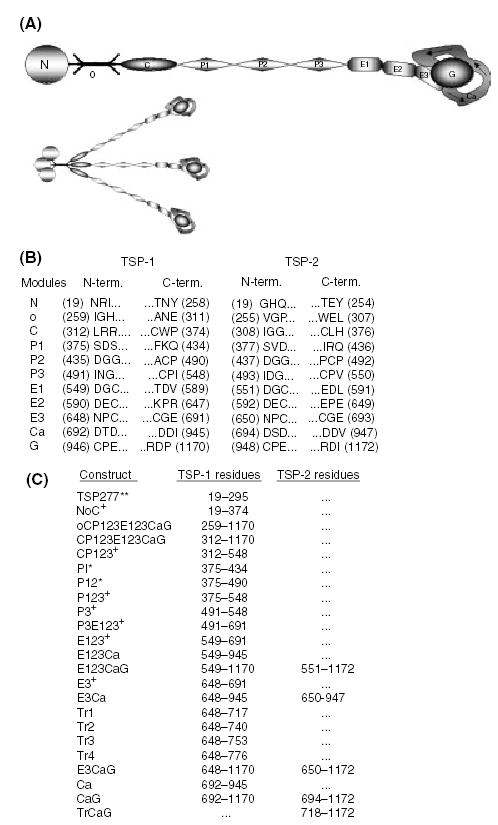
Expression of thrombospondin modules and nomenclature. Modules or groups of modules spanning the human TSP-1 and -2 molecules were expressed in baculovirus. (A) Constructs that include the oligomerization domain are trimeric. The TSP monomer consists of an N-terminal module (N), an oligomerization domain (o), a procollagen module (C), three properdin or type I modules (P), three EGF-like or type II modules (E), several type III repeats or calcium-wire module (Ca), and a lectin-like globular module or G domain (G). This schematic is a representation of TSP in the calcium-replete form, showing the Ca-wire leaving the E3 module and looping around the G domain, forming the C-terminal globe. (B) The three N- and C-terminal residues for each module from TSP-1 and -2 are listed. The residue number is in parentheses. Numbering begins at the initiating methionine, with residue 19 being the first amino acid, and residues 1170 and 1172 from TSP-1 and -2 respectively, being the end of the mature proteins. (C) Tabulation of the recombinant modular arrays and the residues they encompass. The proteins are named based on their modular components. The exceptions are; TSP277, which is the first 277 residues (19–295) of TSP-1; and the E3Ca-1 truncations, Tr1, Tr2, Tr3, and Tr4, which include the E3 module and 1, 2, 3, or four calcium-binding motifs [25] from Ca. Modules derived from TSP-1 are followed by -1, and those derived from TSP-2 are followed by -2. All constructs are produced in coco with no thrombin cleavage site in the linker between the end of the TSP sequence and six his tag unless noted: produced in coco with a thrombin cleavage site in the linker (+), proteins not produced in coco with no tags or linkers (**), and proteins expressed in the GELEX system (*). P3-1 was produced in both coco and GELEX.
Thrombospondin-1 is a major component of platelet alpha granules and because of its accessibility and ease of purification, it is by far the best studied of the TSPs. TSP-1 has been implicated in many different activities both in platelet function and in effects on cells (Table 1). Monoclonal antibodies (mAbs) have been key reagents in such studies. Accurate mapping of the epitopes of these antibodies is a requisite for interpretation of which regions are responsible for the various activities of TSP-1. We have used a baculovirus expression system to produce individual modules or groups of modules spanning the human TSP-1 (hTSP-1) and human TSP-2 (hTSP-2) molecules [2]. We recently showed that A4.1, an important anti-TSP-1 mAb by virtue of its antiangiogenic activity, binds to the most C-terminal EGF-like module (E3), and blocks activities of the properdin modules by steric hindrance [3]. We now report mapping of an additional 13 of the valuable set of anti-TSP-1 mAbs. Based on our results, we assign or reassign the epitopes for four of the 13 mAbs tested, refine the mapping for six, and confirm the epitopes for the remaining three. In addition, we have tested for cross-reactivity of the anti-hTSP-1 mAbs with hTSP-2 and mouse TSP-1 (mTSP-1) and TSP-2 (mTSP-2). The new information is discussed in relation to previous literature of effects of the mAbs and recent structural data [4,5] that allow an appreciation of the proximity of epitopes in the C-terminal region of TSP-1.
Table 1
Activities and epitopes of anti-TSP-1 mAbs. Each mAb, reported activities affected, references, mapping results of this study, and mapping results of previous studies are listed. Abbreviations for previous mapping are: HBD, heparin-binding domain released after proteolytic digestion; 400-kDa, tryptic peptide fragment; Tryptic core, disulfide bonded core after trypsin digestion; 18-kDa, chymotrypsin-generated fragment with N-terminus at residue 1049; 120-kDa TF, non-heparin-binding tryptic fragment; 70-kDa, chymotryptic fragment produced in EDTA; 50-kDa sTSP, 50-kDa chymotryptic fragment of TSP-1 stripped of TGF-β activity produced in EDTA; 120/140-kDa, fragment produced by chymotrypsin in calcium
| mAb | Selected reports of TSP-mediated activities affected | This study | Previous studies |
|---|---|---|---|
| MA-II | Inhibits TSP-1 expression and platelet aggregation/secretion induced by thrombin, and inhibits TSP-1 interaction with immobilized fibrinogen and platelet GPIV (CD36) (Legrand et al. [27]). | N | HBD [8] |
| P12 | Enhances hemagglutination activity of EDTA-treated TSP (Clezardin et al. [6]). | N | 400-kDa [6] |
| ESTs10 | Inhibits attachment of G361, HISM, MG-63 and keratinocyte cells to TSP (Adams and Lawler [9]).
Inhibits attachment of C2C12, G8 and H9c2 cells to TSP (Adams and Lawler [41]). | N | Bacterially expressed 931–1170 [9] |
| TSP127.6 | Antibody selected for based on the ability to block latent TGF-β activation (this report). | P3 | Unpublished |
| 5G11 | Inhibits mesangial cell adhesion to TSR3 (P3-1) (Calzada et al. [3]). | P3 | P3 [3] |
| ESTs12 | Inhibits attachment of G361, HISM and MG-63 cells to TSP, but did not inhibit keratinocyte attachment (Adams and Lawler [9]).
Inhibits attachment of C2C12, G8 and H9c2 cells to TSP (Adams and Lawler [41]). | P3 | P123
379–548 [9] |
| HB8432 | Inhibits mesangial cell attachment and spreading to EGF domains by approximately 50% (Calzada et al. [3]).
Inhibits TSP-stimulated latent TGF-β activation by 32% (Schultz-Cherry and Murphy-Ullrich [14]). | E1,E2 | Tryptic core [10] |
| C6.7 | Inhibits thrombin and A23187-induced aggregation of platelets (Dixit et al. [12]).
Inhibits adhesion of G361 melanoma cells (Prater et al. and Roberts et al. [13,42]). Inhibits TSP-induced chemotaxis of VSMC (Yabkowitz et al. and Nesselroth et al. [43,44]). Inhibits monocyte haptotaxis (Mansfield and Suchard [45]). Inhibits increased TGF-β formation in estrogen-depleted and tamoxifen-treated cells (Harpel et al. [46]). Increases reendothelialization, and decreases the number of VSMC in balloon-injured rat arteries (Chen et al. [47]). | E1,E2 | 18-kDa [12] |
| P10 | Inhibits hemagglutination activity of EDTA-treated TSP (Clezardin et al. [6]).
Inhibits platelet-aggregating activity of osteosarcoma cell line MG63 (Clezardin et al. [48]). | E1,E2 | 120-kDa TF [6] |
| A4.1 | Inhibits adhesion of G361, HISM and MG-63 cells (Adams and Lawler and Prater et al. [9,13]).
Inhibits TSP-mediated antiangiogenesis (Tolsma et al. and Good et al. [49,50]). Inhibits bFGF binding to TSP (Taraboletti [51]). Inhibits β1 integrin-dependent adhesion to the EGF-like and properdin modules (Calzada et al. [3]). Blocks TSP neurite-promoting activity (Osterhout et al. [52]). | E3 | E3 [3] |
| A6.1 | Blocks TSP binding to type V collagen (Galvin et al. [53]).
Inhibits monocyte haptotaxis (Mansfield and Suchard [45]). Blocks PMN adherence to TSP (Suchard et al. [54]). Inhibits adhesion of G361 melanoma cells (Roberts et al. [42]). | Ca 692–717 | 70-kDa [11] |
| mAb133 | Inhibits latent TGF-β activation (Schultz-Cherry and Murphy-Ullrich [14]).
Inhibits increased TGF-β formation in estrogen-depleted and tamoxifen-treated cells (Harpel et al. [46]). | Ca | 50-kDa sTSP [14] |
| MA-I | Inhibits adherence of malaria-parasitized erythrocytes to TSP (Sherwood et al. [55]). | Ca | Bacterially expressed 802–950 [8,27] |
| D4.6 | Inhibits TSP-induced chemotaxis of VSMC (Nesselroth et al. [44]).
Competes with binding of TSP to PMNs (Majluf-Cruz et al. [56]). Sensitive to disulfide interchange catalyzed by protein disulfide isomerase (Hotchkiss et al. [57]). | Ca | 120/140-kDa [11] |
Materials and methods
Monoclonal antibodies
Monoclonal antibodies to TSP-1 included the antibodies P10 and P12 [6], and 5G11 [7] from the 7th Human Leukocyte Differentiation Antigens Workshop; antibodies MA-I and MA-II [8], gifts of Dr Jack Lawler, Harvard Medical School, Boston, MA, USA; antibodies ESTs10 and ESTs12 [9] gifts of Dr N. R. Hunter via Dr Josephine C. Adams, Lerner Research Institute, Cleveland Clinic Foundation; antibody HB8432 [10]; and four antibodies developed in Dr William Frazier’s laboratory, A6.1 and D4.6 [11], C6.7 [12] and A4.1 [12,13], which were gifts from Dr Atul Tandon, Lab Vision/NeoMarkers, Inc., Fremont, CA, USA. MAb133 [14] and TSP127.6 were raised against stripped human platelet TSP-1(sTSP-1) [15] and selected for the ability to inhibit TSP-1-dependent TGF-β activation of bovine aortic endothelial cell-derived latent TGF-β in conditioned media (CM) using the NRK soft agar colony formation assay [14]. A4.1 is a murine IgM, and 5G11 is a rat IgG2a. The other mAbs studied are murine IgG.
Expression of TSP modules using pAcGP67.coco (COCO)
We used the vector pAcGP67.coco (COCO) to express a majority of our TSP-derived constructs [2]. This baculovirus expression system has been used to produce TSP modules that are native, glycosylated, and functional [3,16–19]. The his-tagged recombinant proteins were purified by Ni++-chelate chromatography. Figure 1B lists the residues used as borders for each domain. The recombinant proteins are described based on their modular composition (Fig. 1C).
Expression of TSP277 and GELEX constructs
TSP277 [20] and the GELEX constructs containing the properdin modules [21] were expressed in baculovirus and purified as described.
Expression of full length hTSP-2 without his-tags
Full length hTSP-2 was expressed and purified by Dr Paul A. Tooney using methods previously described for mTSP-1 [22].
Expression of full length mTSP-1 and mTSP-2 without his-tags
Mouse TSP-2 [23] and mTSP-1 [22] were expressed and purified as previously described.
Purification of hTSP-1
Thrombospondin-1 was purified from releasate of thrombin-stimulated platelets by heparin-agarose affinity chromatography and gel filtration on P-300 (Bio-Rad, Richmond, CA, USA) as described [24]. All buffers contained 0.1 mm calcium. For preparations of sTSP-1, the TSP-1 was stripped of TGF-β activity by performing the gel-filtration step at pH 11.0, followed by the dialysis against TBS plus 0.1 mm calcium at pH 7.35 as described [15].
Enzyme-linked immunosorbent assay (ELISA)
Recombinant TSP fragments and hTSP-1 purified from platelets were coated onto 96 well microtiter plates at 10 μg mL−1 in Tris buffer saline (TBS) plus either 0.3 mm or 2 mm calcium. The plates were blocked with 5% non-fat-dried milk and 1% BSA in TBS plus 0.05% tween-20 (TBST). The mAbs were diluted appropriately and incubated with the plate for 2 h. The appropriate species and isotype-specific alkaline phosphatase-conjugated secondary antibody was added for a 1 h incubation. Sigma 104 AP substrate at 1 mg mL−1 in TBS pH 9.0 was added to each well and color development was monitored at 405 nm. A subset of the mAbs was tested in competition ELISA as described [3] to confirm that soluble antigen competed for binding of the mAb to surface adsorbed antigen.
Western blotting
Approximate molar equivalents (10 pmoles) of TSP and TSP-derived proteins were resolved on SDS–PAGE gels. The proteins were subsequently transferred to PVDF membrane. MAb-positive bands were detected by species and isotype-specific, peroxidase-conjugated antibodies and enhanced chemiluminescence technology.
Assay of TSP-1-dependent activation of endothelial cell-derived latent TGF-β
sTSP-1, mAbs, or sTSP-1 + mAbs were added to BAE cells in DMEM with 0.2% FBS for 48 h. Aliquots of CM were harvested and TGF-β activity in the CM was assayed in the NRK soft agar colony formation assay [14] in the presence of EGF.
Results
Experiments performed to characterize 13 anti-TSP-1 mAbs by ELISA (Table 2) and by Western blot (Table 3) are described in the order, N- to C- terminus, of the mapped epitopes. Additional attention is paid to A6.1, which, like A4.1, recognizes an epitope common to human and mTSP-1. All mAbs had been elicited to hTSP-1 from platelets.
Table 2
Results of direct ELISA. Monoclonal antibodies were tested in ELISA against all or most of the following constructs: hTSP-1 (h1), hTSP-2 (h2), mTSP-1 (m1), mTSP-2 (m2), NoC-1, CP123E123CaG-1 (C to G-1), CP123-1, E123-1, and E3CaG-1. The positives (+) ranged from 24% to 130% of the absorbance at 405 nm with full-length hTSP-1, with most constructs at 80–110%. The reaction of A6.1 with hTSP-2 was 15% and is considered a weak positive (w). A negative result (neg) is 5% or less, with most constructs at 0–2%. nd: reactivity of a mAb with a construct was not determined. *: soluble construct competed for binding of mAb to insoluble antigen. Some mAbs were tested against other constructs to more precisely identify the region containing the epitope, as listed in the ‘other select positives’ column
| mAb | h 1 | h 2 | m 1 | m 2 | NoC-1 | C to G-1 | CP123-1 | E123-1 | E3CaG-1 | Other select positives |
|---|---|---|---|---|---|---|---|---|---|---|
| MA-II | + | neg | neg | neg | + | neg | neg | neg | nd | TSP277 |
| P12 | + | neg | neg | neg | + | neg | neg | neg | neg | TSP277 |
| ESTs10 | + | neg | neg | neg | + | neg | neg | neg | neg | TSP277 |
| TSP127.6 | + | neg | neg | neg | neg | + | + | neg | neg | P123-1, P3-1 |
| 5G11 | + | neg | neg | neg | neg | + | + | neg | neg | P3E123-1, P3-1 |
| ESTs12 | + | neg | neg | neg | neg | nd | + | neg | neg | P3-1 |
| HB8432 | + | neg | neg | neg | neg | +* | neg | +* | neg | P3E123-1*, E123CaG-1* |
| C6.7 | + | neg | neg | neg | neg | +* | neg | +* | neg | P3E123-1*, E123CaG-1* |
| P10 | + | neg | neg | neg | neg | +* | neg | +* | neg | P3E123-1*, E123CaG-1* |
| A4.1 | + | neg | + | neg | neg | +* | neg | +* | +* | E3-1* |
| A6.1 | + | w | + | neg | neg | + | neg | neg | + | E3CaG-2 |
| mAb133 | + | neg | neg | neg | neg | + | neg | neg | + | E3Ca-1 |
| MA-I | + | neg | neg | neg | neg | + | neg | neg | + | E3Ca-1, Ca-1 |
| D4.6 | + | neg | + | neg | neg | neg | neg | neg | neg | – |
Table 3
Results of Western blot. The mAbs were examined in Western blot against the panels of potential antigens described in Table 2. Positive bands are designated with (NR) for non-reduced antigen, and (R) for reduced antigen, for weakly positive bands a (w) precedes the NR or R. Negative results are indicated with (neg), and not determined is (nd)
| mAb | h 1 | h 2 | m 1 | m 2 | NoC-1 | C to G-1 | CP123-1 | E123-1 | E3CaG-1 | Other select positives |
|---|---|---|---|---|---|---|---|---|---|---|
| MA-II | nd | nd | nd | nd | NR/R | neg | nd | nd | nd | TSP277 |
| P12 | nd | nd | nd | nd | NR/R | neg | nd | nd | nd | – |
| ESTs10 | nd | nd | nd | nd | NR/R | neg | nd | nd | nd | TSP277 |
| TSP127.6 | NR/wR | nd | nd | nd | neg | nd | NR/R | neg | neg | P3-1 |
| 5G11 | nd | nd | nd | nd | neg | NR/R | NR/R | neg | neg | P3E123-1 |
| ESTs12 | nd | nd | nd | nd | nd | nd | nd | nd | nd | – |
| HB8432 | NR | nd | nd | nd | neg | NR | neg | NR | neg | P3E123-1 |
| C6.7 | NR | nd | neg | nd | neg | NR | neg | NR | neg | P3E123-1 |
| P10 | nd | nd | nd | nd | neg | NR | neg | NR | neg | P3E123-1 |
| A4.1 | NR/R | neg | NR/R | neg | neg | NR/R | neg | NR/R | NR/R | E3-1, E3CaG-2 |
| A6.1 | NR/R | wR | NR/R | wR | neg | NR/R | neg | neg | NR/R | Tr1, Ca-1, CaG-2 |
| mAb133 | NR/R | neg | neg | neg | neg | NR/R | neg | neg | NR/R | E3Ca-1, Ca-1 |
| MA-I | nd | nd | nd | nd | nd | nd | nd | nd | nd | – |
| D4.6 | NR/R | neg | NR/R | neg | neg | NR/R | neg | neg | NR/R | Ca-1, TrCaG-2 |
MA-II
MA-II reacted with hTSP-1, NoC-1 and TSP277 by ELISA, but did not react with CP123E123CaG-1, mTSP-1, hTSP-2, or mTSP-2 (Table 2). MA-II was positive in Western blot with reduced and non-reduced TSP277 and NoC-1 (Table 3). These results and the previous characterization [8] indicate the epitope for MA-II is specific for hTSP-1 and recognizes a sequence difference between the N module of human and mTSP-1s.
P12
In ELISA, P12 was positive with hTSP-1, NoC-1, and TSP277, but was negative with oCP123E123CaG-1, CP123E123CaG-1, and the other TSPs (Table 2). Immuno-blotting of reduced and non-reduced NoC-1 with P12 verified binding to NoC-1 (Table 3). The epitope for P12 therefore is sequence- and species-specific and lies within the N module of hTSP-1.
ESTs10
ESTs10 reacted with NoC-1 and TSP277, both of which are located at the N-terminus of hTSP-1 (Fig. 2A, Tables 2 and and3).3). ESTs10 bound to both reduced and non-reduced protein in Western blotting. By ELISA, ESTs10 did not recognize hTSP-1 lacking N or the other TSPs. These data, therefore, indicate that the epitope for ESTs10 is located in the N module, residues 19–258 of hTSP-1.
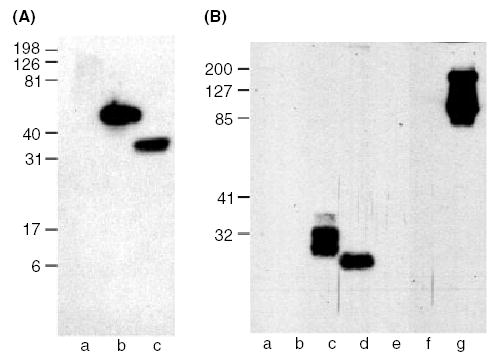
ESTs10 recognizes the N module, and C6.7 recognizes EGF domains 1 and/or 2. Approximate molar equivalents of TSP-derived proteins were resolved in reducing conditions on a 14% gel for ESTs10, and under non-reducing conditions on a 10% SDS–PAGE gel for C6.7. The proteins were transferred to PVDF membrane and immuno-blotted. (A) Western blot of ESTs10, lane (a) CP123E123CaG-1, (b) NoC-1 and (c) TSP277. (B) Western blot of C6.7, lane (a) NoC-1, (b) CP123-1, (c) P3E123-1, (d) E123-1, (e) E3CaG-1, (f) E3Ca-1, and (g) CP123E123CaG-1.
TSP127.6
TSP127.6 is a previously undescribed mAb selected for its ability to inhibit the ability of added sTSP-1 to activate endothelial cell latent TGF-β (Table 4). TSP127.6 mapped to the properdin-like modules (Tables 2 and and3).3). In ELISA and Western blot, TSP127.6 bound to P3-1, P123-1 and CP123-1, but failed to react with P1-1 and P12-1 constructs. These results localize the epitope for TSP127.6 to P3-1 (residues 491–547). TSP127.6 did not react with mTSP-1, and the Western blot signal to hTSP-1 was variably less when the disulfides were reduced.
Table 4
TSP127.6, like mAb133, inhibits TSP-1-dependent activation of endothelial cell-derived latent TGF-β. Results are expressed as mean colonies per well ± SD and are the results of triplicate determinations. The mAbs and sTSP-1 were used at 10 μg mL−1 and 1 μg mL−1, respectively
| Condition | Colonies per well |
|---|---|
| EGF (assay control) | 31 ± 14 |
| EGF + CM | 58 ± 6 |
| EGF + CM + sTSP-1 | 92 ± 6 |
| EGF + CM + sTSP-1 + TSP127.6 | 43 ± 2 |
| EGF + CM + TSP127.6 | 40 ± 6 |
| EGF + CM + sTSP-1 + mAb133 | 47 ± 6 |
| EGF + CM + mAb133 | 37 ± 7 |
5G11
5G11 recognized CP123E123CaG-1, CP123-1, P3E123-1, and P3-1 in ELISA (Table 2). Western blotting was positive with antigen that was non-reduced or reduced and confirmed specificity for the P3 module of TSP-1 (Table 3). 5G11 was specific for hTSP-1, showing no reaction with hTSP-2, mTSP-1, or mTSP-2.
ESTs12
Characterization by ELISA using our recombinant modules confirmed the previous localization of the ESTs12 epitope to residues1 379–548 of hTSP-1 [9], and allowed a more-specific assignment to P3-1, residues 491–547 (Table 2). ESTs12 did not react with mTSP-1, mTSP-2, or hTSP-2. The antibody was not tested in Western blotting.
HB8432
HB8432 reacted with hTSP-1, but not mTSP-1, hTSP-2, or mTSP-2 (Table 2). By both direct and competition ELISAs and Western blotting of non-reduced protein, HB8432 reacted with P3E123-1, E123-1, CP123E123CaG-1, and hTSP-1 (Tables 2 and and3).3). HB8432 did not bind to CP123-1, E3CaG-1, or E3-1. Therefore, the epitope must be in the first two EGF-like modules. Loss of binding upon reduction indicates that the epitope is conformationally determined.
C6.7
In Western blots, C6.7 bound to all non-reduced constructs containing the first and second EGF modules, and there was no binding to E3CaG-1 or E3Ca-1 (Fig. 2B and Table 3). Differential C-mannosylation and O-linked glycosylation of the properdin modules [17,18] likely are the reasons that P3E123-1 (lane c) and CP123E123CaG-1 (lane g) migrate as multiple closely spaced bands as seen in Fig. 2B. CP123E123CaG-1 also exhibits a small amount of protein aggregation. Western blot results were confirmed in direct and competition ELISAs (Table 2). The epitope for C6.7 is located, therefore, in E12 (residues 549–647). The epitope is not present in hTSP-2, mTSP-1, and mTSP-2.
P10
In ELISA, P10 was specific for hTSP-1 and positive with hTSP-1, CP123E123CaG-1, P3E123-1, E123-1, and E123CaG-1, and was negative with CP123-1 and E3CaG-1 (Table 2). Soluble E123-1 competed for binding of P10 to insoluble hTSP-1. In Western blot, P10 was positive with CP123E123CaG-1, E123-1, and P3E123-1, but negative with CP123-1 and E3CaG-1 (Table 3). This reactivity was lost when the antigen was reduced. The epitope for P10 is therefore sequence specific and resides in E12.
A4.1
We recently assigned the A4.1 epitope to the E3 module [3] and describe its reactivity in more detail here. When tested in direct and competition ELISAs, A4.1 bound to hTSP-1, mTSP-1, and all recombinant constructs containing the E3 module of TSP-1 (Table 2). In Western blot, A4.1 recognized E123 and E3 from hTSP-1; E3CaG from hTSP-1 and hTSP-2; CP123E123CaG from hTSP-1, hTSP-1, and mTSP-1 (Table 3). After reduction, there was a loss of A4.1 binding to E3-1 and to the hTSP-2 construct E3CaG-2, but no loss in reactivity with E123-1, E3CaG-1, CP123E123CaG-1, hTSP-1, and mTSP-1 (data not shown). This change in reactivity with certain constructs may be indicative of the importance of surrounding modules to the tertiary structure of the A4.1 epitope in E3.
A6.1
The mAb A6.1 was generated against the reduced and alkylated platelet hTSP-1 and screened for the ability to react differentially with EDTA-treated as opposed to calcium-replete TSP-1 [11]. In ELISA, A6.1 bound to hTSP-1, E3CaG-1, and CP123E123CaG-1, but not to E123-1 or E3-1 (Table 2). In Western blotting, A6.1 was positive with all constructs containing the Ca-wire (Table 3). To localize the epitope further within the wire, we analyzed a set of truncated proteins consisting of E3 and increasingly larger segments of the wire [25] (Fig. 3). This analysis located the epitope for A6.1 to the first calcium-binding motif of Ca. This first motif also contains the Asn to Ser polymorphism at residue 700 that is associated with familial premature coronary artery disease [26], and has been shown to bind one less calcium [25]. A6.1 bound to both the N700 and the S700 proteins. A6.1 also recognizes mTSP-1 in ELISA and Western blots.
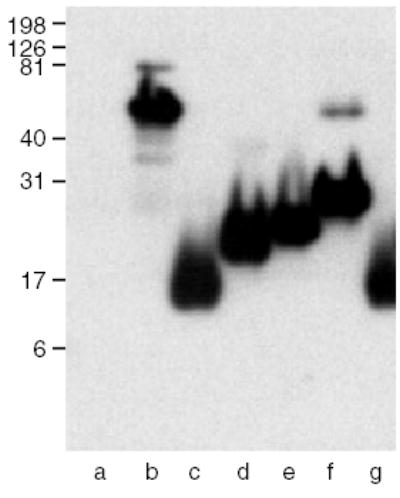
The epitope for A6.1 is in the 1st calcium-binding motif of Ca (residues 692–717). Approximate molar equivalents of TSP-derived proteins were resolved under the reducing conditions on a 14% SDS–PAGE gel, transferred to PVDF membrane and immuno-blotted. Western blot of A6.1, lane (a) E123-1, (b) E3Ca-1, (c) Tr1 (N700), (d) Tr2 (N700), (e) Tr3 (N700), (f) Tr4 (N700), and (g) Tr1 (S700).
MAb133
In ELISA, mAb133 was specific for hTSP-1 and reacted with CP123E123CaG-1, E3CaG-1, and E3Ca-1, but did not bind to CP123-1, P123-1, E123-1, or E3-1 (Table 2). In immuno-blots mAb133 reacted equally well with the reduced and unreduced E3CaG-1, E3Ca-1, and Ca-1, indicating that the epitope is in the calcium-wire, and its exposure is not affected by the presence or absence of the G domain (Table 3).
MA-I
By ELISA, MA-I was specific for hTSP-1 and bound to all constructs tested that included the Ca module (Table 2). This is consistent with the previous assignment of the epitope to residues 802–950 [8,27]. Western blotting was not performed, but previously it was shown that this mAb reacts with non-reduced and reduced protein [8]. The MA-I epitope is, therefore, confirmed to be in the calcium-wire.
D4.6
D4.6 was elicited against the reduced and alkylated platelet hTSP-1, and screened for the ability to react preferentially with EDTA-treated as opposed to calcium-replete TSP-1 [11]. In ELISA, D4.6 bound full-length hTSP-1 and mTSP-1, but did not react with any of our truncated recombinant constructs, hTSP-2, or mTSP-2 (Table 2). However, in Western blot, D4.6 bound to all the constructs from hTSP-1, truncated or full-length, which contained the Ca-wire and the lectin-like globe (Table 3). D4.6 was positive with the hTSP-1-derived E3CaG-1, E3Ca-1 and Ca-1 in Western blot under both non-reducing and reducing conditions. The reaction was much stronger when the antigen was reduced, and when the modules surrounding the Ca-wire were present. D4.6 also bound truncations of the TSP-2 molecule. The effect of having the G domain present was seen to an even greater extent with the TSP-2 proteins. When the antigens were reduced, D4.6 reacted with E3CaG-2, CaG-2, TrCaG-2 (TSP-2 residues 718–1172), and E3Ca-2, but when the antigens were not reduced, the reactivity to E3Ca-2 (lacking the G domain) was lost. D4.6 also bound to mTSP-1 in Western blot. The epitope for D4.6 therefore, is present in the Ca-wire of hTSP-1, hTSP-2, and mTSP-1, but its expression is modulated by reduction and presence of surrounding modules.
Discussion
The localization of the epitopes for the mAbs assayed is summarized in Fig. 4. Three (ESTs10, P12, and MA-II) of the 13 mAbs mapped to the N module; three (5G11, TSP127.6, and ESTs12) mapped to P3; three (C6.7, HB8432, and P10) have epitopes residing in EGF modules 1 and/or 2; and four (A6.1, mAb133, MA-I, and D4.6) mapped to the calcium-wire. We localized A4.1 to E3 in a previous report [3].
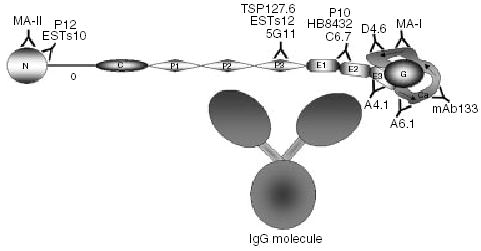
Schematic representation of epitope placement along the TSP-1 molecule. The epitopes for MA-II, P12, and ESTs10 are located in the N module; 5G11, ESTs12, and TSP127.6 in the P3 module; C6.7, HB8432, and P10 in modules E12; A4.1 in the E3 module; and A6.1, mAb133, MAI, and D4.6 in the calcium-wire. The E123CaG portion of the molecule is drawn based on a partial structure of this region of hTSP-1 [4] and the structure of E123CaG from hTSP-2 [5]. The E3CaG complex is approximately 7 × 5.5 nm, with the G domain being approximately 4 × 3 nm, and the EGF-like modules 3 × 1.5 nm. The size of the properdin modules is based on the crystal structure published by Tan [35] and is approximately 5 × 1.2 nm. We have assumed the procollagen module to be similar in proportions to the properdins. The o domain is based on the structure of the coil-coil region of cartilage matrix protein [36] with a length of 5 nm. The N module is about 4 nm in diameter, and is based on the structure of the G modules of laminin [37]. A representation of an IgG molecule based on crystal structures and cryo-electron tomography [38–40] is included to show the relative size compared to TSP-1. The Fc stem (larger globe) is approximately 6.2 × 7 nm, the Fab arms (smaller globes) are approximately 4.7 × 7 nm, and the distances between the three globes are 2–2.5 nm.
The assigned epitopes of A4.1, C6.7, mAb133, and ESTs10 recognized areas of TSP-1 that differ from the original mapping. In each case, the original data can be reconciled with the present results.
A4.1
The basis of the former assignment of the A4.1 epitope to the properdin, rather than EGF-like modules, was the 50- and 70-kDa fragments recovered after chymotryptic digestion of TSP-1 in the presence of EDTA [11,13]. These fragments remain together as trimers and A4.1 recognized both fragments, whereas A6.1 recognized only the 70-kDa fragment [13]. Edman degradation demonstrated that a single N-terminus for both fragments is located at the start of the oligomerization domain at residue 259 [28]. The 50-kDa fragment, therefore, is a product derived from a proteolytic cleavage toward the C-terminal end of the 70-kDa fragment of TSP-1 [13]. The 50-kDa fragment has been assumed to contain the oligomerization domain, the procollagen module, and the properdin modules, and the 70-kDa fragment has been assumed to contain in addition the EGF-like modules [13,28]. The calculated molecular weight for a monomer of a fragment corresponding to oCP123E123 (residues 259–691) is 47.3 kDa. With the mapping of A4.1 to the E3 module [3], it is now clear that the 50-kDa fragment includes the EGF-like modules. The placement of A6.1 in the N-terminal portion of Ca gives further insight into the modular composition of the 50- and 70-kDa fragments. Binding of A6.1 to the most N-terminal repeat of the Ca-wire indicates that the C-terminus of the 70-kDa fragment extends at least 49 residues past the C-terminus of the 50-kDa fragment and likely even further into the Ca-wire. The chymotrypsin cleavage site that results in generation of the 70-kDa fragment into the 50-kDa species, therefore, is between the A4.1 epitope in E3 and the A6.1 epitope in the 1st calcium-binding motif of the Ca-wire.
C6.7
C6.7 had been previously mapped to the lectin-like globe in the C-terminal 18 kDa of TSP-1 [12]. The authors found that C6.7 did not immuno-blot, but was able to immuno-precipitate a fragment present in mild chymotryptic digests of hTSP-1 after the reduction and denaturation [12]. The 18-kDa fragment was sequenced by Edman degradation, and the N-terminus of this fragment was determined to start at residue 1049 [12]. In our hands, C6.7 did react with non-reduced protein in immuno-blots, and the reactivity was lost upon reduction. Thus, C6.7 bound to non-reduced proteins containing EGF-like modules 1 and 2 in both Western blot and ELISA. The assignment of the C6.7 epitope to E12 is not in conflict with the electron micrographs of C6.7 complexed with calcium-replete TSP-1, which depict the antibody binding to the large globular domain [28]. Although the EGF-like modules 1 and 2 are far from the G domain in the primary amino acid sequence, the recent crystal structures of C-terminal portions of TSP-1 [4] and TSP-2 [5] both show the Ca-wire looping around and interacting with the lectin-like globe (Fig. 4). The TSP-2 structure, which includes all three EGF-like repeats, shows that E3 and the Ca-wire interact with the lectin-like globe and are part of the large globular domain [5]. The TSP-2 structure also shows interactions of E2 with the Ca-wire. Thus, E2 is likely close to the large complex of E3, Ca, and G in TSP-1.
mAb133
mAb133 has been reported to recognize a 50-kDa chymotryp-tic fragment of TSP-1 stripped of TGF-β [14]. Using platelet-derived hTSP-1, which had not been stripped of TGF-β, we generated 50- and 70-kDa fragments as previously described [13] and found that mAb133 did react with a band in the 50 kDa region (not shown). However, this band was about 6 kDa larger than the smaller A4.1-reactive fragment on a duplicate blot, and was not visible on a SDS–PAGE gel stained for protein (data not shown). Neither A4.1 nor A6.1 reacted with the mAb133 reactive band. MAb133 did not react with the 70-kDa fragment, a precursor to the 50-kDa [13], whereas A4.1 and A6.1 did (data not shown). MAb133 reacted with all of our Ca-wire containing recombinant proteins in both Western blot and ELISA. Thus, the fragment recognized by mAb133 in chymotryptic digests likely arises from cleavages to release the wire.
ESTs10
The ESTs10 was originally mapped to the C-terminal portion of TSP-1, instead of the N module, using a combination of proteolytic fragments and a bacterially expressed fusion protein encompassing residues 931–1170 [9]. ESTs10 binds well to NoC-1, but most of this binding is lost after cleavage with trypsin (data not shown). Binding of ESTs10 to NoC-1 was also lost after digestion with chymotrypsin using conditions that produce the 25- and 140-kDa fragments from hTSP-1 (data not shown). When full length TSP-1 was digested in these conditions, there was no binding to the 25-kDa fragment, and after prolonged exposure of the film to the blot, there was only very weak binding to a fragment in the 140-kDa range (data not shown). Our results, therefore, indicate that the epitope for ESTs10 is at or very near the trypsin cleavage sites at the junction of the N module and oligomerization sequence and is compatible with original mapping in which the authors did not see binding to the proteolytically derived N-terminal heparin-binding domain, but did find binding to the 140-kDa fragment [9]. ESTs10 did not bind to oCP123E123CaG-1, which begins at Ile259. Trypsin cleaves after Lys262 and Lys264 [29] at the junction between N and the oligomerization domain, but may also cleave after Arg255 in the N module [20]. Inspection of the sequence reveals other potential nearby cleavage sites for trypsin (after Arg228 and Lys230) and chymotrypsin (after Phe217 and Phe219). Degenerative cleavage in this area could explain loss of binding to N, as well as variable binding to the 140-kDa fragment if the epitope is positioned in or near the area containing the proteolytic sites.
Three mAbs, A4.1, A6.1 and D4.6, are to regions completely or nearly completely identical in hTSP-1 and mTSP-1 and, although produced in mice, reacted with both hTSP-1 and mTSP-1. These mAbs also reacted with certain forms of hTSP-2. The other mAbs did not recognize mTSP-1 and recognized regions of TSP-1 in which differences in amino acid sequence between hTSP-1 and mTSP-1 are present.
The 14 mAbs described above have been reported to affect many of the activities of TSP-1 (Table 1). Activities such as cell adhesion, platelet aggregation, antiangiogenesis, cell chemotaxis, activation of latent transforming growth factor-β (TGF-β), and sequestration of basic fibroblast growth factor (bFGF) all have been blocked by various mAbs raised against hTSP-1 (Table 1). The present results, therefore, give additional insight into the possible function of the different TSP-1 regions. Antibodies to the N module indicate that it is involved in cell attachment, interaction with immobilized fibrinogen, and platelet aggregation (Table 1). There is a wealth of potential cell surface receptors to mediate these interactions [30]. Antibodies to the properdin modules interfere with cell attachment, and latent TGF-β activation (Table 1). The IgM A4.1 binds to E3 and indirectly inhibits β1 integrin-dependent adhesion to the properdin modules [3], so it is reasonable to assume that any activities blocked by A4.1 could be attributed to the properdin modules. Thus, the properdin modules could also be responsible for the antiangiogenic, bFGF binding and neurite-promoting activities of TSP-1 blocked by A4.1 (Table 1). Two other functions blocking mAbs, C6.7 and P10, recognize the EGF-like modules, which heretofore have not been an appreciated site of activity in TSP-1. These antibodies, along with HB8432, block TSP-1-mediated cell adhesion, platelet aggregation, chemotaxis, haptotaxis, TGF-β formation, antiangiogenisis, bFGF binding and neurite-promoting activities (Table 1). A similar set of activities has been demonstrated for mAbs that recognize the calcium-wire (Table 1).
Consideration of the structures of IgG and of the likely structure of the C-terminal 60% of TSP-1 [4,5] demonstrates that definitive assignment of activities to a specific region based on the epitope mapping of an IgG, as with the A4.1 IgM, is fraught with uncertainties (Fig. 4). The TSP-1 and immuno-globulin models are based on crystallography, NMR and cryo-electron tomography as described in the figure legend. When these models are displayed side by side, as in Fig. 4, it is apparent that the epitopes in the EGF-like modules and the Ca-wire are closer together than the distance spanned by any two of the three lobes of IgG. Thus, some of the overlapping activities of mAbs that recognize different regions of TSP-1 may result from steric hindrance. Such hindrance would be particularly likely when TSP-1 is interacting with a macromolecule that is as bulky as TSP-1 and IgG. For example, both mAb133 and TSP127.6 were raised and selected for on the basis of the ability to inhibit latent TGF-β activation. Epitope mapping reveals that although these two antibodies can block the same activities (Table 4), their binding sites, TSP127.6 in P3 and mAb133 in Ca, are not in the same region. The KRFK and WSXW sequences in the properdin modules of TSP-1 are critical for TSP-1-mediated activation of the TGF-β latent complex [31–34]. It is possible that TSP127.6 inhibits by binding close to the region of TSP-1 that binds latent TGF-β, whereas mAb133 binds to TSP-1 at a distance from the binding region for latent TGF-β, but in a manner that restricts access of the latent TGF-β to the binding region.
Acknowledgments
This work was supported by NIH grants HL054462 to DFM and HL44575 to JMU. A portion of this investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant No. C06 RR 15490 from the National Center for Research Resources, National Institutes of Health. We would like to thank Drs Blue-leaf A. Hannah, Tina M. Misenheimer, and Kristin G. Huwiler for valuable discussions and reagents; Dr Cheryl Hillery for helpful discussions; Drs Josephine Adams, Jack Lawler, William Frazier, and Atul Tandon for the kind gifts of mAbs; Dr MaryAnn Accavitti-Loper in the UAB Hybridoma Core Facility (P60 AR 20614) for production of mAb133 and TSP127.6; and Mr Antonio Pallero for purification of mAb133 and TSP127.6.
Footnotes
1Residue numbering begins at the initiating methionine. Any reference to residue number from another publication has been converted. The first amino acid of the mature protein is 19.
References
Full text links
Read article at publisher's site: https://doi.org/10.1111/j.1538-7836.2006.01723.x
Read article for free, from open access legal sources, via Unpaywall:
https://onlinelibrary.wiley.com/doi/pdfdirect/10.1111/j.1538-7836.2006.01723.x
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/128691042
Article citations
Thrombospondin-1 Drives Cardiac Remodeling in Chronic Kidney Disease.
JACC Basic Transl Sci, 9(5):607-627, 27 Mar 2024
Cited by: 2 articles | PMID: 38984053
Elevated CD47 is a hallmark of dysfunctional aged muscle stem cells that can be targeted to augment regeneration.
Cell Stem Cell, 29(12):1653-1668.e8, 15 Nov 2022
Cited by: 22 articles | PMID: 36384141 | PMCID: PMC9746883
Supramolecular attack particles are autonomous killing entities released from cytotoxic T cells.
Science, 368(6493):897-901, 07 May 2020
Cited by: 80 articles | PMID: 32381591 | PMCID: PMC7116847
Endothelial MT1-MMP targeting limits intussusceptive angiogenesis and colitis via TSP1/nitric oxide axis.
EMBO Mol Med, 12(2):e10862, 03 Dec 2019
Cited by: 24 articles | PMID: 31793743 | PMCID: PMC7005619
The calcium-binding type III repeats domain of thrombospondin-2 binds to fibroblast growth factor 2 (FGF2).
Angiogenesis, 22(1):133-144, 30 Aug 2018
Cited by: 18 articles | PMID: 30168023
Go to all (33) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe
-
(2 citations)
PDBe - 5G11View structure
Protocols & materials
Related Immune Epitope Information - Immune Epitope Database and Analysis Resource
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Immunochemical analysis of the structure of the signature domains of thrombospondin-1 and thrombospondin-2 in low calcium concentrations.
J Biol Chem, 282(37):27067-27075, 09 Jul 2007
Cited by: 13 articles | PMID: 17620335
Monoclonal antibodies that recognize calcium-dependent structures of human thrombospondin. Characterization and mapping of their epitopes.
J Biol Chem, 261(4):1962-1968, 01 Feb 1986
Cited by: 67 articles | PMID: 2418018
The type 1 repeats of thrombospondin 1 activate latent transforming growth factor-beta.
J Biol Chem, 269(43):26783-26788, 01 Oct 1994
Cited by: 161 articles | PMID: 7929414
[Role of thrombospondin-1 in the development of kidney diseases].
Med Sci (Paris), 29(12):1131-1137, 20 Dec 2013
Cited by: 3 articles | PMID: 24356144
Review
Funding
Funders who supported this work.
NCRR NIH HHS (2)
Grant ID: C06 RR015490
Grant ID: C06 RR 15490
NHLBI NIH HHS (4)
Grant ID: R01 HL054462
Grant ID: R01 HL050061
Grant ID: HL054462
Grant ID: HL44575
NIAMS NIH HHS (1)
Grant ID: P60 AR 20614




