Abstract
Free full text

Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor
Abstract
The main regulator of the human tumor suppresser gene p21(waf1/cip1) is the transcription factor p53, but more recently it has been suggested to be a primary anti-proliferative target for the nuclear receptor VDR in the presence of its ligand 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3). To identify VDR responding regions, we analyzed 20 overlapping regions covering the first 7.1 kb of the p21(waf1/cip1) promoter in MCF-7 human breast cancer cells using chromatin immuno-precipitation assays (ChIP) with antibodies against p53 and VDR. We confirmed two known p53 binding regions at approximate positions −1400 and −2300 and identified a novel site at position −4500. In addition, we found three VDR-associated promoter regions at positions −2300, −4500 and −6900, i.e. two regions showed binding for both p53 and VDR. In silico screening and in vitro binding assays using recombinant and in vitro translated proteins identified five p53 binding sites within the three p53-positive promoter regions and also five 1α,25(OH)2D3 response elements within the three VDR-positive regions. Reporter gene assays confirmed the expected responsiveness of the respective promoter regions to the p53 inducer 5-fluorouracil and 1α,25(OH)2D3. Moreover, re-ChIP assays confirmed the functionality of the three 1α,25(OH)2D3-reponsive promoter regions by monitoring simultaneous occupancy of VDR with the co-activator proteins CBP, SRC-1 and TRAP220. Taken together, we demonstrated that the human p21(waf1/cip1) gene is a primary 1α,25(OH)2D3-responding gene with at least three VDR binding promoter regions, in two of which also p53 co-localizes.
INTRODUCTION
The biologically most active vitamin D metabolite, 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), is essential for mineral homeostasis and skeletal integrity (1), but it also has important roles in the control of the cell growth and differentiation in normal and malignant tissues (2). The 1α,25(OH)2D3 receptor (VDR) is a member of the nuclear receptor superfamily and the only nuclear protein that binds 1α,25(OH)2D3 with high-affinity (Kd = 0.1 nM). One key gene for understanding the anti-proliferative action of 1α,25(OH)2D3 is the cyclin-dependent kinase inhibitor p21(waf1/cip1), which has been suggested first by Jiang et al. (3) to be a 1α,25(OH)2D3 target gene. A VDR binding site, a so-called 1α,25(OH)2D3 response element (VDRE), was suggested to be located at position −750 in relation to the transcriptional start site (TSS) of the p21(waf1/cip1)gene (4). The p21(waf1/cip1) gene is also a transcriptional target of the tumor suppressor protein p53, which is a transcription factor with response elements (REs), at positions −1400 and −2300 of the p21(waf1/cip1) promoter (5). The p53 protein is a master regulator of a prominent transcriptional network that can control the fate of cells in response to stress and can be induced by chemotherapeutic agents, such as 5-fluorouracil (6).
An essential prerequisite for the direct modulation of gene expression by transcription factors, such as VDR and p53, is the location of at least one ligand-activated VDR or phosphorylated p53 molecule close to the basal transcriptional machinery of a primary responding gene. This is traditionally achieved through the specific binding of VDR or p53 to REs within the promoter of the respective gene (7). The DNA-binding domain of the VDR contacts the major groove of a double-stranded hexameric DNA sequence with the consensus sequence RGKTCA (R = A or G, K = G or T). In most cases the heterodimeric partner of VDR is the retinoid X receptor (RXR), another nuclear receptor superfamily member, which also contacts DNA. Therefore, simple VDREs are often formed by a direct repeat of two hexameric core binding motifs spaced by 3 nt (DR3-type VDRE) (8). Additionally, strong DNA-binding of VDR–RXR heterodimers to two hexameric motifs arranged as a direct repeat spaced by 4 nt (DR4-type VDRE) (9) or as an everted repeat with nine intervening nucleotides (ER9-type VDRE) have been described (10). In contrast, p53 binds as a tetramer to the consensus sequence RRRCWWGYYY-N-RRRCWWGYYY (W = A or T, Y = C or T and N can be 0 to 13 bases) (11,12). Although individual REs have been shown to be able to induce transactivation on their own, the observation of multiple VDREs in several primary VDR target genes (13–15) and of two p53-REs within the p21(waf1/cip1) gene (16) suggests that carrying more than one binding site for a given transcription factor in the promoter favors the prominent response of the gene's transcription to the respective regulator. Never-the-less, the average short-term transcriptional response of most primary VDR target genes is only 2-fold or less (17), because most of them are in parallel also under the control of other transcription factors, such as the p21(waf1/cip1) gene by p53.
Ligand-binding to the VDR causes a conformational change within the receptor's ligand-binding domain, which results in the replacement of co-repressor proteins by co-activator (CoA) proteins of the p160-family, such as steroid receptor co-activator 1 (SRC-1) (18), in complex with more general CoAs, such as CREB binding protein (CBP) (19). These CoA complexes have histone acetyltransferase activity, that causes chromatin relaxation (20). In a subsequent step, ligand-activated VDR changes rapidly from interacting with the CoAs of the p160-family to those of mediator complexes, such as thyroid hormone receptor-associated protein 220 (TRAP220), also called Med1 (21). The mediator complex acts as a bridge from activated VDR to the basal transcriptional machinery (22). In this way ligand-activated VDR executes two tasks, the modification of chromatin and the regulation of transcription.
The reported VDRE within the human p21(waf1/cip1) promoter was characterized to be very weak (23) and many studies questioned the p21(waf1/cip1) gene being a primary VDR target gene (17,24,25). In order to challenge these questions, we analyzed in this study the first 7.1 kb of the human p21(waf/cip1) promoter for p53 and VDR binding regions. Whole promoter chromatin immuno-precipitation assays (ChIP) using antibodies against p53 and VDR confirmed two known p53 binding regions at positions −1400 and −2300, identified a novel site at position −4500, found three VDR-associated regions at positions −2300, −4500 and −6900, but could not confirm VDR binding to position −750. In silico/in vitro screening identified five p53 binding sites within the three p53-positive promoter regions and also five VDREs within the three VDR-positive regions. Reporter gene assays confirmed the expected responsiveness of the respective promoter regions to 5-fluorouracil and 1α,25(OH)2D3 and re-ChIP assays confirmed the functionality of the three 1α,25(OH)2D3-reponsive promoter regions by monitoring simultaneous occupancy of VDR with the CoA proteins CBP, SRC-1 and TRAP220. These data demonstrate that the human p21(waf1/cip1) gene is a primary VDR target gene with at least three 1α,25(OH)2D3-responsive promoter regions, in two of which also p53 co-localizes.
MATERIALS AND METHODS
Cell culture
MCF-7 human breast cancer cells were grown in phenol red-free DMEM, supplemented with 5% charcoal-treated fetal bovine serum, 2 mM l-glutamine, 0.1 mg/ml streptomycin and 100 U/ml penicillin, in a humidified 95% air / 5% CO2 incubator. Before mRNA extraction or ChIP assay the cells were treated at a density of 50 to 60% confluency for 0 to 360 min with 300 µM 5-fluorouracil [Sigma-Aldrich, St. Louis, MO, USA, diluted in dimethyl sulfoxide (DMSO)], 10 nM 1α,25(OH)2D3 (kindly provided by Dr Lise Binderup, LEO Pharma, Ballerup, Denmark, diluted in ethanol) or vehicle.
RNA extraction and real-time quantitative RT–PCR
Total RNA was extracted using using the Mini RNA Isolation II kit (Zymo Research, HiSS Diagnostics, Freiburg, Germany) and cDNA synthesis was performed for 1 h at 37°C using 1 µg of total RNA as a template, 100 pmol oligodT18 primer and 40 U reverse transcriptase (Fermentas, Vilnius, Lithuania). Real-time quantitative PCR was performed in an IQ-cycler (BioRad, Hercules, CA, USA) using the dye SybrGreen I (Molecular Probes, Leiden, The Netherlands). Per reaction, 1 U Hot Start Taq polymerase (Fermentas) and 3 mM MgCl2 were used and the PCR cycling conditions were: 40 cycles of 30 s at 95°C, 30 s at 60°C and 30 s at 72°C. Fold inductions were calculated using the formula 2−(ΔΔCt), where ΔΔCt is the ΔCt(stimulus)-ΔCt(solvent),ΔCt is Ct(p21)-Ct(ARP0) and Ct is the cycle at which the threshold is crossed. The gene specific primer pairs (and product sizes) were as follows: p21 gene forward 5′-GCAGACCAGCATGACAGATTT-3′ and reverse 5′-GGATTAGGGCTTCCTCTTGGA-3′ (70 bp) and acidic riboprotein P0 (ARP0, also known as 36B4) control gene forward 5′-AGATGCAGCAGATCCGCAT-3′ and reverse 5′-GTGGTGATACCTAAAGCCTG-3′ (318 bp). PCR product quality was monitored using post-PCR melt curve analysis.
ChIP assays
Nuclear proteins were cross-linked to genomic DNA by adding formaldehyde for 10 min directly to the medium to a final concentration of 1%. Cross-linking was stopped by adding glycine to a final concentration of 0.125 M and incubating for 5 min at room temperature on a rocking platform. The medium was removed and the cells were washed twice with ice-cold phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4 and 8.1 mM Na2HPO4.2H2O). The cells were collected by scraping in ice-cold PBS supplemented with a protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). After centrifugation the cell pellets were resuspended in lysis buffer [1% SDS, 10 mM EDTA, protease inhibitors and 50 mM Tris–HCl (pH 8.1)] and the lysates were sonicated to result in DNA fragments of 300 to 1000 bp in length. Cellular debris was removed by centrifugation and the lysates were diluted 1:10 in ChIP dilution buffer [0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM NaCl, protease inhibitors and 16.7 mM Tris–HCl (pH 8.1)]. Non-specific background was removed by incubating the chromatin resuspension with a salmon sperm DNA/protein A agarose slurry (Upstate Biotechnology, Lake Placid, NY, USA) for 30 min at 4°C with agitation. The samples were centrifuged and the recovered chromatin solutions were incubated with 5 µl of indicated antibodies overnight at 4°C with rotation. IgG (sc-2027) and the antibodies against p53 (sc-6243), GST (sc-138), VDR (sc-1008), CBP (sc-369), SRC-1 (sc-7216), TRAP220 (sc-5334) and phosphorylated RNA polymerase II (Pol II) (sc-13583) were obtained from Santa Cruz Biotechnologies (Heidelberg, Germany). The immuno-complexes were collected with 60 µl of protein A agarose slurry (Upstate Biotechnology) for 2 h at 4°C with rotation. The beads were pelleted by centrifugation for 1 min at 4°C at 100 g and washed sequentially for 5 min by rotation with 1 ml of the following buffers: low salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl and 20 mM Tris–HCl (pH 8.1)], high salt wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl and 20 mM Tris–HCl (pH 8.1)] and LiCl wash buffer [0.25 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris–HCl (pH 8.1)]. Finally, the beads were washed twice with 1 ml TE buffer [1 mM EDTA and 10 mM Tris–HCl (pH 8.0)]. For re-ChIP the immuno-complexes were eluted by adding 100 µl re-ChIP elution buffer (10 mM DTT) at room temperature for 30 min with rotation, the supernatant was diluted 1:40 in ChIP dilution buffer and the antibody against the second protein of interest was added, the new immuno-complexes were allowed to form by incubating at 4°C overnight on a rocking platform, the immuno-complexes were collected by incubating with 120 µl protein A agarose slurry at 4°C for 2 h on a rocking platform and finally washed as indicated above. In both cases the immuno-complexes were then eluted by adding 250 µl elution buffer (1% SDS and 100 mM NaHCO3) and incubation for 15 min at room temperature with rotation. After centrifugation, the supernatant was collected and the cross-linking was reversed by adding NaCl to final concentration of 200 mM and incubating overnight at 65°C. The remaining proteins were digested by adding proteinase K (final concentration 40 µg/ml) and incubation for 1 h at 45°C. The DNA was recovered by phenol/chloroform/isoamyl alcohol (25/24/1) extractions and precipitated with 0.1 volumes of 3 M sodium acetate (pH 5.2) and 2 vol of ethanol using glycogen as a carrier.
PCR of chromatin templates
For each of the 20 regions of the human p21(waf1/cip1) promoter primer pairs were designed (Table 1), optimized and controlled by running PCR with 25 ng genomic DNA (input) as a template. When running immuno-precipitated DNA (output) as a template, the following PCR profile was used: preincubation for 5 min at 95°C, 40 cycles of 30 s at 95°C, 30 s at a primer-specific annealing temperature (see Table 1) and 30 s at 72°C and one final incubation for 10 min at 72°C. The PCR products were separated by electrophoresis through 2.0% agarose. An aliquot of 1 µl SybrGreen (1:2500 dilution) was added to each PCR mix before loading.
Table 1
Genomic PCR primer sequences
| Region no. | Location in p21 promoter | Annealing temperature (°C) | Primer sequences |
|---|---|---|---|
| 1 | +37 to −342 | 60 | 5′-GCTCATTCTAACAGTGCTGTG-3′ |
| 5′-CAAGGAACTGACTTCGGCAG-3′ | |||
| 2 | −324 to −676 | 60 | 5′-CCCGGAAGCATGTGACAATC-3′ |
| 5′-CAGCACTGTTAGAATGAGCC-3′ | |||
| 3 | −677 to −981 | 60 | 5′-GGAGGCAAAAGTCCTGTGTTC-3′ |
| 5′-GGAAGGAGGGAATTGGAGAG-3′ | |||
| 4 | −964 to −1340 | 60 | 5′-CTGAGCAGCCTGAGATGTCAG-3′ |
| 5′-CACAGGACTTTTGCCTCCTG-3′ | |||
| 5 | −1335 to −1688 | 60 | 5′-GAAATGCCTGAAAGCAGAGG-3′ |
| 5′-GCTCAGAGTCTGGAAATCTC-3′ | |||
| 6 | −1670 to −2036 | 60 | 5′-GGAGTCAGATTCTGTGTGTG-3′ |
| 5′-CCTCTGCTTTCAGGCATTTC-3′ | |||
| 7 | −2029 to −2478 | 60 | 5′-CACCACTGAGCCTTCCTCAC-3′ |
| 5′-CTGACTCCCAGCACACACTC-3′ | |||
| 8 | −2456 to −2929 | 60 | 5′-GTGATGCTAGGAACATGAGC-3′ |
| 5′-GATGTGAGGAAGGCTCAGTG-3′ | |||
| 9 | −2910 to −3148 | 56 | 5′-CTGAATACCTGGGACTACAG-3′ |
| 5′-GCTCATGTTCCTAGCATCAC-3′ | |||
| 10 | −3141 to −3558 | 60 | 5′-GACATAGCAGGTGTGATGACC-3′ |
| 5′-GTATTCAGGTGGCTGAGGTG-3′ | |||
| 11 | −3538 to −3941 | 60 | 5′-GAACAGGAAGACCATCCAGG-3′ |
| 5′-GGTCATCACACCTGCTATGTC-3′ | |||
| 12 | −3940 to −4364 | 60 | 5′-GATGCCAACCAGATTTGCCG-3′ |
| 5′-CCTGGCTCTAACAACATCCC-3′ | |||
| 12C | −4149 to −4525 | 60 | 5′-CGCGGTGCTTGGTCTCTATG-3′ |
| 5′-CCTTTCCCAACAAACAAGGGG-3′ | |||
| 13 | −4353 to −4755 | 60 | 5′-CTGAAGGCAGGCAAGACTCG-3′ |
| 5′-CTGGTTGGCATCATCTCGCTG-3′ | |||
| 14 | −4736 to −5268 | 60 | 5′-GCCAGTCATGGTTGTACATGC-3′ |
| 5′-CGAGTCTTGCCTGCCTTCAG-3′ | |||
| 15 | −5248 to −5810 | 60 | 5′-CTTTGCTCCTTCTCCACTCC-3′ |
| 5′-GCATGTACAACCATGACTGGC-3′ | |||
| 16 | −5791 to −6091 | 60 | 5′-CTATGTGCCAAGCTAAGCAC-3′ |
| 5′-GGAGTGGAGAAGGAGCAAAG-3′ | |||
| 17 | −6072 to −6333 | 60 | 5′-CATCAGTTCCCGGTTCTTCTC-3′ |
| 5′-GTGCTTAGCTTGGCACATAG-3′ | |||
| 18 | −6324 to −6731 | 60 | 5′-CAAAGGCACAAAGAGGCCTTC-3′ |
| 5′-GGAACTGATGGAGGCTGGAG-3′ | |||
| 19 | −6715 to −7083 | 60 | 5′-CTAACCTCACAGTACAGGCC-3′ |
| 5′-GCCTCTTTGTGCCTTTGCAC-3′ |
DNA constructs
Full-length cDNAs for human VDR (26) and human RXRα (27) were subcloned into the T7/SV40 promoter-driven pSG5 expression vector (Stratagene, LaJolla, CA, USA). The same constructs were used for both T7 RNA polymerase-driven in vitro transcription/translation of the respective cDNAs and for viral promoter-driven overexpression in mammalian cells. Promoter regions 1 (+20 to −321), 2, 3, 5, 7 (extended, from −1954 to −2620), 12C (extended, from −3818 to −4525) and 19 of the human p21 promoter (for location see Table 1) were cloned by PCR from human genomic DNA and fused with the luciferase reporter gene. (+20 to −321) Point mutations to the promoter region 7 construct were generated using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The location of the mutations are indicated in Figure 6B. All constructs were verified by sequencing.
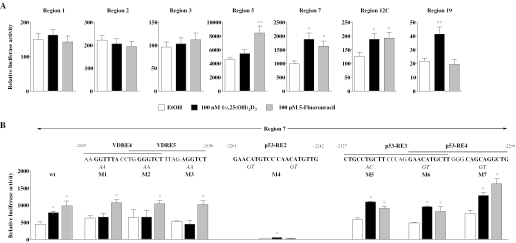
Functionality of p21(waf1/cip1) promoter constructs. Reporter gene assays were performed with extracts from MCF-7 cells that were transiently transfected with luciferase reporter constructs containing regions 1, 2, 3, 7, 12C or 19 of the human p21(waf1/cip1) promoter (A) or wild-type and mutated forms of region 7 (B) and an expression vector for human VDR. The dinucleotides below the RE sequences indicate the point mutations that were introduced into promoter region 7. Cells were treated for 16 h with either solvent, 100 nM 1α,25(OH)2D3 or 100 µM fluorouracil. Relative luciferase activity is shown. Columns represent means of at least three experiments and bars indicate standard deviations. Two-tailed Student's t-tests were performed to determine the significance of the reporter gene induction in reference to solvent controls (*P < 0.05).
Gelshift analysis
Recombinant baculovirus expressed p53 protein (1 U/ng) was purchased from Jena Bioscience (Jena, Germany). In vitro translated VDR and RXR proteins were generated by coupled in vitro transcription/translation using their respective pSG5-based full-length cDNA expression constructs (26) and rabbit reticulocyte lysate as recommended by the supplier (Promega, Madison, WI, USA). Gelshift assays were performed with 100 ng recombinant p53 protein or each 10 ng of in vitro translated VDR and RXRα protein. The proteins were incubated for 15 min in total volume of 20 µl binding buffer [150 mM KCl, 1 mM DTT, 0.2 µg/µl poly(dI-C), 5% glycerol, 10 mM HEPES (pH 7.9)]. Constant amounts (1 ng) of 32P-labeled double-stranded oligonucleotides (50 000 c.p.m.) corresponding to one copy of the putative p53-REs or VDREs were then added and incubation was continued for 20 min at room temperature. Core sequences of the REs are indicated in Figure 4. An double-stranded oligonucleotide with the core sequence GAACTCGGAAAGGCTCAGCTGGCTC served as negative control for p53 binding, while the rat atrial natriuretic factor (ANF) DR3-type VDRE [core sequence AGAGGTCATGAAGGACA (28)] served as reference of the strength of the putative VDREs. Protein–DNA complexes were resolved by electrophoresis through 8% non-denaturing polyacrylamide gels in 0.5× TBE buffer [45 mM Tris, 45 mM boric acid, 1 mM EDTA (pH 8.3)] and quantified on a FLA-3000 reader (Fuji, Tokyo, Japan) using Image Gauge software (Fuji).
000 c.p.m.) corresponding to one copy of the putative p53-REs or VDREs were then added and incubation was continued for 20 min at room temperature. Core sequences of the REs are indicated in Figure 4. An double-stranded oligonucleotide with the core sequence GAACTCGGAAAGGCTCAGCTGGCTC served as negative control for p53 binding, while the rat atrial natriuretic factor (ANF) DR3-type VDRE [core sequence AGAGGTCATGAAGGACA (28)] served as reference of the strength of the putative VDREs. Protein–DNA complexes were resolved by electrophoresis through 8% non-denaturing polyacrylamide gels in 0.5× TBE buffer [45 mM Tris, 45 mM boric acid, 1 mM EDTA (pH 8.3)] and quantified on a FLA-3000 reader (Fuji, Tokyo, Japan) using Image Gauge software (Fuji).

In silico screening for p53-REs and VDREs in the human p21(waf1/cip1) promoter. In silico analysis of the human p21(waf1/cip1) promoter indicated five putative p53-REs in promoter regions 5, 7 and 12 and eight putative VDREs (RE1 to RE8) in promoter regions 3, 4, 7, 12 and 19. The decameric p53 binding sites and the hexameric nuclear receptor binding sites of each RE core sequence are shown in bold and their location relative to the TSS is indicated.
Transfection and luciferase reporter gene assays
MCF-7 cells were seeded into 6-well plates (105 cells/ml) and grown overnight in phenol red-free DMEM supplemented with 5% charcoal-stripped fetal bovine serum. Plasmid DNA containing liposomes were formed by incubating a reporter plasmid and the expression vector for human VDR (each 1 µg) with 10 µg N-[1-(2,3-Dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP, Roth, Karlsruhe, Germany) for 15 min at room temperature in a total volume of 100 µl. After dilution with 900 µl phenol red-free DMEM, the liposomes were added to the cells. Phenol red-free DMEM supplemented with 500 µl 15% charcoal-stripped fetal bovine serum was added 4 h after transfection. At this time, 100 µM 5-fluorouracil, 100 nM 1α,25(OH)2D3 or solvent were also added. The cells were lysed 16 h after onset of stimulation using the reporter gene lysis buffer (Roche Diagnostics) and the constant light signal luciferase reporter gene assay was performed as recommended by the supplier (Canberra-Packard, Groningen, The Netherlands). The luciferase activities were normalized with respect to protein concentration and induction factors were calculated as the ratio of luciferase activity of ligand-stimulated cells to that of solvent controls.
RESULTS
Induction of p21(waf1/cip1) mRNA expression by 5-fluorouracil and 1α,25(OH)2D3
In MCF-7 human breast cancer cells the expression of p21(waf1/cip1) mRNA was monitored by real-time quantitative RT–PCR in relation to the control gene ARP0 over a time period of 0 to 360 min (Figure 1). Within this time the p53 inducer 5-fluorouracil provided a steady increase of p21(waf1/cip1) mRNA expression up to a level of 5.2-fold after 360 min of stimulation (Figure 1A). The natural VDR ligand 1α,25(OH)2D3 also induced p21(waf1/cip1) mRNA expression, but weaker and more transiently than 5-flurouracil, showing a maximum of 1.6-fold induction after 120 min of stimulation (Figure 1B). The response of the p21(waf1/cip1) gene to 1α,25(OH)2D3 is comparable to that of the primary VDR target gene cyclin C [showing a maximum 1.8-fold induction after 2 h (14)]. Moreover, both genes show a high basal expression, which is ~10 000-fold higher than that of the most responsive 1α,25(OH)2D3 target, the 24-hydroxylase (CYP24) gene [compare ref. (14) and data not shown].
000-fold higher than that of the most responsive 1α,25(OH)2D3 target, the 24-hydroxylase (CYP24) gene [compare ref. (14) and data not shown].
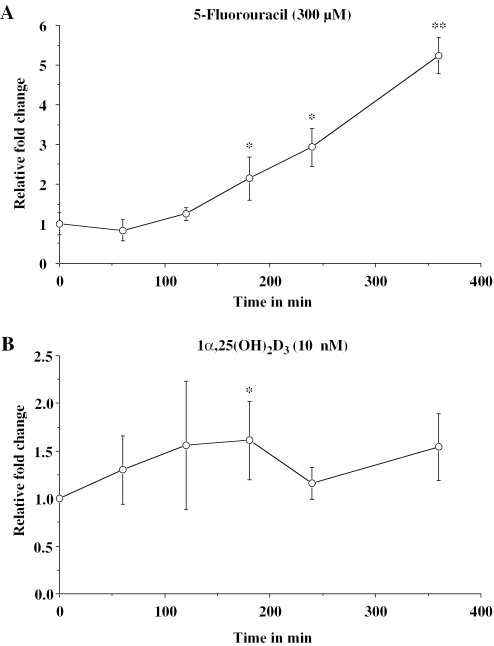
Time course of p21(waf1/cip1) mRNA expression. Real-time quantitative RT–PCR was used to determine p21(waf1/cip1) mRNA expression relative to the control gene ARP0 in MCF-7 human breast cancer cells in response to 300 µM 5-fluorouracil (A) or 10 nM 1α,25(OH)2D3 (B) over a time period of 360 min. Data points indicate the means of at least three independent cell treatments and the bars represent standard deviation. Two-tailed Student's t-tests were performed to determine the significance of the mRNA induction in reference to solvent controls (*P < 0.05, **P < 0.01).
Whole p21(waf1/cip1) promoter screening for p53 binding
In order to localize novel p53 and VDR binding sites (and to challenge already known REs) within the human p21(waf1/cip1) promoter, we designed 19 overlapping primer pairs that cover evenly the first 7.1 kb of chromosomal DNA upstream of the TSS (Figure 2A). To minimize the number of primers in repetitive sequences, we employed the web-based CENSOR server screening service (29) to identify repetitive sequences in the promoter sequence. We determined that the repetitive sequence content of this segment of human chromosome 6 is 24.2%. Where possible the remaining 73.8% unique sequence was used to design the overlapping PCR primer pairs, but for regions 9, 10, 14, 15, 17 and 18 one of the primers is located in repetitive sequence (Table 1). Moreover, due to unspecific binding of IgG to promoter region 12 (Figure 2B), we designed an alternative primer pair detecting region 12C (Figure 2A).
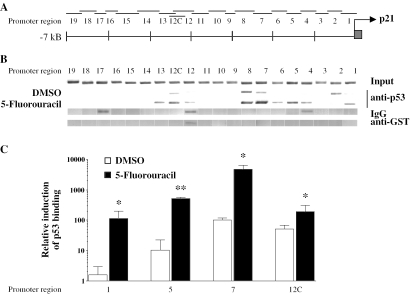
Location of p53 protein within chromatin on human p21(waf1/cip1) promoter. Nineteen overlapping regions evenly cover the first 7.1 kb of the human p21(waf1/cip1) promoter (A) Due to IgG binding to region 12 an alternative region 12C was chosen (Table 1). Chromatin was extracted from MCF-7 cells that had been treated for 360 min with solvent (DMSO) or 300 µM fluorouracil. ChIP assays using an antibody against p53 were performed (B) Precipitations with IgG and anti-GST antibody served as controls for the specificity of the p53 detection. Representative agarose gels of the input lane confirm the comparable detection sensitivity of the 20 different promoter regions. The induction of p53 binding by 5-fluorouracil to promoter regions 1, 5, 7 and 12C was determined by quantitative real-time PCR (C) Columns indicate the means of at least three independent cell treatments and the bars represent standard deviations. A one-tailed Student's t-tests was performed to determine the significance of the increase in p53 binding in fluorouracil-treated samples in reference to solvent controls (*P < 0.05, **P < 0.01).
Chromatin was extracted from MCF-7 cells that were grown overnight in the presence of 5% charcoal-treated fetal bovine serum, stimulated for 360 min with solvent (DMSO) or 300 µM 5-fluorouracil and then cross-linked for 10 min in the presence of formaldehyde. ChIP assays were performed using an antibody against p53 and representative agarose gels of the PCR products are shown (Figure 2B). Comparable detection sensitivity for the 20 different p21(waf1/cip1) promoter regions was demonstrated by representative PCR products that were obtained using DNA liberated from the recovered chromatin by reverse cross-linking (input lane). Immunoprecipitations with IgG and anti-glutathione S-transferase (GST) antibodies served as negative controls. PCR products obtained with p53-enriched chromatin visualized the localization of p53 to the first 7.1 kb of the p21(waf1/cip1) promoter. We found induction of p53 binding to promoter regions 1, 5, 7 and 12C. The binding to region 4 may be unspecific (see IgG control lane), while the binding to regions 2, 6 and 13 may represent flanking effects of the ChIP method.
The relative binding of p53 to regions 1, 5, 7 and 12C was also determined by quantitative real-time PCR (Figure 2C). Strongest induction (70-fold) was found on the TSS (region 1), followed by region 5 (48-fold) and region 7 (46-fold), while on region 12C only a 3.7-fold induction of p53 binding could be detected. Interestingly, regions 7 and 12C showed significantly higher basal association of p53 than regions 1 and 5. For regions 5 and 7 this confirms the findings of a previous study (30), further downstream regions of the p21(waf1/cip1) promoter have not been studied before. Taken together, we could confirm the expected binding of p53 to promoter regions 5 and 7 and could show for the first time binding of p53 protein to region 12C. We presume the association of p53 with the TSS region (region 1) is most likely a consequence of chromatin looping of distal regions (e.g. regions 5, 7 and 12C) to bring them in contact with Pol II.
Whole p21(waf1/cip1) promoter screening for VDR binding
ChIP assays were performed using an antibody against VDR on chromatin that was extracted from MCF-7 cells after stimulation for 0, 15, 30, 60, 120 and 180 min with 10 nM 1α,25(OH)2D3. Representative agarose gels of the PCR products obtained with VDR-enriched chromatin visualized the time-dependent localization of VDR to the first 7.1 kb of the p21(waf1/cip1) promoter (Figure 3). We found significant VDR binding to promoter regions 7, 12C and 19. The VDR binding to the TSS (region 1) is again most likely the sign of complexes of Pol II with VDR-associated with distal promoter regions. We consider signals at regions 2 and 13 as flanking effects and the apparent association of VDR to regions 4, 12 and 17 as unspecific, since these two regions also bind IgG (see Figure 2B). In summary, as an essential condition for a primary VDR target gene, the TSS of the human p21(waf1/cip1) gene associates with VDR. Moreover, the first 7.1 kb of the promoter contain three distal VDR binding sites in regions 7, 12C and 19.
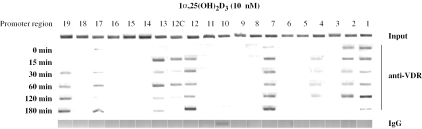
Ligand-modulated VDR binding to the human p21(waf1/cip1) promoter. Chromatin was extracted from MCF-7 cells that had been treated for indicated time periods with 10 nM 1α,25(OH)2D3. ChIP assays using an antibody against VDR were performed. The VDR and RXR association of the 20 regions of the human p21(waf1/cip1) promoter was monitored for the six treatment times. Precipitations with IgG served as controls for the specificity of the VDR detection. Representative agarose gels of at least three independent cell treatments are shown.
Identification of p53-REs and VDREs on the human p21(waf1/cip1) promoter
The ChIP assays with anti-p53 and anti-VDR antibodies (Figures 2 and and3)3) suggest that the human p21(waf1/cip1) promoter contains multiple responsive regions for both p53 and VDR. In order to challenge this prediction, we performed in silico screening of the first 7.1 kb of the p21(waf1/cip1) promoter for p53 consensus sequence RRRCWWGYYY-N-RRRCWWGYYY and for DR3-, DR4- and ER9-type VDREs [consensus sequence for each half-site RGKTCA allowing one mismatch per half-site (23)]. We found five putative p53-REs, of which three are located in promoter region 7 and one each in regions 5 and 12 (Figure 4). Moreover, we found eight putative VDREs, of which four have a DR3-type structure, three are DR4-type REs and one is an ER9-type RE. Each two of these VDREs were found in regions 4, 7 and 19, while each one was found in regions 3 and 12. The DR3-type VDRE in region 3 has been published earlier (4). No putative RE was found in region 1.
In vitro binding of p53 and VDR–RXR heterodimers to putative REs
To confirm further the physical association between p53 and the identified regions gelshift experiments were performed on double-stranded oligonucleotides containing the five putative p53-REs sequences, using recombinant baculovirus expressed p53 protein (Figure 5A). An oligonucleotide with mutated p53 binding sites served as negative control. Strong p53 binding was observed to the known p53 binding site within region 7 (p53-RE2), while the p53-REs 3, 4 (region 7) and 5 (region 12) showed only 10–12% of this binding. The p53-RE1 (region 5) showed only very week complex formation with p53 protein (3%); this may reflect the fact that this sequence appears to diverge significantly from the established consensus p53 binding sequence (12). The negative control did not show any p53 binding at all.
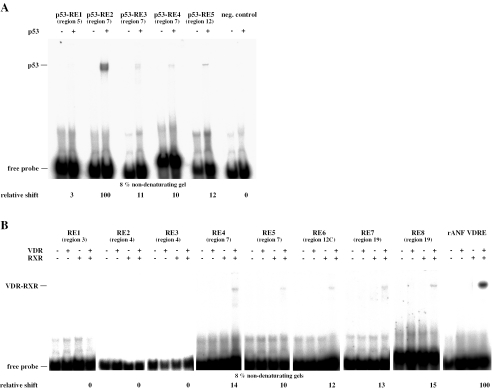
In vitro analysis of p53-REs and VDREs within the human p21(waf1/cip1) promoter. Gelshift experiments were performed with recombinant p53 protein (A) and in vitro translated VDR and RXRα alone or in combination (B) and in the presence of 32P-labeled REs. An oligonucleotide with mutated binding sites was used as negative control for p53 binding. Protein–DNA complexes were resolved from free probe through non-denaturing 8% polyacrylamide gels. Representative gels are shown. The relative amount of p53 and VDR–RXR heterodimer binding was quantified on a FLA-3000 reader in relation the known p53 binding site p53-RE2 and the rat ANF DR3-type VDRE, respectively. NS indicates non-specific complexes.
To assess basal association between the eight putative VDREs the gelshift assays were performed with in vitro translated VDR and RXR proteins, either alone or in combination (Figure 4B) under conditions identical to our earlier DR3-type VDRE comparative study (23). The DR3-type VDRE of the rat ANF gene promoter (28) served as a positive control. In reference to this control, REs 4, 5 (region 7), 6 (region 12C) and 7, 8 (region 19) showed 10–15% of VDR–RXR complex formation. In contrast, REs 1, 2 and 3 in regions 3 and 4, showed no binding of VDR–RXR heterodimers. Taken together, our in silico/in vitro scanning for REs in the human p21(waf1/cip1) promoter provided one strong p53-RE, four weaker p53-REs and five VDREs in regions 5, 7, 12C and 19. In contrast, under our stringent gelshift conditions no binding of VDR–RXR heterodimers to the described VDRE (RE1) in region 3 could be detected.
Functionality of p53 and VDR binding site containing regions of the p21(waf1/cip1) promoter
PCR fragments representing regions 1, 2, 3, 5, 7, 12C and 19 of the human p21(waf1/cip1) promoter were fused with the thymidine kinase promoter driving the firefly luciferase reporter gene. The reporter gene constructs were transfected into MCF-7 cells, stimulated for 16 h with 100 nM 1α,25(OH)2D3 or 100 µM 5-fluorouracil and relative reporter gene activity was measured (Figure 6). Promoter regions 7, 12C and 19 showed statistically significant induction by 1α,25(OH)2D3 (1.5- to 1.9-fold), while regions 5, 7 and 12C were also 1.5- to 1.9-fold inducible by 5-fluorouracil (Figure 6A). Regions 1, 2 and 3 were inducible by neither of the two stimuli. Interestingly, the basal activity of the reporter gene constructs varied significantly with low activity of region 19, intermediate activity of regions, 1, 2, 3 and 12C and strong activity of regions 5 and 7 suggesting varying amounts of other transcription factors associating with these regions.
In order to analyze the contribution of the different REs within promoter region 7, we introduced point mutants into the complex VDRE 4/5 and p53-REs 2, 3 and 4 as indicated in Figure 6B. Reporter gene assays in MCF-7 cells indicated that all three half-sites of VDREs 4 and 5 are critical for the response of the promoter region to 1α,25(OH)2D3 (see mutants 1, 2 and 3 in Figure 6B) and that the p53-RE2 is essential for the response to 5-fluorouracil (mutant 4). Moreover, mutation of the p53-RE2 reduced the basal activity by a factor of 20. Interestingly, the mutated promoter region was still responsive to 1α,25(OH)2D3. In contrast, mutations in p53-REs 3 and 4 did not affect the responsiveness of the promoter construct to 5-flurouracil or 1α,25(OH)2D3 (mutants 5, 6 and 7).
In summary, the functional assays confirmed the results from the ChIP scanning, in silico screening and in vitro binding studies. Namely that regions 5, 7 and 12C contain functional p53 binding sites and regions 7, 12C and 19 have functional VDREs. The known p53-RE2 was confirmed to be of central importance for the basal activity of the p21(waf1/cip1) promoter and the complex VDRE was shown to mediate the response of promoter region 7 to 1α,25(OH)2D3. In contrast, there is neither direct p53 nor VDR binding in response to their inducers within the proximal p21(waf1/cip1) promoter (regions 1, 2 and 3).
Co-localization of VDR and its partner proteins
An additional test for the functionality of the VDREs in the chromatin context of MCF-7 cells was performed by re-ChIP experiments in response to 0, 15, 30, 60, 120 and 180 min treatment with 10 nM 1α,25(OH)2D3 (Figure 7). Initial rounds of immuno-precipitation were performed with the VDR antibody and then the precipitated material was interrogated further with antibodies against CBP, SRC-1 and TRAP220. This approach was undertaken to enrich for chromatin templates that were associated with both VDR and its partner proteins at the same time. Single ChIP assays with anti-phosphorylated Pol II antibodies served as reference for the transcriptional activity of the test promoter regions. From the double-fractionated chromatin template, VDR was found to associate in a 1α,25(OH)2D3-stimulated and time-dependent fashion with CBP, SRC-1 and TRAP220 on the p21(waf1/cip1) promoter regions 1 (TSS), 7, 12C and 19, but not on the negative control region 3. The association of VDR with its partner proteins showed individual profiles. On regions 7 and 19 there is rather constant association of VDR with CBP, while on region 12C and the TSS there was a peak after 15 to 60 min of ligand treatment. The respective VDR-CBP co-localization on the TSS was also rather constant, but showed a maximum at time point 15 min. Some association of VDR with SRC-1 was observed at nearly all time points with maxima at 0 and 60 min and a minimum at 120 min for region 7, a maximum between 15 and 60 min for region 12C and maxima for time points 0, 30 and 180 min for region 19. These rather divergent VDR-SRC-1 association profiles were integrated at the TSS with maxima at 15 and 120 min. The VDR-TRAP220 association was found to peak at 60 min for region 7, and at 60 to 180 min for regions 12C and 19. The TSS showed some VDR-TRAP220 association already after 15 min, but the main interaction was observed at 60 to 180 min. For regions 7 and 12C the activity of phosphorylated Pol II was highest at 60 min, while for region 19 rather constant activity was detected and even region 3 showed low level of constant association of Pol II. The TSS integrates these activities by high basal association with phosphorylated Pol II over all time points with a slight maximum at 60 min.
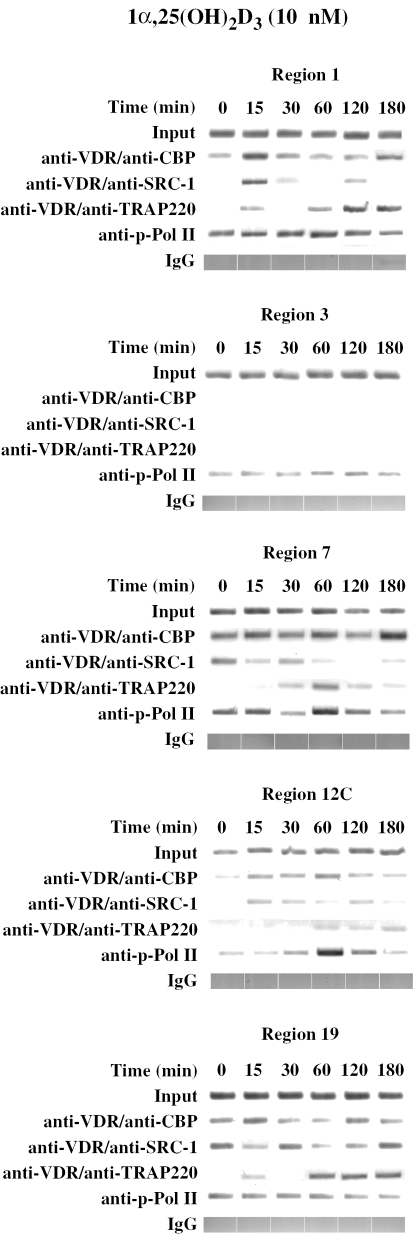
VDR complexes on 1α,25(OH)2D3-responsive p21(waf1/cip1) promoter regions. Chromatin was extracted from MCF-7 cells that had been treated for indicated time periods with 10 nM 1α,25(OH)2D3. Re-ChIP experiments were performed with a first immuno-precipitation with anti-VDR and a second precipitation with anti-CBP, anti-SRC-1 and anti-TRAP220 antibodies as indicated. Regular ChIP assays with anti-phosphorylated Pol II antibody serve as a positive control and precipitations with IgG as specificity controls. The associations of VDR and its partner proteins were monitored on the human p21(waf1/cip1) promoter regions 1 (TSS), 3 (negative control), 5, 7, 12C and 19. Representative agarose gels are shown.
Taken together, re-ChIP assays confirmed 1α,25(OH)2D3-dependent functionality of promoter regions 7, 12C and 19, by demonstrating individual association profiles with different CoA proteins. Equally the data generally supported the concept of initial, rapid recruitment of SRC family members CoAs, followed subsequently by members of the mediator complex.
DISCUSSION
In recent years 1α,25(OH)2D3 and its low-calcemic analogues have emerged as promising anti-cancer agents. However, the mechanisms of the anti-proliferative, pro-differentiating and pro-apoptotic effects of VDR ligands vary and are cell-specific. Therefore, they cannot be explained by a single molecular pathway in all cell types, but the status of the p53 gene, being mutated in over 50% of human tumors (31), may be of critical impact in most cases. The p53 protein is well known as the master regulator of the p21(waf1/cip1) gene, which in turn for the last decade has appeared to be the most promising 1α,25(OH)2D3 target concerning cell growth regulatory effects of the nuclear hormone. In this study we have undertaken whole promoter ChIP assays covering the first 7.1 kb of the human p21(waf1/cip1) to characterize the critical p53 and VDR binding sites (for summary see Table 2). For each of the two transcription factors we found three binding regions within the promoter. Interestingly, two of these, regions 7 and 12C at approximate positions −2300 and −4500 relative to the TSS, were identical for both, i.e. they contain both p53-REs and VDREs. This leaves only region 5 at position −1400 unique for p53 binding and region 19 at position −6900 restricted to VDR binding. Functional studies confirmed the responsiveness of the respective regions to the p53 inducer 5-fluorouracil and VDR activation by 1α,25(OH)2D3, respectively.
Table 2
Summary of the characterization of the human the p21(waf1/cip1) promoter by ChIP, gelshift and reporter gene assays
| Region no. | p53 ChIP | VDR ChIP | Functional p53-RE | Functional VDRE | Response to 5-fluorouracil | Response to 1α,25(OH)2D3 | CoA binding in re-ChIP |
|---|---|---|---|---|---|---|---|
| 1 (TSS) | + | + | − | − | − | − | + |
| 3 | − | − | − | − | − | − | − |
| 5 | + | − | + | − | + | − | ND |
| 7 | + | + | + | + | + | + | + |
| 12/12C | + | + | + | + | + | + | + |
| 19 | − | + | − | + | − | + | + |
The p53 binding regions 5 and 7 have been well characterized by others by a range of approaches including ChIP assays (16,30,32). However, due to the fact that most analyses of the p21(waf1/cip1) promoter were restricted to the first 3 kb, the novel p53 binding site at position −4000 (region 12) have been overlooked so far. In vitro gelshift analysis indicated a 10-fold lower binding affinity of this novel site, compared to the perfect site at position −2300 (region 7), and quantitative real-time PCR from released chromatin template in ChIP assays detected 10-fold less induction of p53 binding to that p21(waf1/cip1) promoter region. In contrast, this novel p53 binding region demonstrated significant induction after 5-fluorouracil treatment in reporter gene assays and rather high basal association with p53 in quantitative ChIP assays.
Binding of p53 to the p21(waf1/cip1) promoter in the absence of genotoxic signals have also been observed previously (32). Since the p21(waf1/cip1) promoter contains one high- and several low-affinity sites, it may support a hypothesis that affinity dictates the choice between cell cycle arrest and apoptosis, with high-affinity sites regulating cell cycle arrest and low-affinity sites regulating pro-apoptotic effects (33). In cells with a wild-type p53 gene the novel p53-RE at position −4000 may be of minor impact for the response of the p21(waf1/cip1) gene to cellular stress mediated by p53, but still critical for the induction of apoptosis. Moreover, in cases of p53 gene mutations the elevated p53 protein association to region 12 and the location of a functional VDRE may become of higher impact. The same holds true for region 7, which contains a very potent p53-REs and a complex VDRE. Therefore, we hypothesize that high basal p53 binding paired with closely located VDREs may represent a integrated system to assure sufficient p21(waf1/cip1) mRNA expression even in cases where regular p53 signaling is impaired by mutations.
In cells, such as the MCF-7 breast cancer cells that were used in this study, having a wild-type p53 gene, the basal p21(waf1/cip1) mRNA expression appear elevated, compared to other VDR target genes, such as CYP24. The drastic loss of basal activity of promoter region 7 by mutagenesis of its potent p53-RE supports this view. Moreover, possibly reflecting this treatment with 1α,25(OH)2D3 results only in a minor 1.6-fold further elevation of mRNA levels. This may indicate that cells with wild-type p53 gene have already sufficiently high p21(waf1/cip1) protein levels, so that they need not take advantage of the 1α,25(OH)2D3 signaling pathway to further increase their p21(waf1/cip1) expression. This may also explain the varied observations concerning the response of the p21(waf1/cip1) gene to 1α,25(OH)2D3, found by various groups. The adjacent functional response elements for p53 and VDR suggest that these factors co-operate in normal breast epithelial cells, perhaps to regulate p21(waf1/cip1) mRNA levels, such that in 1α,25(OH)2D3-replete environments the p53 activation of the gene is very readily undertaken to bring about cell cycle control and/or programmed cell death. Anyway, the demonstration of functional 1α,25(OH)2D3-responding promoter regions confirms that the p21(waf1/cip1) is a primary 1α,25(OH)2D3-responding gene.
The CYP24 gene probably is the most responsive primary VDR target gene, because its protein product leads to the degradation of 1α,25(OH)2D3 and therefore the eventual extinction of the transcriptional signal (13). Therefore, it makes sense that VDR–RXR heterodimers are the central transcription factors on the CYP24 promoter and that the basal expression of the gene in the absence of an activating signal for VDR–RXR heterodimers is very low (14). Thus the CYP24 gene appears to be somewhat of a special example of VDR gene regulation. In contrast, the basal expression of other VDR target genes, such as cyclin C, PPARδ and p21, is 10 000- to 100
000- to 100 000-times higher as that of the CYP24 gene (14,34) suggesting that also other transcription factors than VDR–RXR heterodimers contribute to the gene's activity. Therefore, the stimulation of the gene's mRNA expression is elevated from a high level only 1.5- to 3.0-fold, which still represents the production of more mRNA molecules than in case of a 1000-fold stimulation of the CYP24 gene. The promoters for the genes CYP24 and cyclin C contain four VDREs (13,14) and the insulin-like growth factor binding proteins 1, 3 and 5 genes each carry three VDREs in their promoter (15). Thus the number of VDREs within a promoter does not correlate with the inducibility of a VDR target gene. The p21 promoter contains three 1α,25(OH)2D3-responsive regions (7, 12C and 19, see also Table 2), each of which show 1α,25(OH)2D3-dependent recruitment of CoA proteins to VDR-bound chromatin regions. Moreover, each of the three promoter regions contains at least one VDRE. This confirms again the rule of the multiple VDREs in primary 1α,25(OH)2D3-responsive genes. Please note that the previously suggested VDRE [RE1 (4)] is not binding VDR–RXR heterodimers and is located in the non-1α,25(OH)2D3-responsive promoter region 3 (position −750).
000-times higher as that of the CYP24 gene (14,34) suggesting that also other transcription factors than VDR–RXR heterodimers contribute to the gene's activity. Therefore, the stimulation of the gene's mRNA expression is elevated from a high level only 1.5- to 3.0-fold, which still represents the production of more mRNA molecules than in case of a 1000-fold stimulation of the CYP24 gene. The promoters for the genes CYP24 and cyclin C contain four VDREs (13,14) and the insulin-like growth factor binding proteins 1, 3 and 5 genes each carry three VDREs in their promoter (15). Thus the number of VDREs within a promoter does not correlate with the inducibility of a VDR target gene. The p21 promoter contains three 1α,25(OH)2D3-responsive regions (7, 12C and 19, see also Table 2), each of which show 1α,25(OH)2D3-dependent recruitment of CoA proteins to VDR-bound chromatin regions. Moreover, each of the three promoter regions contains at least one VDRE. This confirms again the rule of the multiple VDREs in primary 1α,25(OH)2D3-responsive genes. Please note that the previously suggested VDRE [RE1 (4)] is not binding VDR–RXR heterodimers and is located in the non-1α,25(OH)2D3-responsive promoter region 3 (position −750).
In conclusion, our study provides insight into the regulation of the human p21(waf1/cip1) gene by 1α,25(OH)2D3. Further we demonstrate that whole promoter ChIP screening with anti-VDR and anti-p53 antibodies is suitable for RE identification and that within 7.1 kb of the p21(waf1/cip1) promoter there are five VDREs and five p53-REs, being located, often in clusters, at positions −1400, −2300, −4500 and −6900. This may help to understand the physiological function of these pathways in the process of normal mammary gland renewal and anti-proliferative effects of 1α,25(OH)2D3 in p53 in cancer cells.
Acknowledgments
The authors would like to thank Dr Lise Binderup for 1α,25(OH)2D3 and Dr Christian Frank for support in in silico analysis. Grants from the Academy of Finland, the Finnish Cancer Organization and the Juselius Foundation (to C.C.), the British Association for Cancer Research (to C.M.B.) and the BBSRC (to M.J.C.) supported this research. The authors declare they have no conflict of interest. Funding to pay the Open Access publication charges for this article was provided by Academy of Finland.
Conflict of interest statement. None declared.
REFERENCES
Articles from Nucleic Acids Research are provided here courtesy of Oxford University Press
Full text links
Read article at publisher's site: https://doi.org/10.1093/nar/gkj460
Read article for free, from open access legal sources, via Unpaywall:
https://academic.oup.com/nar/article-pdf/34/2/543/7129291/gkj460.pdf
Citations & impact
Impact metrics
Citations of article over time
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1093/nar/gkj460
Article citations
Crosstalk between BER and NHEJ in XRCC4-Deficient Cells Depending on hTERT Overexpression.
Int J Mol Sci, 25(19):10405, 27 Sep 2024
Cited by: 0 articles | PMID: 39408734 | PMCID: PMC11476898
Vitamin D in Cancer Prevention and Treatment: A Review of Epidemiological, Preclinical, and Cellular Studies.
Cancers (Basel), 16(18):3211, 20 Sep 2024
Cited by: 0 articles | PMID: 39335182 | PMCID: PMC11430526
Review Free full text in Europe PMC
Downregulation of Histone H3.3 Induces p53-Dependent Cellular Senescence in Human Diploid Fibroblasts.
Genes (Basel), 15(5):543, 25 Apr 2024
Cited by: 0 articles | PMID: 38790171 | PMCID: PMC11121134
FOXO1 promotes cancer cell growth through MDM2-mediated p53 degradation.
J Biol Chem, 300(4):107209, 21 Mar 2024
Cited by: 5 articles | PMID: 38519029 | PMCID: PMC11021968
The Influence of Single Nucleotide Polymorphisms on Vitamin D Receptor Protein Levels and Function in Chronic Liver Disease.
Int J Mol Sci, 24(14):11404, 13 Jul 2023
Cited by: 3 articles | PMID: 37511164 | PMCID: PMC10380285
Go to all (153) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Regulation of the human cyclin C gene via multiple vitamin D3-responsive regions in its promoter.
Nucleic Acids Res, 33(8):2440-2451, 29 Apr 2005
Cited by: 37 articles | PMID: 15863722 | PMCID: PMC1087898
The impact of chromatin organization of vitamin D target genes.
Anticancer Res, 26(4a):2637-2645, 01 Jul 2006
Cited by: 15 articles | PMID: 16886674
Review
The human peroxisome proliferator-activated receptor delta gene is a primary target of 1alpha,25-dihydroxyvitamin D3 and its nuclear receptor.
J Mol Biol, 349(2):248-260, 07 Apr 2005
Cited by: 119 articles | PMID: 15890193




