Abstract
Free full text

Regulation of Gross Chromosomal Rearrangements by Ubiquitin and SUMO Ligases in Saccharomyces cerevisiae
Abstract
Gross chromosomal rearrangements (GCRs) are frequently observed in many cancers. Previously, we showed that inactivation of Rad5 or Rad18, ubiquitin ligases (E3) targeting for proliferating cell nuclear antigen (PCNA), increases the de novo telomere addition type of GCR (S. Smith, J. Y. Hwang, S. Banerjee, A. Majeed, A. Gupta, and K. Myung, Proc. Natl. Acad. Sci. USA 101:9039-9044, 2004). GCR suppression by Rad5 and Rad18 appears to be exerted by the RAD5-dependent error-free mode of bypass DNA repair. In contrast, Siz1 SUMO ligase and another ubiquitin ligase, Bre1, which target for PCNA and histone H2B, respectively, have GCR-supporting activities. Inactivation of homologous recombination (HR) proteins or the helicase Srs2 reduces GCR rates elevated by the rad5 or rad18 mutation. GCRs are therefore likely to be produced through the restrained recruitment of an HR pathway to stalled DNA replication forks. Since this HR pathway is compatible with Srs2, it is not a conventional form of recombinational pathway. Lastly, we demonstrate that selection of proper DNA repair pathways to stalled DNA replication forks is controlled by the Mec1-dependent checkpoint and is executed by cooperative functions of Siz1 and Srs2. We propose a mechanism for how defects in these proteins could lead to diverse outcomes (proper repair or GCR formation) through different regulation of DNA repair machinery.
Transmission of genetic information without deleterious alterations is one of the most important tasks for a cell to achieve in every cell cycle. To cope with this task, cells have evolved systems that survey, alert, and repair potentially lethal DNA damage (53, 54). However, in situations where such systems are impaired, DNA damage accumulates and causes genetic changes. Accumulation of genetic changes, which is defined as a genomic instability, is frequently observed in various types of genetic disorders, including cancers (31, 67). Genomic instability has been documented as a preceding step for multiple inactivations of tumor suppressor genes and activations of proto-oncogenes (26, 34, 37). One type of genomic instability observed frequently in many cancers is gross chromosomal rearrangement (GCR). GCR includes translocations, deletions of chromosome arms, interstitial deletions, inversions, amplifications, chromosome end-to-end fusion, and aneuploidy (26). Although little is known about the causes and origin of GCR in cancer cells, recent studies on genes mutated in inherited cancer predisposition syndromes have demonstrated that proteins functioning in DNA damage responses, DNA repair, and DNA recombination play crucial roles in the suppression of spontaneous and/or DNA damage-induced GCRs (12, 25, 44).
To understand the mechanisms by which GCR is suppressed and which proteins are required to generate GCRs in the absence of correct DNA repair, several quantitative assays were developed in Saccharomyces cerevisiae (7, 20, 40). A yeast GCR assay that can measure the rate of accumulation of different classes of genome rearrangements has been used to study pathways for GCR. This assay can detect interstitial deletions or nonreciprocal translocations with microhomology, nonhomology, or divergent homology (referred to as homeology) at the rearrangement breakpoint; chromosome fusions; and deletion of a chromosome arm combined with addition of a new telomere (referred to as de novo telomere addition). Through extensive genetic analysis and screening, seven pathways that suppress and four pathways that are required for the formation of GCRs have been identified. Seven pathways for the suppression of GCRs include the following: (i) at least three different cell cycle checkpoints that function during DNA replication (3, 20, 24, 32, 40, 41, 65), (ii) recombination pathways whose genetic requirements resemble those of break-induced replication (BIR) (38), (iii) a pathway that suppresses de novo telomere additions (38), (iv) at least two pathways for proper chromatin assembly during DNA replication (42), (v) pathways that prevent chromosome ends from being joined to each other and to broken DNAs (6, 38, 47, 52), (vi) a mismatch repair pathway that prevents recombination between divergent DNA sequences (39), and (vii) pathways that detoxify reactive oxygen species (20, 21, 60). Four pathways required for the formation of GCRs are the following: (i) telomerase and its accessory proteins for de novo telomere addition (38, 47, 52), (ii) mitotic checkpoint and mitotic exit network (43), (iii) the Rad1-Rad10 endonuclease complex (22), and (iv) ligase 4 and Lif1 (38).
To further extend our knowledge of GCR suppression mechanisms, we recently screened the entire yeast nonessential open reading frames and identified 10 additional genes (ALO1, CDC50, CSM2, ELG1, ESC1, MMS4, RAD5, RAD18, TSA1, and UFO1), mutations of which increased the GCR rate (60).
In the yeast Saccharomyces cerevisiae, DNA repair genes are classified into three epistasis groups (13). The RAD3 epistasis group functions in nucleotide excision repair (51) and has only limited implications in the suppression of GCRs. The RAD52 epistasis group directs double-strand break (DSB) repair mainly through homologous recombination (HR) (64). We have shown that BIR, a type of HR, plays an important role in the suppression of GCRs (38). The RAD6 epistasis group mediates postreplication repair (PRR), which resolves stalled DNA replication forks (4). PRR can be divided into two major pathways, namely, translesion synthesis (TLS) and error-free mode of bypass. When DNA replication machinery encounters a damaged DNA template, ubiquitin ligase (E3) Rad18 along with the ubiquitin-conjugating enzyme (E2) Rad6 monoubiquitinates proliferating cell nuclear antigen (PCNA) on lysine 164 (18). PCNA is a homotrimeric protein, which functions to load different DNA polymerases or DNA repair machinery on DNA (4). Monoubiquitinated PCNA switches a replicative DNA polymerase to nonessential TLS DNA polymerases, such as DNA polymerase ζ encoded by REV3/REV7 or DNA polymerase η encoded by RAD30 (the RAD18-dependent pathway) (10, 50). In certain conditions, Rad5 (E3) along with the Ubc13-Mms2 (E2 and E2 variant, respectively) complex adds a noncanonical lysine 63 (K63)-linked polyubiquitin chain to the monoubiquitinated lysine residue of PCNA. Polyubiquitinated PCNA recruits the error-free mode of bypass (the RAD5-depedent pathway), which presumably involves template switching to the undamaged nascent sister chromatid (29, 58). Since the monoubiquitination of PCNA by Rad18 is required for the further polyubiquitination by Rad5, the RAD5-dependent pathway is also dependent on RAD18. Furthermore, the same lysine 164 of PCNA is also alternatively modified with small ubiquitin-like modifier (SUMO) catalyzed by Siz1 SUMO ligase (18). Although the biological significance of SUMOylation of PCNA is yet to be fully characterized, it has been suggested that the SUMOylated PCNA physically recruits Srs2 to stalled DNA replication forks and suppresses the unscheduled recombination (17, 18, 46, 49, 62).
Accumulating evidence suggests roles of PRR proteins in genomic stability. For instance, the targeted mutation of the mouse RAD18 gene in embryonic stem cells increased genomic instability, including sister chromatid exchange, homologous recombination, and illegitimate recombination (66). Mutations of XPV, the mammalian homolog of RAD30 that encodes the TLS polymerase η, were frequently found in xeroderma pigmentosum variant syndrome (23, 36). However, the molecular mechanisms of GCR suppression by PRR and GCR formation in the absence of PRR are poorly understood. It is also unclear how PRR communicates with other DNA repair pathways to suppress genomic instability.
In the present study, we demonstrate that the RAD5-dependent error-free mode of bypass PRR pathway is central to suppressing GCR formation. In the absence of the error-free mode of bypass, GCRs are generated through the illegitimate recruitment of a recombination pathway, which appears to be different from conventional recombination pathways. Siz1 cooperates with Srs2 to execute switching between the PRR and HR repair pathways. The uncoordinated regulation of these proteins due to the defective signaling by improper PCNA modifications may lead to GCR in the absence of Rad5 and/or Rad18.
MATERIALS AND METHODS
General genetic methods.
Methods for the construction and propagation of gene-disrupted strains were described previously (7, 40). The sequences of primers used to generate gene-knockout cassettes and to confirm correct disruption are available upon request. All S. cerevisiae strains used in this study were derived from the S288c strain RDKY3615 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3). Genotypes of each strain used for the GCR assay are listed in Table Table11.
TABLE 1.
Saccharomyces cerevisiae strains used in this study
 |
Construction of pol30-119(K164R) and srs2(K41R) strains.
The strain that has a pol30-119(K164R) mutation integrated at the POL30 genomic locus was generated by transforming RDKY3615 with a SacII fragment of plasmid pCH1654 (1, 17) and designated as YKJM2624. UV-sensitive clones were selected, and the integration of mutation was confirmed by genomic DNA sequencing. The srs2(K41R) mutation was introduced into the strain RDKY3023 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8) by the pop-in and pop-out method using the BglII-linealized plasmid pHK286 (27), and the resulting strain, YKJM2897, was selected by testing methyl methanesulfonate (MMS) sensitivity. The presence of mutation was further confirmed by the presence of a restriction enzyme StyI site at the mutation site and DNA sequencing. URA3 was then incorporated at the HXT13 locus to create YKJM3034.
Characterization of spontaneous GCR rates.
All GCR rates were determined by fluctuation analysis using the method of the median with at least two independent clones. The average GCR rates from at least two or more independent experiments using either 5 or 11 cultures for each clone are reported as previously described (7, 30, 40). The pif1-m2 mutation inactivates the telomerase-inhibitory activity of Pif1. Since the pif1-m2 mutation shows a synergistic increase in GCR rates with strains carrying many GCR mutator mutations (38), some mutations were examined in the pif1-m2 background.
Sensitivity to MMS.
Cells in the exponential phase were serially diluted, and 5 μl of cells was spotted on different plates. After 2 to 3 days of incubation at 30°C, pictures were taken.
RESULTS
Rad5-dependent PRR is a major pathway to suppress GCRs during DNA replication.
In a previous study, we identified a mutation in either RAD5 or RAD18 that enhanced GCR formation in the genome-wide screening (60). Similar to previous observations, a mutation in either the RAD5 or the RAD18 gene increased the de novo telomere addition type of GCR formation rate 68- or 65-fold compared to the wild type (Table (Table2).2). The rad5 rad18 strain showed an 83-fold increase in the GCR rate, which is comparable to that in the rad5 or rad18 strain (Table (Table2).2). Since the error-free mode of bypass depends on both Rad5 and Rad18, the high GCR rates in the rad5 and rad18 strains seem to be the consequences of defects in the error-free mode of bypass rather than TLS. To confirm this idea, different genes encoding downstream DNA polymerases in the RAD6 epistasis group were mutated either in the wild type or in the pif1-m2 strain, and the GCR rate of each strain was determined. REV1 (deoxycytidyl transferase), REV3, REV7 (heterodimer subunits of Polζ), RAD30 (Polη), and POL32 (a subunit of Polδ) encode DNA polymerases, which bypass DNA damage by TLS in the PRR pathway (10, 50). Mutations in these genes had no significant effect on the GCR formation rate (Table (Table2).2). Even simultaneous deletion of various polymerases showed a minimum increase in the GCR rates (Table (Table2).2). Therefore, the GCR increases in the rad5 and rad18 strains are caused by the defect in the error-free mode of bypass. Rad6 is a ubiquitin-conjugating enzyme (E2), which functions with various ubiquitin ligases (E3s), such as Ubr1 (9), Bre1 (72), and Rad18 (2). Ubc13 and Mms2 are an E2 and an E2 variant, respectively, and form an E2 heterodimer complex for Rad5 (19). A mutation in RAD6, UBC13, or MMS2 did not increase the GCR formation rate and did not affect the GCR rate caused by the pif1-m2 mutation (Table (Table22).
TABLE 2.
Defects in proteins functioning in PRR caused different effects in the rate of GCR formation
| Relevant genotypea | Wild type
| pif1-m2
| ||
|---|---|---|---|---|
| Strain number | GCR rate (CANr-5FOAr)b | Strain number | GCR rate (CANr-5FOAr)b | |
| Wild type | RDKY3615 | 3.5 × 10−10 (1) | RDKY4343 | 4.8 × 10−8 (137) |
| rad5Δ | YKJM1385 | 2.4 × 10−8 (68) | YKJM1387 | 2.2 × 10−7 (629) |
| rad18Δ | YKJM1389 | 2.3 × 10−8 (65) | YKJM1391 | 2.5 × 10−7 (714) |
| rev1Δ | YKJM2168 | 8.3 × 10−10 (2) | YKJM1588 | 6.5 × 10−8 (185) |
| rev3Δ | YKJM1558 | 6.4 × 10−10 (2) | YKJM1569 | 5.0 × 10−8 (143) |
| rev7Δ | YKJM2170 | >1.2 × 10−9 (3) | YKJM2231 | 4.5 × 10−8 (130) |
| rad30Δ | YKJM121 | 5.8 × 10−10 (2) | YKJM1552 | 3.7 × 10−8 (106) |
| pol32Δ | YKJM1560 | 6.5 × 10−10 (2) | YKJM1567 | 6.8 × 10−8 (194) |
| ubc13Δ | YKJM1585 | 1.3 × 10−9 (4) | YKJM1579 | 4.6 × 10−8 (130) |
| mms2Δ | YKJM2135 | >2.6 × 10−10 (1) | YKJM2229 | 3.4 × 10−8 (98) |
| rad6Δ | YKJM1573 | 6.1 × 10−10 (2) | YKJM1566 | 2.3 × 10−8 (66) |
Rad5 and Rad18 cooperate with the Mec1-dependent checkpoint to suppress the Tel1-dependent de novo telomere addition type of GCR.
We have identified seven pathways that suppress and four pathways required for GCR formation. To identify which pathway(s) interacts with RAD5 and RAD18, mutations in selected genes from each pathway were tested for their effect on GCR rates (Table (Table3).3). Mec1 is a major transducer kinase that mediates DNA-damage or S-phase checkpoint signals (11). The GCR rates of mec1 rad5 and mec1 rad18 double mutants were comparable to that of a mec1 single mutant (Table (Table3).3). This result is consistent with the fact that stalled DNA replication forks activate the Mec1-dependent replication checkpoint, which in turn causes cell cycle arrest to resolve the stalled DNA replication forks by PRR (33). The sml1 mutation, which is necessary to maintain viability of mec1 cells, was included in all strains carrying the mec1 mutation. Tel1 is another transducer kinase functioning in cell cycle checkpoint redundantly with Mec1. The tel1 mutation inactivates the de novo telomere addition type of GCR (38). An additional tel1 mutation in the rad5 or rad18 strain significantly reduced GCR rates caused by the rad5 or rad18 mutation (Table (Table3).3). Since the primary GCR structure generated from the rad5 or rad18 strain is de novo telomere addition (60), the reduction of GCR rates by the tel1 mutation is likely due to the decrease in the activity for de novo telomere addition. Consistent with this idea, mutations that inactivate de novo telomere addition activity (est2 and yku70) (38) also significantly reduced the GCR rate observed in the rad5 or rad18 strain (Table (Table3).3). Furthermore, the GCR rates of rad5 and rad18 mutants were synergistically increased up to ~600- to 700-fold compared to the wild type when the pif1-m2 mutation that enhances de novo telomere addition activity was added (Table (Table3).3). From these observations, we concluded that the error-free mode of bypass suppresses the Tel1-dependent de novo telomere addition pathway in cooperation with the Mec1-dependent checkpoint.
TABLE 3.
rad5 and rad18 mutations interact differently with other mutations affecting GCR formation
| Relevant genotypea | Wild type
| rad5Δ
| rad18Δ
| |||
|---|---|---|---|---|---|---|
| Strain number | GCR rate (CANr-5FOAr)b | Strain number | GCR rate (CANr-5FOAr)b | Strain number | GCR rate (CANr-5FOAr)b | |
| Wild type | RDKY3615 | 3.5 × 10−10 (1) | YKJM1385 | 2.4 × 10−8 (68) | YKJM1389 | 2.3 × 10−8 (65) |
| mec1Δ sml1Δ | RDKY3735 | 4.6 × 10−8 (131) | YKJM1508 | 5.2 × 10−8 (149) | YKJM1506 | 5.8 × 10−8 (166) |
| tel1Δ | RDKY3731 | 2.0 × 10−10 (1) | YKJM1514 | 1.4 × 10−9 (4) | YKJM1493 | 3.0 × 10−9 (9) |
| est2Δ | RDKY4347 | 1.2 × 10−10 (0.3) | YKJM1564 | 5.9 × 10−10 (2) | YKJM1562 | >3.4 × 10−10 (1) |
| pif1-m2 | RDKY4343 | 4.8 × 10−8 (137) | YKJM1387 | 2.2 × 10−7 (629) | YKJM1391 | 2.5 × 10−7 (714) |
| yku70Δ | RDKY3639 | 1.4 × 10−9 (4) | YKJM1496 | 1.2 × 10−9 (3) | YKJM2137 | 5.5 × 10−9 (16) |
| mre11Δ | RDKY3633 | 2.2 × 10−7 (629) | YKJM1500 | 2.0 × 10−7 (571) | YKJM1498 | 2.3 × 10−7 (657) |
Mre11 is a component of the MRX (Mre11/Rad50/Xrs2) complex. The deletion of MRE11 causes defects in various aspects of DNA metabolism, including checkpoints, HR, nonhomologous end joining, and telomere maintenance (8, 48, 64). mre11 mutations also increase GCR formation (7, 59). The combinations of an mre11 mutation with rad5 or rad18 caused GCR rates comparable to that caused by the mre11 mutation. This result suggests that Mre11 also cooperates with Rad5 and Rad18 to suppress spontaneous GCRs during DNA replication.
GCRs are generated by the restrained recruitment of the recombination repair pathway.
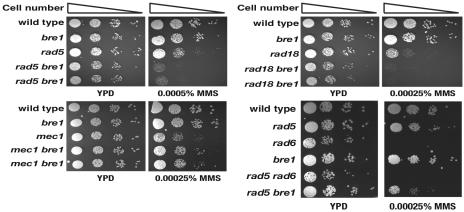
In the absence of the error-free mode of bypass, the conventional HR pathway repairs DNA damage that stalls the replication fork (56). Reflecting their reciprocal roles in PRR, combinations of the rad51 or rad52 mutation sensitized the rad5 or rad18 strain to an alkylating agent, MMS (Fig. (Fig.1).1). Given the important role of the error-free mode of bypass in suppressing GCR formation, we hypothesized that the GCRs observed in the rad5 and rad18 strains could be generated through an HR pathway. To test this hypothesis, an additional gene in the RAD52 epistasis group was mutated either in the rad5 strain or in the rad18 strain, and the GCR rates were measured (Table (Table4).4). In accordance with our hypothesis, an additional mutation in RAD51, RAD52, RAD54, RAD55, or RAD57 reduced the elevated GCR formation rate caused by the rad5 or rad18 mutation to the GCR rate close to that of the wild type. In contrast, an additional rad59 mutation, which inactivates a different branch of the HR pathway (64), resulted in a synergistic increase in the GCR rate of the rad5 or rad18 strain (Table (Table4).4). These observations suggest that, in the absence of RAD5 or RAD18, GCRs are generated through an HR pathway involving Rad51, Rad52, Rad54, Rad55, and Rad57 but not through a Rad59-dependent recombination pathway.
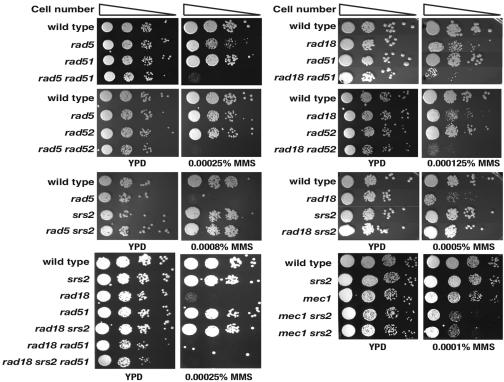
A mutation of recombination genes (rad51 and rad52) increases the MMS sensitivity of the rad5 and rad18 strains, while the srs2 mutation rescued them.
TABLE 4.
GCR formation enhanced by rad5 or rad18 mutation is decreased by an inactivation affecting homologous recombination
| Relevant genotypea | Wild type
| rad5Δ
| rad18Δ
| |||
|---|---|---|---|---|---|---|
| Strain number | GCR rate (CANr-5FOAr)b | Strain number | GCR rate (CANr-5FOAr)b | Strain number | GCR rate (CANr-5FOAr)b | |
| Wild type | RDKY3615 | 3.5 × 10−10 (1) | YKJM1385 | 2.4 × 10−8 (68) | YKJM1389 | 2.3 × 10−8 (65) |
| rad51Δ | RDKY3636 | 3.5 × 10−9 (10) | YKJM1883 | >2.2 × 10−9 (6) | YKJM1901 | >1.8 × 10−9 (5) |
| rad52Δ | RDKY4421 | 3.5 × 10−8 (100) | YKJM1903 | >2.4 × 10−9 (7) | YKJM1905 | >3.1 × 10−9 (9) |
| rad54Δ | RDKY4473 | 1.9 × 10−9 (5) | YKJM2025 | 2.6 × 10−9 (8) | YKJM2033 | 1.7 × 10−9 (5) |
| rad55Δ | RDKY5203 | 1.9 × 10−9 (5) | YKJM2594 | >3.0 × 10−9 (9) | YKJM2596 | >3.0 × 10−9 (9) |
| rad59Δ | RDKY4423 | 7.5 × 10−9 (21) | YKJM2029 | 5.6 × 10−8 (159) | YKJM2037 | 5.0 × 10−8 (142) |
| srs2Δ | YKJM0315 | >3.1 × 10−10 (0.9) | YKJM1907 | >3.1 × 10−10 (0.9) | YKJM1909 | >2.9 × 10−10 (0.8) |
| srs2-K41R | YKJM3034 | >1.9 × 10−9 (5) | YKJM3037 | >1.2 × 10−9 (3) | YKJM3038 | >1.1 × 10−9 (3) |
| pol30-119 | YKJM2624 | 1.0 × 10−9 (3) | YKJM3209 | 8.3 × 10−10 (2) | YKJM2742 | 8.4 × 10−10 (2) |
Srs2 helicase inhibits recombination by disrupting the Rad51 single-stranded DNA nucleoprotein filaments (27, 70). The srs2 mutation can almost completely suppresses MMS sensitivity caused by the inactivation of the error-free mode of bypass in an HR-dependent manner (Fig. (Fig.1)1) (5, 68). It has been suggested that Srs2 might create a DNA intermediate preferred by the error-free mode of bypass, thereby suppressing the HR pathway during DNA replication. If the GCRs generated in the rad5 or rad18 strain were caused through an HR pathway, an additional mutation of srs2 in the rad5 or rad18 strain would enhance GCR formation. Unexpectedly, however, an additional srs2 mutation in rad5 and rad18 strains reduced GCR formation to levels even slightly lower than that of the wild type (Table (Table4).4). Essentially, an identical effect was observed with a strain carrying a point mutation that inactivates the helicase activity of Srs2 [srs2(K41R)]. One possible interpretation of this paradoxical observation is that, in the absence of Rad5 or Rad18, GCRs are generated through the restrained recruitment of the HR pathway to substrates modified by Srs2. In other words, there is a type of GCR-supporting recombination pathway that is different from the conventional HR pathways and compatible with the helicase activity of Srs2. Furthermore, we also want to point out that the inactivation of Srs2 activates the conventional HR pathways, which would suppress GCR formation. The reduction of GCR formation by the srs2 mutation through the activation of conventional HR pathways is supported by the enhanced MMS sensitivity, which is comparable to the MMS sensitivity of the rad18 rad51 strain, when an additional rad51 mutation is combined to the rad18 srs2 strain (Fig. (Fig.1).1). The GCR rate of the rad18 srs2 rad51 strain was also lower than that of the rad18 strain (Table (Table4).4). Based on these results, we concluded that, in the absence of Rad5 or Rad18, GCRs are generated through the restrained recruitment of an HR pathway to substrates modified by Srs2.
SUMOylation of PCNA by the E3 SUMO ligase Siz1 is required for GCR formation.
Since the rad5 or rad18 mutation increased the GCR formation rate, a logical extension of this observation would be to ask whether the mutation changing the lysine in PCNA [pol30-119(K164R)] ubiquitinated by Rad5 and Rad18 could cause a similar increase in GCR formation. However, to our surprise, the pol30-119(K164R) mutation did not increase the GCR formation rate and even reduced the GCR rates observed in the rad5 and rad18 strains to the wild-type level (Table (Table4).4). The E3 SUMO ligase Siz1 and E3 ubiquitin ligases Rad18-Rad5 compete for the same lysine (K164) of PCNA. The different modifications have been suggested to drive different pathways (Fig. (Fig.2)2) (18). We therefore hypothesized that in the absence of proper ubiquitination, the SUMOylation of PCNA would still be required for GCR formation. In accordance with this hypothesis, a siz1 mutation reduced the elevated GCR rates caused by rad5 and rad18 to the wild-type level, similar to the pol30-119 mutation (Table (Table5).5). We also found that the siz1 mutation significantly diminished the enhanced GCR formation by the mec1 mutation to the level of the wild type and moderately reduced the increased GCR rate of the rfa1-t33 strain but did not affect those of the rad27, mre11, and pif1-m2 strains (Table (Table5).5). These observations strongly suggest that SUMOylation is specifically required for GCRs caused by defects in the Mec1-dependent checkpoint and PRR. Furthermore, a siz1 mutation fully rescued the MMS sensitivity of the rad5, rad18, or mec1 strain and partially rescued that of the rfa1-t33 strain but failed to rescue that of the rad27 or mre11 strain (Fig. (Fig.2).2). The clear correlation of sensitivity to MMS and GCR rates in these strains suggests that these phenotypes are controlled by the same underlying mechanism. Unlike the siz1 mutation, however, the srs2 mutation enhanced the MMS sensitivity of the mec1 strain, suggesting that Siz1 and Srs2 also have separate functions (Fig. (Fig.11).
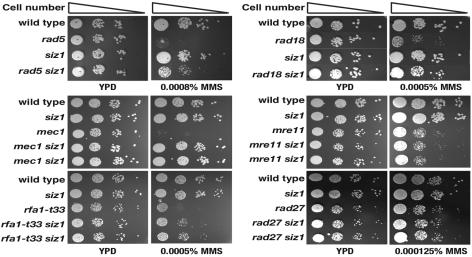
The siz1 mutation rescues the MMS sensitivity of strains carrying rad5, rad18, mec1, or rfa1-t33 mutations but not that of strains carrying mre11 or rad27 mutations.
TABLE 5.
The siz1 mutation decreases GCR rates from a subset of GCR mutator strains
| Relevant genotypea | Wild type
| siz1Δ
| ||
|---|---|---|---|---|
| Strain number | GCR rate (CANr-5FOAr)b | Strain number | GCR rate (CANr-5FOAr)b | |
| Wild type | RDKY3615 | 3.5 × 10−10 (1) | YKJM2179 | 1.3 × 10−9 (4) |
| rad5Δ | YKJM1385 | 2.4 × 10−8 (68) | YKJM2182 | 1.1 × 10−9 (3) |
| rad18Δ | YKJM1389 | 2.3 × 10−8 (65) | YKJM2185 | >1.2 × 10−9 (4) |
| mec1Δ sml1Δ | RDKY3735 | 4.6 × 10−8 (131) | YKJM2959 | 1.1 × 10−9 (3) |
| rfa1-t33 | RDKY3617 | 2.7 × 10−7 (771) | YKJM2961 | 1.3 × 10−7 (377) |
| rad27Δ | RDKY3630 | 3.4 × 10−7 (971) | YKJM2957 | 4.3 × 10−7 (1,229) |
| mre11Δ | RDKY3633 | 2.2 × 10−7 (629) | YKJM2963 | 1.8 × 10−8 (514) |
| pif1-m2 | RDKY4343 | 4.8 × 10−8 (137) | YKJM2227 | 3.7 × 10−8 (107) |
Bre1, another Rad6-coupled E3 ubiquitin ligase, is indispensable for GCR formation.
To examine the roles of E2 ubiquitin-conjugating enzymes coupled with Rad5 and Rad18, the effect of a mutation in RAD6, UBC13, or MMS2 was investigated. Unexpectedly, single mutations in these genes did not increase the GCR formation rate either in the wild type or in the pif1-m2 background (Table (Table2).2). Furthermore, an additional rad6 mutation abolished the enhanced GCR rates in the rad5 or rad18 strain (Table (Table6).6). Since all known functions of Rad6 are exclusively reliant on its E2 activity (63), the lack of GCR increase with the rad6 mutation suggests that a separate E3 ligase with Rad6 might have a supportive role for GCR formation.
TABLE 6.
The bre1 mutation decreases GCR rates from a subset of GCR mutator strains
| Relevant genotypea | Wild type
| bre1Δ
| ||
|---|---|---|---|---|
| Strain number | GCR rate (CANr-5FOAr)b | Strain number | GCR rate (CANr-5FOAr)b | |
| Wild type | RDKY3615 | 3.5 × 10−10 (1) | YKJM2233 | >9.9 × 10−10 (3) |
| rad5Δ | YKJM1385 | 2.4 × 10−8 (68) | YKJM2626 | 7.4 × 10−10 (2) |
| rad18Δ | YKJM1389 | 2.3 × 10−8 (65) | YKJM2628 | >2.6 × 10−9 (7) |
| mec1Δ sml1Δ | RDKY3735 | 4.6 × 10−8 (131) | YKJM2377 | 7.4 × 10−9 (21) |
| pif1-m2 | RDKY4343 | 4.8 × 10−8 (137) | YKJM2291 | 1.0 × 10−8 (29) |
Bre1 is another known Rad6-associated E3 ubiquitin ligase which ubiquitinates histone H2B (45). Since histone H2B is implicated in PRR response (35), we examined whether Bre1 could be the E3 ligase that functions for GCR formation (Table (Table6).6). Indeed, the bre1 mutation significantly reduced the elevated GCR rates caused by the rad5 or rad18 mutation and mildly reduced those in the mec1 and pif1-m2 strains. These results suggest that Bre1 might be generally required for GCR formation, probably by allowing DNA repair machinery access to DNA lesions through the modulation of chromatin structure. In accordance with this idea, an additional bre1 mutation sensitized rad5 and rad18 strains to MMS compared to a respective single mutant (Fig. (Fig.3).3). Since the rad5 bre1 strain is still slightly more resistant to MMS than the rad6 strain, Rad18 and Bre1 function together with Rad6 for MMS damage. However, such synergistic sensitization to MMS was not observed in the mec1 strain (Fig. (Fig.3).3). Although there is no other E3 ligase currently known for Ubc13 and Mms2, the lack of a GCR rate increase by these mutations could be due to a similar effect.
DISCUSSION
DNA replication errors have been suggested as a major cause of spontaneous GCRs (16, 26, 28, 55). When DNA replication machinery encounters DNA damage, PCNA is either mono- or polyubiquitinated and these modifications recruit divergent repair machinery (18). Failure of recruiting proper repair machinery would result in the persistent arrest or collapse of the DNA replication fork, which is subsequently converted into a DSB in the next round of DNA replication. Therefore, it is conceivable that defects in sensing DNA replication arrest or recruiting the proper repair machinery can be causative in spontaneous formation of GCRs. Consistently, defects in earlier steps of the DNA replication checkpoint, such as rfc5-1 and mec1, increased the de novo telomere addition type of GCRs (40). Our present study clearly demonstrates that defects in the selection step of repair pathways for stalled DNA replication forks also increase the same type of GCRs. The suppressive effect on GCR formation by E3 ubiquitin ligases Rad5 and Rad18 appears to depend on the error-free mode of bypass (Fig. (Fig.4A),4A), because the GCR rate of the rad5 rad18 double mutant was comparable to those of respective single mutants, and deletions of any known TLS polymerases did not increase the GCR formation rate (Table (Table22).
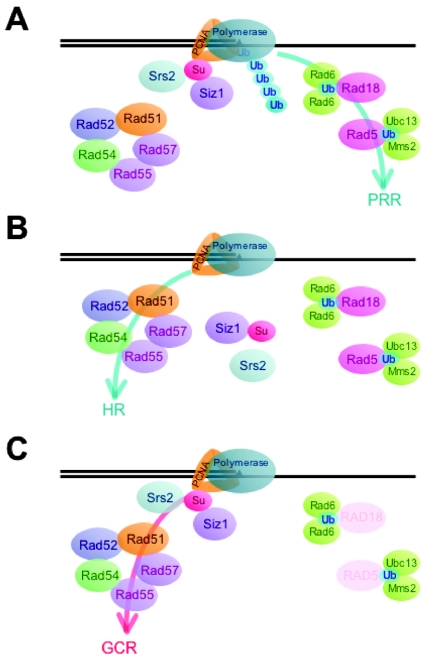
A model for GCR formation caused by the rad5 and/or rad18 mutations. (A) When the replication fork is stalled, a homotrimeric PCNA (shown as an orange triangle) is modified by K63-linked multiubiquitination, which effectively recruits the Rad5/Rad18-dependent error-free mode of bypass PRR pathway (blue arrow), while a PCNA modification with SUMO by Siz1 and subsequent recruitment of a helicase Srs2 suppress the conventional HR pathway. (B) If PRR cannot resolve the replication fork arrest, the coordinated suppression of Srs2 and Siz1 allows the conventional HR pathway to repair the stalled replication fork (blue arrow). (C) In the absence of Rad18 or Rad5, in rare events beside the conventional HR pathway repair, disturbance of the HR-mediated repair process by the helicase activity of Srs2 and SUMOylation of PCNA leads to the generation of GCRs (red arrow). Mutations of srs2, siz1, or pol30-119(K164R) (in PCNA) in the rad5 or rad18 strain eliminate the inhibition of the conventional HR pathway, which in turn facilitates the correct repair through the conventional HR pathway and thus diminishes GCR formation. The mutation of any genes in the HR pathways (Rad51, Rad52, Rad54, Rad55, or Rad57) alleviates both the conventional HR and GCR pathways. Ub and Su represent ubiquitin and SUMO, respectively.
In the absence of Rad5 or Rad18, persistent replication block could induce the activation of the Mec1-dependent checkpoint, which allows efficient recruitment of the conventional HR pathway by suppressing the antirecombinational effect of Srs2 (Fig. (Fig.4B)4B) (33). In rare cases, however, Srs2 might create DNA structure favorable to a GCR-promoting recombination pathway (Fig. (Fig.4C)4C) while suppressing the recruitment of the conventional HR pathway. This model is supported by the observation that the elimination of either the helicase activity of Srs2 or the recombination proteins resulted in a significant reduction in GCRs caused by the rad5 or rad18 mutation (Table (Table4).4). Although Srs2 is generally recognized as an antirecombination factor, it has been suggested that Srs2 may also have a prorecombination effect in certain conditions (14). Synergistic sensitivity to MMS caused by mutations in a recombination gene and rad5 or rad18 (Fig. (Fig.1)1) (62) suggests that a certain recombination pathway may be functional to resolve stalled DNA replication forks even in the presence of Srs2 in the rad5 or rad18 strain. Therefore, we believe that both Srs2 and a certain type of recombination pathway are required for GCR formation (Fig. (Fig.4C).4C). Since cells with the srs2 mutation cannot efficiently resume cell cycle progression after mitotic arrest, it could be also possible that the srs2 strain simply failed to recover from cell cycle arrest. It could contribute to the reduction of the GCR rates (69).
Despite the obvious importance of HR for the suppression of genomic instability, a mutation in most recombination genes does not induce a GCR formation rate as high as that induced by mutations in other strong GCR mutator genes (7, 38). This could be explained, at least in part, by our present observations that a recombination pathway is required for GCR formation in certain conditions. Simultaneous elimination of recombination and PRR could result in the elimination of both proper DNA repair and misrepair that leads to GCR. Spontaneous unrepaired DNA damage therefore might be accumulated and cause cell death, which reflects the absence of GCR formation. In accordance with this hypothesis, strains carrying mutations in both a recombination gene (rad51 or rad52) and a PRR gene (rad5 or rad18) had synergistically increased sensitivity to MMS (Fig. (Fig.1).1). If the recombination proteins are directly involved in the generation of GCRs, what could be their role(s)? Breakpoint structures caused by the rad5 or rad18 mutation were predominantly de novo telomere addition (60). In rare events during the resolution of stalled DNA replication forks, recombination proteins could cause DSBs. Such DSBs in turn may make de novo telomere addition machinery accessible for GCR formation. The Rad51-depedent pathway and Rad59-dependent pathway compete for similar DNA structures. In contrast to Rad51, the Rad59-depedent pathway seems to suppress mainly the GCR formation pathway by competing with the Rad51-depednent pathway in this process.
Previously, we demonstrated that the BIR type of recombination repair is important for the suppression of GCRs (38). The genetic interaction between HR and PRR also implies that the enhanced GCR rate by the rad52 mutation through the inactivation of BIR was reduced by either the rad5 or the rad18 mutation (Table (Table4).4). PRR might function to generate GCR in the absence of BIR. Alternatively, it could be the increase of genomic instabilities, including GCR, by simultaneous inactivations in both PRR and BIR, which results in cell death and apparent reduction of GCR formation.
PCNA is modified with SUMO during the early S phase of the cell cycle or by treatment of genotoxic stresses at the lethal level (18). Recently, it has been shown that PCNA SUMOylation promotes PRR by recruiting Srs2 (17, 18, 46, 49, 62). The absence of polyubiquitination in PCNA due to the inactivation of Rad5 or Rad18 might lead to persistent SUMOylation in PCNA (Fig. (Fig.4C).4C). We demonstrated that the lysine 164 of PCNA, a PCNA SUMOylation site, is required for GCR formation in the absence of Rad5, Rad18, or Mec1 (Tables (Tables44 and and5).5). However, it is unclear whether the SUMOylation of PCNA is enough to redirect the role of Srs2 from HR suppression to GCR formation. Identical genetic interactions of Siz1 and Srs2 in the rad5 or rad18 strain for GCR formation and MMS sensitivity suggest that Srs2 and Siz1 are in the same pathway, at least for GCR formation and PRR by MMS damage. Indeed, a strong genetic and biochemical interaction between SUMOylation and Srs2 has been observed (46, 49, 61). However, we need to point out that Srs2 and Siz1 have different roles, because the srs2 mutation enhances MMS sensitivity in the mec1 strain, which is opposite to what we observed for the siz1 mutation (Fig. (Fig.11).
The siz1 mutation reduced GCR rates increased by rad5, rad18, mec1, or rfa1-t33 mutations but not by mutations in RAD27, MRE11, or PIF1 (Table (Table5).5). Notably, the resistance to MMS caused by an additional siz1 mutation was also observed only in mutants where GCR was suppressed by the siz1 mutation (Fig. (Fig.2).2). Therefore, the SUMOylation by Siz1 is at least required to produce GCR and sensitize to MMS in strains carrying rad5, rad18, mec1, and rfa1-t33 mutations but not in ones carrying rad27, mre11, or pif1 mutations. Rad5, Rad18, Mec1, and Rpa1 proteins function upstream to sense DNA damage and transfer signal to downstream DNA repair proteins, while Rad27, Mre11, and Pif1 have their own roles in DNA metabolisms, including DNA repair and telomere maintenance. Therefore, SUMOylation may be specifically required for DNA damage tolerance and GCR formation derived from defects in damage-sensing steps but not in DNA repair machinery itself. Finally, because accumulating evidence suggests that a variety of proteins involved in DNA repair undergo modification with SUMO (71, 73), it might be possible that other proteins functioning in GCR, such as Ku and Rad52, could be targets by SUMOylation for suppression and/or generation of GCRs.
Mutations in ubiquitin-conjugating enzymes (mms2, ubc13, and rad6) did not increase the GCR rate significantly (Table (Table2).2). Since all known functions of Rad6 have been associated with its E2 activity (63), the lack of GCR could be explained by a GCR-supportive activity of Rad6, which is likely to be conveyed through the interaction with E3(s) other than Rad18. Our data strongly support that Bre1 is, at least, one for such activity (Table (Table6).6). Since Bre1-Rad6 modifies histone H2B, histone modification by Bre1-Rad6 may be required to make GCR formation machinery accessible to DNA damage. The absence of histone modifications could cause cells to die due to the accumulation of DNA damage resulting from the inaccessibility of any repair machinery. It is slightly different from the GCR suppression effect of siz1 mutation, since the siz1 mutation activates the conventional HR, which repairs DNA damage properly to reduce GCR, while the bre1 mutation blocks both DNA repair and GCR machinery. Therefore, although both mutations reduced GCR formation, the siz1 mutation reduced MMS sensitivity due to DNA repair capability by the conventional HR, but the bre1 mutation enhanced MMS sensitivity due to the lack of any possibility to handle DNA damage (Fig. (Fig.22 and and3).3). The requirement for histone modifications in GCR formation was suggested by the fact that the mutation of the phosphorylation site in the histone H2AX reduced the GCR rate in a cac1 strain that is defective in a chromatin assembly factor (42). Similarly, Ubc13 and Mms2 might have another E3 ligase(s) other than Rad5, which is required to generate GCRs similar to Rad6 and Bre1. The inactivation of this GCR favored pathway could counterbalance the GCR rate similar to wild type when UBC13 or MMS2 was mutated (Table (Table22).
The K63-linked polyubiquitination and SUMOylation have been implicated in regulations of the DNA damage responses by altering functions of their target proteins (15, 57). Reflecting the complexity of these modifications, many components of ubiquitin and SUMO modification systems and their target sites appear to have multiple, sometimes counteracting roles in GCR formation. Our present data dissected in depth multiple levels of regulations in the recruitment of different DNA metabolism pathways to stalled DNA replication forks that lead to divergent outcomes: proper DNA repair or GCR formation.
Acknowledgments
We thank C. Chen (Dana Faber Cancer Research Institute) for helpful discussion; H. Klein (New York University), R. Kolodner (Ludwig Institute for Cancer Research), and L. Prakash (University of Texas at Galveston) for strains and plasmids; and D. Bodine (NHGRI), J. Puck (NHGRI), P. Schwartzberg (NHGRI), and members of the Myung laboratory for comments on the manuscript. K.M. especially thanks E. Cho.
This research was supported in part by the Japan Society for the Promotion of Science (to A.M.) and by the intramural research program of the National Human Genome Research Institute, National Institutes of Health (to K.M.).
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.26.4.1424-1433.2006
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1367189?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Article citations
Mec1-independent activation of the Rad53 checkpoint kinase revealed by quantitative analysis of protein localization dynamics.
Elife, 12:e82483, 06 Jun 2023
Cited by: 3 articles | PMID: 37278514 | PMCID: PMC10259420
Non-Recombinogenic Functions of Rad51, BRCA2, and Rad52 in DNA Damage Tolerance.
Genes (Basel), 12(10):1550, 29 Sep 2021
Cited by: 4 articles | PMID: 34680945 | PMCID: PMC8535942
Review Free full text in Europe PMC
Non-recombinogenic roles for Rad52 in translesion synthesis during DNA damage tolerance.
EMBO Rep, 22(1):e50410, 02 Dec 2020
Cited by: 12 articles | PMID: 33289333 | PMCID: PMC7788459
Mgs1 function at G-quadruplex structures during DNA replication.
Curr Genet, 67(2):225-230, 25 Nov 2020
Cited by: 6 articles | PMID: 33237336 | PMCID: PMC8032586
Review Free full text in Europe PMC
The HLTF-PARP1 interaction in the progression and stability of damaged replication forks caused by methyl methanesulfonate.
Oncogenesis, 9(12):104, 07 Dec 2020
Cited by: 9 articles | PMID: 33281189 | PMCID: PMC7719709
Go to all (49) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Functional and physical interaction of yeast Mgs1 with PCNA: impact on RAD6-dependent DNA damage tolerance.
Mol Cell Biol, 26(14):5509-5517, 01 Jul 2006
Cited by: 45 articles | PMID: 16809783 | PMCID: PMC1592726
The Saccharomyces cerevisiae Rad6 postreplication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants.
Mol Cell Biol, 29(19):5226-5237, 27 Jul 2009
Cited by: 19 articles | PMID: 19635810 | PMCID: PMC2747975
SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase.
Nature, 436(7049):428-433, 01 Jun 2005
Cited by: 419 articles | PMID: 15931174
PCNASUMO and Srs2: a model SUMO substrate-effector pair.
Biochem Soc Trans, 35(pt 6):1385-1388, 01 Dec 2007
Cited by: 7 articles | PMID: 18031227
Review
Funding
Funders who supported this work.