Abstract
Free full text

A Dirichlet process model for detecting positive selection in protein-coding DNA sequences
Abstract
Most methods for detecting Darwinian natural selection at the molecular level rely on estimating the rates or numbers of nonsynonymous and synonymous changes in an alignment of protein-coding DNA sequences. In some of these methods, the nonsynonymous rate of substitution is allowed to vary across the sequence, permitting the identification of single amino acid positions that are under positive natural selection. However, it is unclear which probability distribution should be used to describe how the nonsynonymous rate of substitution varies across the sequence. One widely used solution is to model variation in the nonsynonymous rate across the sequence as a mixture of several discrete or continuous probability distributions. Unfortunately, there is little population genetics theory to inform us of the appropriate probability distribution for among-site variation in the nonsynonymous rate of substitution. Here, we describe an approach to modeling variation in the nonsynonymous rate of substitution by using a Dirichlet process mixture model. The Dirichlet process allows there to be a countably infinite number of nonsynonymous rate classes and is very flexible in accommodating different potential distributions for the nonsynonymous rate of substitution. We implemented the model in a fully Bayesian approach, with all parameters of the model considered as random variables.
Natural selection leaves a detectable signature in comparisons of protein-coding DNA sequences; a bias in the ratio of the rates of nonsynonymous and synonymous substitutions is unambiguous evidence for natural selection. Purifying selection causes the rate of nonsynonymous substitution to be smaller than the rate of synonymous substitution. In fact, the predominant pattern found in analysis of alignments of protein-coding DNA is that nonsynonymous substitutions have a lower rate than synonymous substitutions. This finding is consistent with natural selection acting to eliminate deleterious mutations that change the protein to a less functional form. However, natural selection can also act to increase the probability that a nonsynonymous mutation is fixed in the population. Positive, or direction, selection causes the relative rates of nonsynonymous to synonymous substitutions to be >1. Examples of positive selection have been found in many genes but perhaps most famously in human major histocompatibility complex (MHC) (1), HIV-1 envelope (env) gene (2), sperm lysins (3), and primate stomach lysozymes (4).
Although seemingly simple, detecting positive natural selection from alignments of protein-coding DNA is recognized as a formidable statistical problem. Many methods have been proposed to detect the footprint of natural selection, all of which are based on measuring the relative rates or numbers of nonsynonymous and synonymous substitutions (2, 5–20). Most of the methods assume a constant rate of nonsynonymous and synonymous substitutions across the sequence. These methods are ill-suited for detecting positive natural selection when only a few of the amino acid positions in a gene are under the influence of positive natural selection, with the others under purifying selection. Applying a method that assumes a constant rate of nonsynonymous change across the sequence potentially masks the signature of positive natural selection that is present at only a few positions in the alignment (21). In cases where such methods have been successful, many sites are typically under strong positive selection (e.g., MHC). More recently, Nielsen and Yang (2) developed a method that allows the rate of nonsynonymous substitution to vary across the sequence. The method of Nielsen and Yang has proven useful for detecting positive natural selection in sequences where only a few sites are under directional selection.
The Nielsen and Yang (2) approach assumes that the nonsynonymous/synonymous rate ratio (dN/dS ratio) at a site is a random variable drawn from some probability distribution. The goal is to estimate parameters of the model from alignments of protein-coding DNA and, by using these parameter estimates, identify amino acids under positive selection. Their model is quite complicated and contains many parameters to estimate. Some of the parameters account for the fact that the sequences are related to one another through some unknown phylogenetic tree. Other parameters account for biases in the substitution process, such as an increased rate of mutation for transitions. The remaining parameters, however, describe how dN/dS varies across the sequence. Nielsen and Yang showed not only that it is practical to efficiently estimate these parameters from alignments of protein-coding DNA sequences but also that one can use an empirical Bayesian approach to detect specific amino acid residues that are under the influence of positive natural selection.
How should variation in the rate of nonsynonymous substitution across a sequence be modeled? Population genetics theory is largely silent on this issue. For the most part, we lack information on the distribution of selection coefficients for new mutations and the demography of the populations under consideration, making it difficult to predict a distribution for the rate of nonsynonymous substitution (22, 23). The original Nielsen and Yang (2) approach assumed that a site could be in one of three categories, each of which differed in its dN/dS rate ratio. With probability p1, a site is in category 1, which has dN/dS = 0; with probability p2, the site is in category 2, which has dN/dS = 1; and with probability p3, the site is in a category that has dN/dS > 1 (p1 + p2 + p3 = 1). Nielsen and Yang (2) also considered a model that allowed dN/dS to vary continuously on the interval (0, 1); a gamma distribution, truncated on the interval (0. 1) was used to model amino acid sites that are acting neutrally or under the influence of purifying natural selection. Under this model, a site has dN/dS drawn from a truncated gamma distribution with probability p1 or has dN/dS > 1 with probability p2. Later, Yang et al. (12) systematically explored more ways to model variation in dN/dS across a sequence. They considered a total of 13 models. The “M10” model from Yang et al. (12), for example, assumes that, with probability p1, the dN/dS rate ratio is drawn from a beta distribution on the interval (0, 1) and, with probability p2, the dN/dS rate ratio is drawn from an offset gamma distribution. Even though many of the models considered in Nielsen and Yang (2) and in Yang et al. (12) describe dN/dS as varying continuously, in practice, these continuous distributions are discretized to allow the likelihood to be calculated (the likelihood is averaged over the different discrete categories). At least as currently implemented, then, all of these models describing how the rate of nonsynonymous substitution varies across the sequence are discrete.
We take a different approach to modeling variation in dN/dS across sites, allowing sites to be in one of a number of classes, with each class having a different dN/dS ratio. The prior probability distribution for the number of classes and the dN/dS for each class is described by a Dirichlet process prior. The Dirichlet process prior provides a flexible way to model situations in which the data elements are drawn from a mixture of simpler parametric distributions. For typical mixture models, the number of mixture components is assumed to be known, and determining the correct number of components a priori for a particular model is difficult. For the Dirichlet process prior, however, the number of mixture components is countably infinite, obviating the need to determine the correct number of mixture components. Here we apply the Dirichlet process to the problem of detecting positive selection in protein-coding DNA sequences. Instead of assuming that the dN/dS rate ratio for a site is drawn from a particular parametric distribution, as is the case for Nielsen and Yang (2) and Yang et al. (12), we assume that a site is assigned to a category with a particular dN/dS value. The number of selection categories and the dN/dS value for each category are both considered random variables in our model. The Dirichlet process has been used in one other case for an evolutionary problem: Lartillot and Philippe (24) used the Dirichlet process to model variation in the substitution process across alignments of amino acid sequences. Similarly, Pond and Frost (25) described a flexible discretization scheme that is able to fit a wide range of rate distributions. However, this scheme still requires that the maximum number of rate classes be specified a priori.
We estimate the parameters of the model in a Bayesian framework. Calculating the joint posterior probability distribution of the parameters involves summation over all possible phylogenetic trees relating the sequences and, for each tree, integration over all possible combinations of parameter values. We use Markov chain Monte Carlo (MCMC) to approximate the posterior probability distribution of the parameters and apply the method to six alignments of protein-coding DNA sequences (12, 17, 26, 27).
Results and Discussion
The Dirichlet process prior has two components: One is a parameter, usually called α, that influences the probability that data elements find themselves in the same cluster. In other words, the parameter controls the “clumpiness” of the process. The other component of a Dirichlet process prior is a probability distribution that describes the probability of the parameter assigned to each cluster. (The Dirichlet process prior is described in more detail in Materials and Methods.) We examined the robustness of the inferences of positive selection to three different choices for α, choosing α such that the prior mean of the number of components (k) was E(k) ≈ 1, E(k) = 5, and E(k) = 10; we did this to keep the number of selection classes to a manageable number because the likelihood calculations become too computationally cumbersome when k is large (e.g., k > 25). An alternative that we did not explore is to place a prior probability distribution on α. Often in problems using a Dirichlet process prior, a gamma hyperprior is placed on α.
Tables 11–6 compare the posterior and prior probability distributions for the number of mixture components (selection categories) for each of the six alignments. For all of the alignments we examined, there is very little posterior probability for k = 1 and k = 2, even when there may be a substantial amount of prior probability for those cases. The data are very difficult to explain with only a few selection classes.
Table 1.
Prior and posterior probabilities for the number of selection classes, k, for HIV-1 vif
| k | E(k) ≈ 1 E(k|X) = 3.04 | E(k) = 5.00 E(K|X) = 6.33 | E(k) = 10.00 E(k|X) = 10.21 | |||
|---|---|---|---|---|---|---|
| f(k) | f(k|X) | f(k) | f(k|X) | f(k) | f(k|X) | |
| 1 | 0.9434 | 0.0000 | 0.0139 | 0.0000 | 0.0000 | 0.0000 |
| 2 | 0.0550 | 0.0000 | 0.0648 | 0.0000 | 0.0004 | 0.0000 |
| 3 | 0.0015 | 0.9634 | 0.1439 | 0.0334 | 0.0026 | 0.0002 |
| 4 | 0.0000 | 0.0363 | 0.2040 | 0.1096 | 0.0096 | 0.0027 |
| 5 | 0.0000 | 0.0002 | 0.2086 | 0.1929 | 0.0256 | 0.0134 |
| 6 | 0.0000 | 0.0000 | 0.1647 | 0.2292 | 0.0528 | 0.0371 |
| 7 | 0.0000 | 0.0000 | 0.1050 | 0.1967 | 0.0879 | 0.0751 |
| 8 | 0.0000 | 0.0000 | 0.0556 | 0.1288 | 0.1216 | 0.1225 |
| 9 | 0.0000 | 0.0000 | 0.0251 | 0.0667 | 0.1432 | 0.1561 |
| 10 | 0.0000 | 0.0000 | 0.0098 | 0.0286 | 0.1460 | 0.1630 |
| >10 | 0.0000 | 0.0000 | 0.0047 | 0.0143 | 0.3828 | 0.4085 |
The prior and posterior probabilities for the number of selection classes are f(k) and f(k|X), respectively.
Table 2.
Prior and posterior probabilities for the number of selection classes, k, for HIV-1 pol
| k | E(k) ≈ 1 E(k|X) = 3.22 | E(k) = 5.00 E(k|X) = 5.99 | E(k) = 10.00 E(k|X) = 9.58 | |||
|---|---|---|---|---|---|---|
| f(k) | f(k|X) | f(k) | f(k|X) | f(k) | f(k|X) | |
| 1 | 0.9285 | 0.0000 | 0.0157 | 0.0000 | 0.0001 | 0.0000 |
| 2 | 0.0690 | 0.0006 | 0.0686 | 0.0000 | 0.0006 | 0.0000 |
| 3 | 0.0025 | 0.7837 | 0.1459 | 0.0313 | 0.0033 | 0.0006 |
| 4 | 0.0001 | 0.2096 | 0.2014 | 0.1518 | 0.0114 | 0.0055 |
| 5 | 0.0000 | 0.0062 | 0.2036 | 0.2316 | 0.0285 | 0.0252 |
| 6 | 0.0000 | 0.0001 | 0.1611 | 0.2467 | 0.0557 | 0.0607 |
| 7 | 0.0000 | 0.0000 | 0.1041 | 0.1688 | 0.0891 | 0.1018 |
| 8 | 0.0000 | 0.0000 | 0.0566 | 0.0979 | 0.1199 | 0.1404 |
| 9 | 0.0000 | 0.0000 | 0.0265 | 0.0467 | 0.1388 | 0.1615 |
| 10 | 0.0000 | 0.0000 | 0.0108 | 0.0161 | 0.1405 | 0.1599 |
| >10 | 0.0000 | 0.0000 | 0.0057 | 0.0093 | 0.3797 | 0.3349 |
The prior and posterior probabilities for the number of selection classes are f(k) and f(k|X), respectively.
Table 3.
Prior and posterior probabilities for the number of selection classes, k, for HIV-1 env
| k | E(k) ≈ 1 E(k|X) = 2.16 | E(k) = 5.00 E(k|X) = 6.06 | E(k) = 10.00 E(k|X) = 10.61 | |||
|---|---|---|---|---|---|---|
| f(k) | f(k|X) | f(k) | f(k|X) | f(k) | f(k|X) | |
| 1 | 0.9505 | 0.0000 | 0.0124 | 0.0000 | 0.0000 | 0.0000 |
| 2 | 0.0483 | 0.8532 | 0.0613 | 0.0039 | 0.0003 | 0.0000 |
| 3 | 0.0012 | 0.1350 | 0.1415 | 0.0430 | 0.0020 | 0.0001 |
| 4 | 0.0000 | 0.0117 | 0.2057 | 0.1328 | 0.0082 | 0.0014 |
| 5 | 0.0000 | 0.0001 | 0.2129 | 0.2147 | 0.0232 | 0.0085 |
| 6 | 0.0000 | 0.0000 | 0.1682 | 0.2344 | 0.0502 | 0.0265 |
| 7 | 0.0000 | 0.0000 | 0.1060 | 0.1820 | 0.0867 | 0.0600 |
| 8 | 0.0000 | 0.0000 | 0.0550 | 0.1069 | 0.1233 | 0.1024 |
| 9 | 0.0000 | 0.0000 | 0.0240 | 0.0510 | 0.1476 | 0.1399 |
| 10 | 0.0000 | 0.0000 | 0.0090 | 0.0221 | 0.1513 | 0.1635 |
| >10 | 0.0000 | 0.0000 | 0.0039 | 0.0091 | 0.3844 | 0.4688 |
The prior and posterior probabilities for the number of selection classes are f(k) and f(k|X), respectively.
Table 4.
Prior and posterior probabilities for the number of selection classes, k, for human influenza virus hemagglutinin
| k | E(k) ≈ 1 E(k|X) = 2.24 | E(k) = 5.00 E(k|X) = 5.50 | E(k) = 10.00 E(k|X) = 9.46 | |||
|---|---|---|---|---|---|---|
| f(k) | f(k|X) | f(k) | f(k|X) | f(k) | f(k|X) | |
| 1 | 0.9383 | 0.0000 | 0.0149 | 0.0000 | 0.0000 | 0.0000 |
| 2 | 0.0598 | 0.7744 | 0.0673 | 0.0048 | 0.0005 | 0.0000 |
| 3 | 0.0018 | 0.2146 | 0.1460 | 0.0775 | 0.0029 | 0.0020 |
| 4 | 0.0000 | 0.0107 | 0.2037 | 0.1963 | 0.0103 | 0.0062 |
| 5 | 0.0000 | 0.0003 | 0.2064 | 0.2670 | 0.0267 | 0.0244 |
| 6 | 0.0000 | 0.0000 | 0.1625 | 0.2124 | 0.0540 | 0.0580 |
| 7 | 0.0000 | 0.0000 | 0.1037 | 0.1355 | 0.0883 | 0.1156 |
| 8 | 0.0000 | 0.0000 | 0.0553 | 0.0620 | 0.1208 | 0.1564 |
| 9 | 0.0000 | 0.0000 | 0.0252 | 0.0301 | 0.1412 | 0.1720 |
| 10 | 0.0000 | 0.0000 | 0.0100 | 0.0104 | 0.1436 | 0.1571 |
| >10 | 0.0000 | 0.0000 | 0.0050 | 0.0038 | 0.3820 | 0.2962 |
The prior and posterior probabilities for the number of selection classes are f(k) and f(k|X), respectively.
Table 5.
Prior and posterior probabilities for the number of selection classes, k, for Japanese encephalitis virus env
| k | E(k) ≈ 1 E(k|X) = 2.01 | E(k) = 5.00 E(k|X) = 3.58 | E(k) = 10.00 E(k|X) = 5.44 | |||
|---|---|---|---|---|---|---|
| f(k) | f(k|X) | f(k) | f(k|X) | f(k) | f(k|X) | |
| 1 | 0.9344 | 0.0000 | 0.0149 | 0.0000 | 0.0001 | 0.0000 |
| 2 | 0.0635 | 0.9849 | 0.0669 | 0.1832 | 0.0006 | 0.0223 |
| 3 | 0.0021 | 0.0146 | 0.1445 | 0.3497 | 0.0032 | 0.1089 |
| 4 | 0.0000 | 0.0005 | 0.2017 | 0.2627 | 0.0110 | 0.1863 |
| 5 | 0.0000 | 0.0000 | 0.2052 | 0.1365 | 0.0281 | 0.2201 |
| 6 | 0.0000 | 0.0000 | 0.1627 | 0.0500 | 0.0556 | 0.2066 |
| 7 | 0.0000 | 0.0000 | 0.1049 | 0.0139 | 0.0897 | 0.1348 |
| 8 | 0.0000 | 0.0000 | 0.0568 | 0.0032 | 0.1213 | 0.0721 |
| 9 | 0.0000 | 0.0000 | 0.0263 | 0.0008 | 0.1406 | 0.0316 |
| 10 | 0.0000 | 0.0000 | 0.0106 | 0.0000 | 0.1421 | 0.0117 |
| >10 | 0.0000 | 0.0000 | 0.0054 | 0.0000 | 0.3774 | 0.0057 |
The prior and posterior probabilities for the number of selection classes are f(k) and f(k|X), respectively.
Table 6.
Prior and posterior probabilities for the number of selection classes, k, for vertebrate β-globin
| k | E(k) ≈ 1 E(k|X) = 3.03 | E(k) = 5.00 E(k|X) = 5.65 | E(k) = 10.00 E(k|X) = 8.95 | |||
|---|---|---|---|---|---|---|
| f(k) | f(k|X) | f(k) | f(k|X) | f(k) | f(k|X) | |
| 1 | 0.9462 | 0.0000 | 0.0132 | 0.0000 | 0.0000 | 0.0000 |
| 2 | 0.0525 | 0.0000 | 0.0630 | 0.0000 | 0.0004 | 0.0000 |
| 3 | 0.0014 | 0.9716 | 0.1422 | 0.0524 | 0.0024 | 0.0008 |
| 4 | 0.0000 | 0.0280 | 0.2039 | 0.1811 | 0.0092 | 0.0094 |
| 5 | 0.0000 | 0.0004 | 0.2100 | 0.2687 | 0.0250 | 0.0323 |
| 6 | 0.0000 | 0.0000 | 0.1664 | 0.2343 | 0.0524 | 0.0816 |
| 7 | 0.0000 | 0.0000 | 0.1060 | 0.1474 | 0.0881 | 0.1395 |
| 8 | 0.0000 | 0.0000 | 0.0560 | 0.0732 | 0.1227 | 0.1767 |
| 9 | 0.0000 | 0.0000 | 0.0250 | 0.0307 | 0.1449 | 0.1797 |
| 10 | 0.0000 | 0.0000 | 0.0097 | 0.0086 | 0.1477 | 0.1493 |
| >10 | 0.0000 | 0.0000 | 0.0047 | 0.0035 | 0.3815 | 0.2276 |
The prior and posterior probabilities for the number of selection classes are f(k) and f(k|X), respectively.
We also examined the marginal posterior probability of ω = dN/dS for each site. Fig. 1 shows the probability distribution for four arbitrarily chosen sites from the HIV-1 env alignment. Three things are apparent: First, and as expected, different sites for the same data set have different probability distributions for ω. Second, the distribution for ω varies depending on the value of the parameter used in the analysis. The distributions were similar when the prior expectation of the number of selection classes was 5 or 10 but were different when the prior expectation was 1. In short, when the model attempts to explain the data with too few selection categories, the marginal posterior probability distribution for ω at a site becomes a compromise between the necessities for that site and others that are grouped into the same selection category. Third, and perhaps more importantly, the marginal posterior probability distribution of ω for a site can be used to directly calculate the probability that the site experiences positive selection. The probability that ω > 1 is the probability that the site is under positive selection. This quantity can be directly calculated from the output of the MCMC procedure as the fraction of the time the site had ω > 1. Fig. 2 shows the probability that each site is under positive selection for all of the analyses of this paper. There is strong concordance between the sites that we find to be under positive selection with the sites that Yang et al. (12) found to be under positive selection. Yang et al. (12) found that analysis of different probability distributions describing how ω varies across a sequence would typically result in a set of sites that always were found to be under positive selection and another set of sites that were inferred to be under positive selection for some models but not others. In general, the Dirichlet process prior picks out the sites that were consistently found to be under positive selection by Yang et al. (12) but assigns lower probabilities to these same sites of being under positive selection. For example, the Yang et al. (12) analysis of the HIV-1 env alignment consistently found sites 28, 66, and 87 to be under positive selection (with probabilities of 0.99 or greater), and sites 26 and 51 to be under positive selection with probability 0.99 under the M12 model. (The M12 model is a mixture of two normal distributions with a discrete category with ω = 0.) Our analysis of the HIV-1 env alignment finds sites 26, 28, 51, 66, 83, and 87 to be under positive selection, all having a probability of >0.95 and having ω > 1. Our analysis does not condition on the maximum likelihood values of the parameters (the tree, branch lengths, and substitution model parameters) as is the case of the Nielsen and Yang (2) approach. It is likely that the accommodation of uncertainty in the model parameters causes the probabilities of sites being in particular categories to be dampened relative to approaches that do not account for parameter uncertainty.
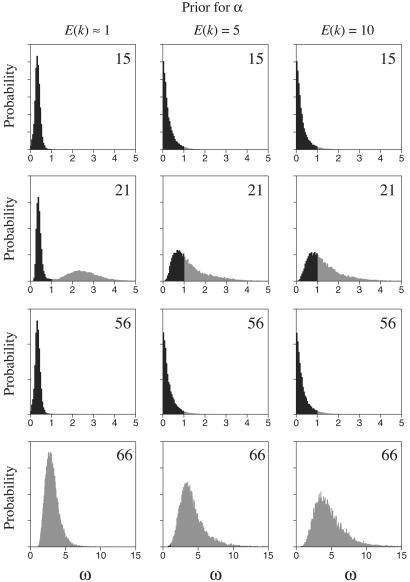
The posterior probability density distribution of ω, the dN/dS rate ratio, for four sites of the HIV-1 env alignment. The lightly shaded portion of each distribution is the part that has ω > 1.
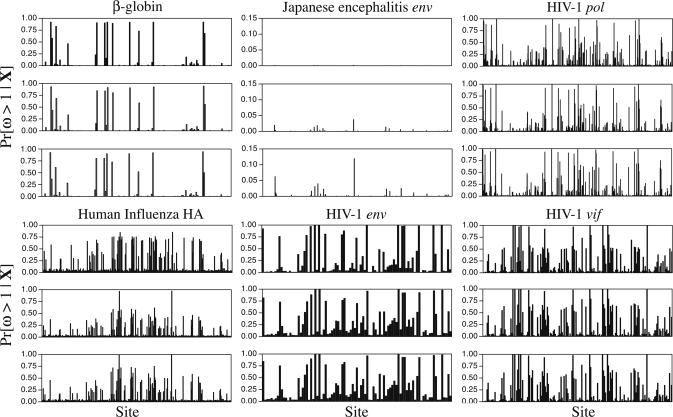
The posterior probabilities of sites being under positive selection for each of the analyses of the six alignments of this study. The graphs are grouped by alignment, with each group consisting of three graphs. The top graph of each group has E(k) ≈ 1, the middle graph has E(k) = 5, and the bottom graph has E(k) = 10.
Table 7 lists sites that had a high probability of being under positive selection for all six genes. For the most part, the same sites are found to be under positive selection regardless of the value of the concentration parameter used in the analysis. For example, sites 26, 28, 51, 66, 83, and 87 of the HIV-1 env alignment were inferred to be under positive selection regardless of the value of α assumed in the analysis. Sites 24, 68, 69, and 76 had a probability >0.95 of being under positive selection when E(k) ≈ 1 but not when E(k) = 5 or E(k) = 10. However, the probability of those sites being under positive selection was just below the 0.95 threshold. (Sites 24, 68, 69, and 76 had probabilities ranging between 0.88 and 0.93 of having ω > 1 when the expected number of selection categories was set to 5 or 10.)
Table 7.
Sites potentially under positive selection
| Data | E(k) | Sites with probability >0.95 of being under positive selection |
|---|---|---|
| Vertebrate β-globin | 1 | – |
| 5 | – | |
| 10 | – | |
| Japanese encephalitis virus env | 1 | – |
| 5 | – | |
| 10 | – | |
| Human influenza virus hemagglutinin | 1 | – |
| 5 | 226, 135 | |
| 10 | 226, 135 | |
| HIV-1 env | 1 | 28, 66, 26, 87, 51, 83, 76, 69, 68, 24 |
| 5 | 28, 66, 26, 87, 83, 51 | |
| 10 | 28, 66, 26, 87, 83, 51 | |
| HIV-1 pol | 1 | 67, 347, 478, 779, 568, 761 |
| 5 | 67, 347, 779, 478, 3, 568 | |
| 10 | 67, 347, 779, 478, 3, 568 | |
| HIV-1 vif | 1 | 33, 167, 33, 127, 39, 109, 122, 47, 92, 37 |
| 5 | 33, 167, 127, 31, 37, 109, 39, 122, 92, 47, 63 | |
| 10 | 33, 127, 167, 31, 37, 109, 122, 39, 92, 47 |
Methods for detecting the presence of positive natural selection in protein-coding DNA have become an important tool in studies of molecular evolution. The recent advances that allow the nonsynonymous/synonymous rate ratio to vary across the sequence have opened up the possibility of detecting specific amino acid residues that are functionally important, displaying an elevated dN/dS rate ratio. The method we describe here represents an important extension of existing methods by allowing a more flexible description of how dN/dS varies across a sequence and by accounting for uncertainty in parameters of the model when making inferences of positive selection.
Materials and Methods
Data.
We assume an alignment of protein-coding DNA sequences is available. The alignment is contained in the matrix X = {xij}, where i = 1, 2, …, s and j = 1, 2, …, c; s is the number of sequences, and c is the number of codons in each sequence. The information at the jth site is contained in the vector xj = (x1j, x2j, …, xsj)′.
Phylogenetic Model.
The s species are related to one another through an unknown phylogeny, represented as a bifurcating tree. Possible phylogenies are denoted τ1, τ2, …, τB(s), where B(s) is the number of possible phylogenetic trees for s species. We assume a time-reversible model of nucleotide substitution (see below), which means that the phylogenetic trees can be arbitrarily rooted without changing the likelihood. Therefore, we assume unrooted phylogenetic trees throughout this study. The number of unrooted phylogenetic trees for s species is B(s) = (2s − 5)!/[2s−3(s − 3)!] (28). Each unrooted tree has 2s − 3 branches. Each branch on the tree has a length, ν. The set of branch lengths for the jth tree is denoted νj = (ν1, ν2, …, ν2s−3).
We assume that substitutions occur on the phylogenetic tree according to a continuous-time Markov process. The instantaneous rate of change from codon i to codon j is contained in a matrix, Q, the entries of which are given by

where ω is the nonsynonymous/synonymous rate ratio, κ is the transition/transversion rate ratio, and πj is the stationary frequency of codon j (2, 14, 15). The rate matrix, Q, is 61 × 61 in dimension; although there are 64 possible codon triplets, the three termination codons of the universal code are excluded from the state space, leaving the 61 sense codons as the states of the substitution process. The diagonal elements of the rate matrix Q are specified such that each row sums to 0. We rescale the matrix such that the average rate of synonymous substitution is 1; this means that the branch lengths are in terms of expected number of synonymous substitutions per codon site. This scaling is different from the usual for phylogenies, where the rate matrix is rescaled such that the branch lengths are in terms of expected number of substitutions per site. Finally, note that the substitution process is time-reversible because πiqij = πjqji for all i ≠ j. Practically speaking, this means that we can use unrooted phylogenies instead of rooted trees when calculating the likelihood on the tree (29). The transition probabilities of the process are the probability of finding the process in codon j conditioned on the process starting in codon i and run over a branch of length ν. The transition probabilities can be calculated as P(ν) = eQν.
Each of the c sites can be in one of k selection categories, where each category differs in its dN/dS value. The information on the category membership is contained in an association vector, z. For example, for the alignment of four sites given above, one possible assignment of sites to selection categories is z = (1, 1, 2, 3). Here, k = 3, and the first two sites are in the same category. The allocation vector describes a partition of the sites into different classes, each class with its own dN/dS value. The number of ways to partition a set of c sites into k classes is described by the Stirling numbers of the second kind (30):
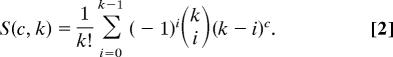
The sites for the example alignment of c = 4 sites can be assigned to k = 3 categories in a total of S(4, 3) = 6 ways: z = (1, 1, 2, 3); z = (1, 2, 1, 3); z = (1, 2, 3, 1); z = (1, 2, 2, 3); z = (1, 2, 3, 2); and z = (1, 2, 3, 3). We label possible partitions of the sites into classes by using the restricted growth function notation of Stanton and White (31), where elements are sequentially numbered with the constraint that the index numbers for two sites are the same if they are found in the same category. The number of sites in the ith category is denoted ηi. For the example given above, η1 = 2, η2 = 1, and η3 = 1.
Likelihood.
Assuming that substitutions are independent across sites, the likelihood for an alignment can be calculated as the product of the site likelihoods:
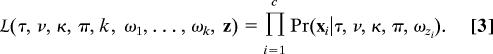
We used the Felsenstein (29) pruning algorithm to calculate likelihoods for specific combinations of parameter values.
Statistical Model.
We estimate parameters in a Bayesian framework. All parameters of the model are considered random values with their own prior and posterior probability distributions. The joint posterior probability of all of the parameters of the model is
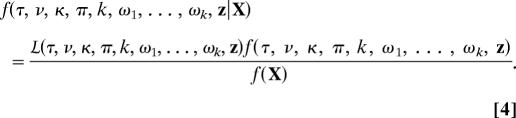
The posterior probability [f(· |X)] is equal to the product of the likelihood [ (·) or, equivalently, f(X| ·)] and the prior probability distribution [f(·)] of the parameters divided by the marginal likelihood [f(X)].
(·) or, equivalently, f(X| ·)] and the prior probability distribution [f(·)] of the parameters divided by the marginal likelihood [f(X)].
Bayesian analysis requires that a prior probability distribution be specified for the parameters. Here we assume that the parameters take the prior probability distributions shown in Table 8.
Table 8.
The prior probability distributions assigned to parameters of the phylogenetic model
| Parameter | Symbol | Prior |
|---|---|---|
| Tree topoology | τ | All trees have equal prior probability |
| Branch lengths | ν | Branch lengths are independent exponential (10) random variables |
| Codon frequencies | π | Flat Dirichlet distribution |
| Transition/transversion ratio | κ | Ratio of two identically distributed exponential random variables |
| dN/dS rate ratio | ω | Ratio of two identically distributed exponential random variables |
| Category information | z,k | Dirichlet process |
| Dirichlet process parameter | α | Fixed such that E(k) is small [E(k) ≈ 1, E(k) = 5, E(k) = 10] |
The model, as described, is fairly typical of codon models implemented to date (32). However, the Dirichlet process prior is new to the problem of detecting positive natural selection and requires more explanation. The Dirichlet process prior is widely used in clustering problems as a nonparametric alternative, where the data elements are drawn from a mixture distribution with a countably infinite number of components (33, 34). [Ewens and Tavaré (35) describe the relationship between the Dirichlet process prior and the Ewen’s sampling formula from population genetics.] The Dirichlet process prior has two parameters: a concentration parameter, which, loosely speaking, determines the degree to which data elements are grouped together, and a base measure, G0(·), which describes the probability distribution of the parameter value associated with each cluster. The model can be described as follows: First, the number of classes, k, and an allocation variable, z, are randomly drawn from the probability distribution
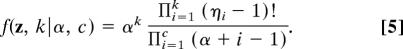
Once the sites have been partitioned into k categories, a ω = dN/dS value is assigned to each category by drawing randomly from the following probability distribution k times, G0(ωi) = f(ωi) = [1/(1 + ωi)2] which is the probability density of the ratio of two identically distributed exponential random variables. Here, ωi is the dN/dS rate ratio for the ith selection category. The probability of having k categories is obtained by summing over all possible partitions for k categories, and is
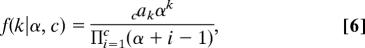
where nak is the absolute values of the Stirling numbers of the first kind. The expected number of categories is
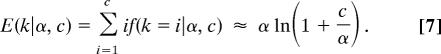
Finally, the probability of finding two sites grouped together into the same cluster is f(zi = zj|α, c) = 1/(1 + α)].
MCMC.
We approximated posterior probabilities by using MCMC (36, 37). The general idea is to construct a Markov chain that has as its state space the parameters of the statistical model and a stationary distribution that is the posterior probability of the parameters. Samples drawn from the Markov chain when at stationarity are valid, albeit dependent, draws from the posterior probability distribution of the parameters (38). The fraction of the time the Markov chain visits any particular combination of parameter values is an approximation of the joint posterior probability of those parameters.
We wrote a computer program that implements the MCMC method for the phylogenetic model described above. For each iteration of the Markov chain, the program randomly picks a parameter to change, proposes a new value for the parameter, and then accepts or rejects the proposed state as the next state of the chain by using the Metropolis–Hastings formula. Most of the proposal mechanisms have been described elsewhere. We use the LOCAL mechanism of Larget and Simon (39) to simultaneously change the tree and branch lengths (τ, ν). We update the codon frequencies (π) by using a Dirichlet proposal mechanism and change the transition/transversion rate ratio (κ) and the category dN/dS values (ω) by using a rate multiplier [i.e., we multiply the old parameter value by eλ(u−½) to obtain a new value for the parameter, where u is a uniform(0, 1) random number and λ is a tuning parameter]. Finally, we update the allocation vector by using a Gibbs sampling algorithm for nonconjugate models proposed by Neal (40). We implement the method of Neal (40) as follows: First, we randomly choose a site, i, and remove it from the set of k selection classes currently in computer memory. If the site is in a selection class by itself (i.e., ηzi for the class was 1), then the entire class is deleted, and k is decreased by 1. Otherwise, ηzi for the selection class that the site is associated with is decreased by 1. We then construct five new (temporary auxiliary) classes by drawing the dN/dS value for each from the prior probability distribution. These auxiliary classes represent components that have no other sites associated with them. Gibbs sampling is performed to update site i but with respect to the distribution that includes the temporary auxiliary parameters. The site is assigned to the jth previously existing class with probability Zη−if(xi|ωj) and to the jth of five auxiliary classes with probability Z(α/5)f(xi|ωj), where Z is the appropriate normalizing constant. After this update, all dN/dS values not associated with a selection class are discarded.
All analyses were run for a total of 2 million update cycles. Chains were thinned, with samples taken every 100 cycles. Samples taken during the first 100,000 cycles were discarded as the burn-in phase. The MCMC procedure was repeated, resulting in two sets of samples for each gene. Convergence was assessed by using the program tracer (http://evolve.zoo.ox.ac.uk/software.html?id=tracer).
Data Analysis.
We analyzed six alignments of protein-coding DNA sequences. These alignments included: (i) β-globin gene sequences from s = 17 vertebrates (12); (ii) s = 23 env gene sequences from Japanese encephalitis virus (12, 26); (iii) s = 28 sequences of the HA1 domain of the hemagglutinin gene of human influenza virus A (17); (iv) s = 13 sequences of the HIV-1 env gene V3 region (27); (v) HIV-1 pol gene sequences from s = 23 isolates (12); and (vi) s = 29 HIV-1 vif gene sequences (12).
Acknowledgments
J.P.H. was supported by National Institutes of Health Grant GM-069801 and National Science Foundation Grants DEB-0445453 and CACTTOL-22035A.
Abbreviation
| MCMC | Markov chain Monte Carlo. |
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0508279103
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1458866?pdf=render
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.0508279103
Article citations
Expectation-Maximization enables Phylogenetic Dating under a Categorical Rate Model.
Syst Biol, 73(5):823-838, 01 Oct 2024
Cited by: 0 articles | PMID: 38970346
Molecular characterization and evolution of the resident population of some alfalfa mosaic virus (AMV) isolates in Egypt.
BMC Microbiol, 23(1):261, 18 Sep 2023
Cited by: 0 articles | PMID: 37723462 | PMCID: PMC10506327
Impact of transient acquired hypermutability on the inter- and intra-species competitiveness of Pseudomonas aeruginosa.
ISME J, 17(11):1931-1939, 04 Sep 2023
Cited by: 3 articles | PMID: 37666975 | PMCID: PMC10579334
Genes and sites under adaptation at the phylogenetic scale also exhibit adaptation at the population-genetic scale.
Proc Natl Acad Sci U S A, 120(11):e2214977120, 10 Mar 2023
Cited by: 4 articles | PMID: 36897968 | PMCID: PMC10089192
A Bayesian Mutation-Selection Framework for Detecting Site-Specific Adaptive Evolution in Protein-Coding Genes.
Mol Biol Evol, 38(3):1199-1208, 01 Mar 2021
Cited by: 8 articles | PMID: 33045094 | PMCID: PMC7947879
Go to all (48) article citations
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Not so different after all: a comparison of methods for detecting amino acid sites under selection.
Mol Biol Evol, 22(5):1208-1222, 09 Feb 2005
Cited by: 1423 articles | PMID: 15703242
Estimating absolute rates of synonymous and nonsynonymous nucleotide substitution in order to characterize natural selection and date species divergences.
Mol Biol Evol, 21(7):1201-1213, 10 Mar 2004
Cited by: 50 articles | PMID: 15014159
Evolutionary models accounting for layers of selection in protein-coding genes and their impact on the inference of positive selection.
Mol Biol Evol, 28(12):3297-3308, 20 Jun 2011
Cited by: 24 articles | PMID: 21690564
Analysis of selection in protein-coding sequences accounting for common biases.
Brief Bioinform, 22(5):bbaa431, 01 Sep 2021
Cited by: 15 articles | PMID: 33479739
Review
Funding
Funders who supported this work.
NIGMS NIH HHS (2)
Grant ID: R01 GM069801
Grant ID: GM-069801





