Abstract
Free full text

Cooperative Regulation of Extracellular Signal-Regulated Kinase Activation and Cell Shape Change by Filamin A and β-Arrestins
Abstract
β-Arrestins (βarr) are multifunctional adaptor proteins that can act as scaffolds for G protein-coupled receptor activation of mitogen-activated protein kinases (MAPK). Here, we identify the actin-binding and scaffolding protein filamin A (FLNA) as a βarr-binding partner using Son of sevenless recruitment system screening, a classical yeast two-hybrid system, coimmunoprecipitation analyses, and direct binding in vitro. In FLNA, the βarr-binding site involves tandem repeat 22 in the carboxyl terminus. βarr binds FLNA through both its N- and C-terminal domains, indicating the presence of multiple binding sites. We demonstrate that βarr and FLNA act cooperatively to activate the MAPK extracellular signal-regulated kinase (ERK) downstream of activated muscarinic M1 (M1MR) and angiotensin II type 1a (AT1AR) receptors and provide experimental evidence indicating that this phenomenon is due to the facilitation of βarr-ERK2 complex formation by FLNA. In Hep2 cells, stimulation of M1MR or AT1AR results in the colocalization of receptor, βarr, FLNA, and active ERK in membrane ruffles. Reduction of endogenous levels of βarr or FLNA and a catalytically inactive dominant negative MEK1, which prevents ERK activation, inhibit membrane ruffle formation, indicating the functional requirement for βarr, FLNA, and active ERK in this process. Our results indicate that βarr and FLNA cooperate to regulate ERK activation and actin cytoskeleton reorganization.
Cytoskeletal reorganization is fundamental for cell shape change, signaling, locomotion, and many other important dynamic cellular processes. The MAPK JNK, p38, and ERK play key roles in the regulation of cytoskeletal dynamics (21), as many extracellular signals that promote a change in cell shape converge at MAPK, which in turn phosphorylate downstream targets involved in actin cytoskeleton regulation. For example, ERK can phosphorylate myosin light chain kinase (27, 37), calpain (18), focal adhesion kinase (22), and ribosomal S6 kinase (15), which all play important roles in cytoskeletal reorganization and cell migration.
Initially appreciated for their roles in G protein-coupled receptor desensitization and endocytosis, β-arrestins are now considered multifunctional adaptor molecules, with over 20 binding partners, including trafficking proteins, nonreceptor tyrosine kinases, guanine nucleotide exchange factors, and MAPK components, identified to date (28, 29). There exist two isoforms of βarr, βarr1 and βarr2. These share a high degree of homology (~80%) and several biological functions, such as their abilities to bind to agonist-occupied GPCRs, preventing G protein-mediated signal transduction, and to bridge receptors with components of the clathrin-dependent endocytic machinery (clathrin and AP-2). With regard to MAPK, βarr can act as scaffolds for GPCR-mediated activation of ERK1/2 (12, 33), JNK3 (35), and p38 (44). βarr bind multiple components of MAPK cascades to promote efficient activation of MAPK, additionally redirecting them to extranuclear compartments (12, 33, 35). The βarr-mediated cytoplasmic sequestration of active ERK, for example, results in both direct and indirect effects within the cell. First, by redirecting active ERK from the nucleus to the cytoplasm, βarr inhibits activation of the transcription factor Elk-1 (45, 46), a nuclear ERK target, and functionally results in a decrease in proliferation (12, 45). Second, the targeting of active ERK to specific cytoplasmic compartments presumably permits the kinase to phosphorylate a specific subset of substrates. Despite the increasing number of observations indicating that both βarr and MAPK are required for GPCR-dependent chemotaxis and/or cell shape regulation (16, 17, 23, 44), the specific cytoskeletal regulatory pathways involved remain to be elucidated and likely implicate additional key βarr partners.
The filamin family of actin-binding proteins are large scaffolding molecules that integrate cell signaling events and cell shape change (43) and would be attractive candidates to provide a link between βarr-mediated signaling events and cytoskeletal regulation. These proteins are located in the periphery of the cytoplasm, where they cross-link actin filaments into three-dimensional networks and link them to cellular membranes. There are three isoforms of FLN, namely, FLNA, FLNB, and FLNC; these share around 70% homology and are structurally similar. FLNA and FLNB are widely expressed in human tissues, whereas FLNC is expressed predominantly in muscle (46). Structurally, FLNA is a 280-kDa dimer composed of an amino-terminal actin-binding domain; 24 repeat regions, each composed of around 96 amino acids; and two hinge regions that give the molecule flexibility. Dimerization occurs via the carboxy-terminal repeat 24 of FLNA. Both the actin-binding and dimerization domains are required for the branching of actin filaments. FLNA interacts with a number of proteins with roles in signaling and cytoskeletal reorganization and is regulated by phosphorylation. Of particular note is the fact that FLNA binds to several members of the GPCR family, including dopamine D2/D3 (30, 31), calcium-sensing receptor (5, 20), and μ-opioid receptor (39), as well as other classes of receptors, such as the insulin receptor (19). FLNA also interacts with the Rho family GTPases Rho, Rac, Cdc42, and GTP-bound RalA (38). The p21-activated kinase 1, a downstream effector of Rac1 and Cdc42, binds and phosphorylates FLNA and is reciprocally activated by FLNA in a two-way regulatory interaction (47). Finally, like βarr, FLNA has also been implicated in MAPK signaling induced by a variety of extracellular stimuli. Indeed, FLNA can interact with the MAPK kinases MEK1 and MKK4 (34) and is phosphorylated by ribosomal S6 kinase (49), an ERK target.
Using a cytoplasmic yeast two-hybrid system (Sos recruitment system) we identified FLNA as a βarr-binding partner and have assessed here its role in GPCR-induced βarr-mediated ERK activation and cytoskeletal rearrangement. Our data indicate that FLNA and βarr cooperate to orchestrate cell shape change via a signaling pathway that involves ERK activation.
MATERIALS AND METHODS
Abbreviations.
The following abbreviations are used in this paper: AT1AR, angiotensin II type 1a receptor; βarr, β-arrestin(s); CaR, calcium-sensing receptor; FLN, filamin; FLNA, filamin A; GPCR, G protein-coupled receptor; Gal4AD, Gal4 activation domain; LexABD, LexA DNA binding domain; M1MR, muscarinic M1 receptor; Sos, Son of sevenless; SRS, Sos recruitment system; GEF, guanine nucleotide exchange factors; JNK, Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase(s); MAP2K, MAPK kinase(s); mGAP, mammalian GTPase-activating protein; HA, hemagglutinin; Ura, uracil; GST, glutathione S-transferase; GFP, green fluorescent protein; YFP, yellow fluorescent protein; IgG, immunoglobulin G; ACh, acetylcholine; siRNA, small interfering RNA; RNAi, RNA interference; PBS, phosphate-buffered saline; P-ERK1/2, phospho-ERK1/2; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; and SEM, standard error of the mean.
cDNA expression constructs. (i) Yeast two-hybrid expression vectors.
The plasmid pSosGly was generated by inserting an adaptor coding for five glycine residues in frame with and 3′ to the carboxy terminus of the hSos cDNA sequence, using the NcoI and AatII sites contained in pSos (Stratagene), to reduce structural interference between the two proteins in the chimera upon expression. Subsequently, full-length βarr2 (amino acids 1 to 410) was amplified by PCR and introduced downstream of and in frame with the SosGly fusion by use of βssHII and SalI sites to create pSosGlyβarr2. A series of pSosGlyβarr2 C-terminal truncations was created by introducing stop codons at various places in the βarr2 cDNA within pSosGlyβarr2 by use of a QuikChange mutagenesis kit (Stratagene). By use of this methodology, pSosGlyβarr2ΔC4 (coding for amino acids 1 to 392 of βarr2), pSosGlyβarr2ΔC3 (amino acids 1 to 380), pSosGlyβarr2ΔC2 (amino acids 1 to 359), pSosGlyβarr2ΔC1 (amino acids 1 to 337), and pSosGlyβarr2ΔC (amino acids 1 to 316) were generated.
Plasmids for expression of FLNA(22-24), containing amino acids 2363 to 2647 of FLNA, fused to the Gal4AD or LexABD were constructed by excising FLNA(22-24) from pMyr-FLNA(22-24) obtained in the SRS screen (see below) by use of EcoRI and XhoI sites and insertion into EcoRI/XhoI sites of pGAD-GE or EcoRI/SalI sites of pLex10, respectively. To generate βarr1 and βarr2 fused to the Gal4AD, full-length βarr were amplified by PCR and cloned into pGAD-GE using XhoI/XbaI sites. Similarly, βarr2 was amplified by PCR and inserted into the SalI site of pLex10 to create a LexABD-βarr2 hybrid. pLex-Ras and pGAD-Raf have been described previously (42) and were kindly provided by S. Benichou (Institut Cochin, Paris, France).
(ii) Bacterial expression vectors.
To allow the production of a bacterial FLNA fusion protein for use in pulldown experiments, pHis6-TAT-HA-FLNA(22-24) was generated by cloning the EcoRI/XhoI fragment of FLNA(22-24) into pTAT-HA (a kind gift from S. Dowdy, HHMI, Washington University School of Medicine) between KpnI and XhoI restriction sites. βarr cDNAs were amplified by PCR and cloned into pGEX-5X-1 (Amersham) by use of PspAI/XhoI (for βarr1) or SalI/NotI (for βarr2) sites to allow the production of GST-βarr fusion proteins.
(iii) Mammalian expression vectors.
pMyc-FLNA(22-24) and pFLAG-FLNA(22-24) were generated by excising FLNA(22-24) cDNA from pMyr-FLNA(22-24) by use of EcoRI and XhoI sites and inserting it in frame to and downstream of Myc and FLAG tags in pCMVTag3B and pCMVTag2B (Stratagene), respectively, by use of the same sites. cDNAs corresponding to amino acids 2363 to 2554, 2363 to 2522, 2386 to 2647, and 2427 to 2647 of FLNA were amplified by PCR and cloned between EcoRI and XhoI sites in pCMVTag3B to create pMyc-FLNA(2363-2554), pMyc-FLNA(2363-2522), pMyc-FLNA(2386-2647), and pMycFLNA(2427-2647), respectively. pMyc-FLNA(22-24) alanine-scanning mutants 1 to 9 (see Fig. Fig.4)4) were generated using a QuikChange mutagenesis kit.
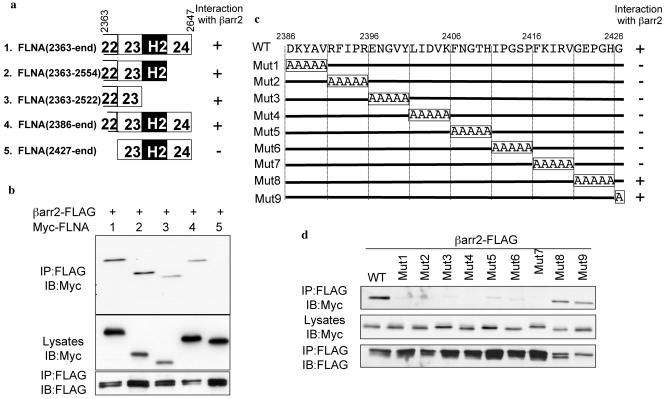
Mapping of the interaction domains of βarr and FLNA. (a) Schematic representation of FLNA(22-24) truncations indicating interaction with βarr2. (b) βarr2-FLAG was expressed with Myc-tagged FLNA(22-24) truncation mutants as indicated. The amounts of the various Myc-FLNA constructs contained in βarr2-FLAG immunoprecipitates (IP) were assessed by Western blotting (IB) with an anti-Myc antibody at a 1:1,000 dilution. (c) Schematic representation of FLNA(22-24) alanine-scanning mutagenesis indicating interaction with βarr2. WT, wild type; Mut1 to Mut9, mutations 1 to 9. (d) βarr2-FLAG was expressed with Myc-tagged alanine-substituted mutants of FLNA(22-24) (mutants 1 to 9) as indicated; the amounts of Myc-tagged constructs contained in βarr2-FLAG immunoprecipitates were assessed by Western blotting. (e) Schematic representation of wild-type (FL) and N- and C-terminal βarr2 truncations (C and N, respectively) indicating interaction with FLNA(22-24). (f) FLAG-FLNA(22-24) or empty control vector was expressed with Myc-tagged wild-type (FL) and N- and C-terminal βarr2 truncations in COS cells. The amounts of the various Myc-βarr2 constructs contained in FLAG-FLNA(22-24) immunoprecipitates were assessed by Western blotting.
Full-length βarr1 (amino acids 1 to 418) and full-length βarr2 were amplified by PCR; βarr1 was cloned into pCMVTag2A and βarr2 to pCMVTag3A by use of EcoRV and SalI sites to generate pFLAG-βarr1 and pMyc-βarr2, respectively. The N-terminal domain of βarr2 (amino acids 1 to 185) and the C-terminal domain of βarr2 (amino acids 186 to 410) were amplified by PCR and cloned either between the EcoRI and XhoI sites (for the N-terminal domain) or between the BamHI and SalI sites (for the C-terminal domain) of pCMVTag3B to create pMyc-βarr2N(1-185) and pMyc-βarr2C(186-410), respectively. The integrity of all constructs was verified by nucleotide sequencing (MWG or Institut Cochin sequencing facility). Sequences of all primers used are available on request.
pcDNA3.1-HA-M1MR was obtained from the Guthrie cDNA Resource Center and pEYFP-tubulin from Clontech. The plasmids for pcDNA3.1-HA-AT1AR, pCMV5-βarr1, pcDNA1-AT1AR-DRY/AAY, and full-length pCMV-FLNA expression plasmid were gifts of M. G. Caron, R. J. Lefkowitz (HHMI, Duke University Medical Center), L. Hunyady (Semmelweis University, Budapest, Hungary), and C. J. Loy (National University of Singapore, Singapore), respectively. R. Seger (The Weizmann Institute of Science, Israel) provided the pMEKK97A and pGFP-ERK2 constructs. pβarr2-FLAG has been described previously (41).
SRS library screening.
Library screening was performed sequentially. First, cdc25H yeast cells (Stratagene) were cotransformed with pSosGlyβarr2ΔC1 bait plasmid (containing amino acids 1 to 337 of βarr2) and a plasmid (pYes2-GAP, a kind gift of A. Aronheim, Rappaport Faculty of Medicine, Haifa, Israel) containing mGAP to reduce Ras GTPase false positives obtained in the screen (3). Subsequently, pSosGlyβarr2ΔC1/mGAP-containing cdc25H yeast cells were transformed with a human thymus cDNA library fused to a myristylation signal (Stratagene) under the control of the galactose-induced pGal1 promoter. Following library transformation, yeast cells were plated onto Leu−/Ura−/Trp− agar plates containing 2% galactose (to induce library expression) and 1% raffinose and placed at 25°C for 40 h before being shifted to 37°C (nonpermissive temperature). Clones that grew at 37°C after 5 to 7 days were scored as potential βarr interactors, and plasmid DNA was subsequently extracted and sequenced. By use of this approach, approximately 1.5 × 106 clones were screened. To determine the specificity of the interactions, prey plasmids were retransformed into pSosGlyβarr2Δ1/mGAP cdc25H yeast. Three days after transformation, yeast cells growing at 25°C on Leu−/Ura−/Trp− plates containing 2% glucose were subsequently patched onto Leu−/Ura−/Trp− plates containing either 2% glucose or 2% galactose-1% raffinose and assessed for growth at 37°C 3 to 5 days later. Clones that grew at 37°C in the presence of galactose (prey induced) but not glucose (prey repressed) were considered bona fide interactors.
Classical yeast two-hybrid assay.
The L40 yeast reporter strain containing a LexA-inducible gene, HIS3, was cotransformed with pLexABD and pGal4AD hybrid expression vectors and plated on selective medium. Transformants were subsequently assayed for histidine auxotrophy as previously described (42).
Cell culture.
COS and Hep2 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (GIBCO), penicillin, and streptomycin (100 IU of penicillin and 100 μg of streptomycin/ml; Sigma) at 37°C in a humidified 5% CO2 atmosphere. Human A7 and M2 melanoma cells were a generous gift of T. P. Stossel and Y. Ohta (Brigham and Women's Hospital, Harvard Medical School). The M2 melanoma cells are FLNA deficient. The A7 cell line was derived from M2 melanoma cells through the stable expression of a plasmid encoding full-length FLNA (11). M2 and A7 cells were maintained in minimal essential medium with Earle's salt supplemented with 8% newborn calf serum, 2% fetal calf serum, penicillin, and streptomycin at 37°C in a humidified 5% CO2 atmosphere. In addition, A7 cells were subcultured in the presence of 0.5 mg/ml G418 (GIBCO) to maintain the presence of the FLNA-containing plasmid.
Immunoprecipitation and immunoblotting.
Approximately 3 × 106 COS cells growing in 100-mm dishes were transiently cotransfected with 1 μg FLAG epitope-tagged βarr or FLNA expression constructs using GENEJuice (Novagen) and 1 μg of plasmids encoding Myc fusion or wild-type proteins, as indicated in the figure legends. Forty-eight hours posttransfection, cells were lysed in 1 ml of cold glycerol lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol, 100 μM Na3VO4, 1 mM NaF, supplemented with protease inhibitors) and clarified by centrifugation at 13,000 × g for 20 min at 4°C. Immunoprecipitations were performed on 500 μg of cell lysates by use of 20 μl of a 50% slurry of monoclonal M2 anti-FLAG-affinity agarose, with constant agitation overnight at 4°C. Following incubation, immune complexes were washed four times with lysis buffer, and immunoprecipitated proteins were detected by Western blot analysis. Additionally, cell lysates (25 μg) were subjected to Western blot analysis to confirm expression of transfected constructs. Coimmunoprecipitation experiments with full-length FLNA or endogenous proteins were performed as described above with the following modifications. Cells were lysed in 1 ml of lysis buffer 1 (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 100 μM Na3VO4, 1 mM NaF, supplemented with protease inhibitors) and clarified by centrifugation at 13,000 × g for 20 min at 4°C. Following immunoprecipitation, immune complexes were washed twice with lysis buffer 1, twice in lysis buffer 2 (50 mM Tris [pH 7.5], 500 mM NaCl, 0.1% NP-40, 0.05% sodium deoxycholate), and twice in lysis buffer 3 (50 mM Tris [pH 7.5], 0.1% NP-40, 0.05% sodium deoxycholate). Coimmunoprecipitation experiments with FLAG-βarr1, GFP-ERK2, and FLNA(22-24) were performed as previously described (45).
Western blotting for FLAG fusion proteins was performed using rabbit polyclonal anti-FLAG antibody (Sigma) at a 1:1,000 dilution. Coprecipitated Myc fusion, βarr1, and full-length FLNA proteins were detected by use of rabbit polyclonal anti-Myc (Santa Cruz) at a 1:1,000 dilution, anti-βarr1 (a kind gift of J. Benovic, University of Philadelphia) at a 1:1,000 dilution, and monoclonal FLNA (Chemicon) antibodies at a 1:1,000 dilution, respectively. Horseradish peroxidase-conjugated polyclonal donkey anti-rabbit or anti-mouse IgG was used as a secondary antibody.
ERK activation experiments were performed essentially as previously described (46). COS cells growing in six-well plates (approximately 3.5 × 105 cells/well) were transfected with plasmids encoding HA-M1MR, HA-AT1AR, or AT1AR-DRY/AAY (250 ng/well) and GFP-ERK2 (125 ng/well) with or without FLAG-βarr1 or -2 (375 ng/well) and with or without Myc-FLNA(22-24) (125 ng/well). Following overnight serum starvation, cells were stimulated with either 100 μM acetylcholine or 1 μM angiotensin II for 5 min and subsequently lysed in 300 μl cold glycerol lysis buffer/well. To determine expression of transfected proteins and phospho-ERK, aliquots of clarified whole-cell lysates (25 μg) were subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-ERK1/2 (Upstate) at a 1:20,000 dilution, anti-phospho-ERK1/2 (Cell Signaling) at a 1:2,000 dilution, or anti-Myc and anti-FLAG rabbit polyclonal antibodies as described above. Horseradish peroxidase-conjugated polyclonal donkey anti-rabbit IgG was used a secondary antibody.
A7 and M2 cell lysates (30 μg) were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with anti-FLNA mouse monoclonal antibody and anti-βarr (A2CT, a kind gift from Robert J. Lefkowitz, HHMI, Duke University Medical Center) at a 1:5,000 dilution, anti-MEK1 (Cell Signaling) at a 1:1,000 dilution, and anti-ERK1/2 rabbit polyclonal antibodies. Horseradish peroxidase-conjugated polyclonal donkey anti-rabbit or anti-mouse IgG was used as a secondary antibody. Blots were developed using either an ECL kit (Amersham) or Supersignal chemiluminescence reagent (Pierce).
Immunofluorescence.
A7 and M2 cells were seeded onto coverslips in six-well plates and used for immunofluorescence 2 days later. Cells were fixed and processed for fluorescence microscopy as previously described (40).
Hep2 cells were transfected and processed for immunofluorescence essentially as previously detailed (25) with some modifications. Briefly, cells were grown to 60 to 70% confluence in 100-mm dishes (approximately 7 × 106 cells) prior to transfection by electroporation in HEBS buffer (20 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 6 mM d-glucose) using two 450-V, 125-μF pulses (Gene Electropulser II; Bio-Rad) and 0.5 μg of relevant cDNAs in the presence or absence of 20 μg of relevant siRNA. Forty-eight or 72 (for siRNA transfections) hours posttransfection, cells were treated as described in the figure legends. Following treatment, cells were fixed in 4% paraformaldehyde-PBS for 20 min at 4°C and quenched for 10 min in 0.27% NH4Cl-0.37% glycine in PBS at room temperature. A 1% bovine serum albumin-0.2% saponin-PBS mixture was subsequently used to block and permeabilize fixed cells. Primary antibody incubations with rat anti-HA (3F10; Roche) or rabbit anti-βarr (A2CT) at a 1:100 dilution, anti-MEK1 (Santa Cruz) at 1:250 dilution, or a mouse anti-FLNA at a 1:200 dilution were performed for 1 h at room temperature. Cells were subsequently washed with 1% bovine serum albumin-0.2% saponin-PBS and incubated with Alexa Fluor donkey anti-rat (647), anti-rabbit (594), or anti-mouse (488) immunoglobulin (Molecular Probes) at a 1:800 dilution for 45 min at room temperature. After being washed, coverslips were mounted on slides in 90% glycerol (Sigma)-3% N-propyl-galate (Sigma)-PBS. Where indicated, coverslips were stained with Alexa Fluor 594 phalloidin (Molecular Probes) according to the manufacturer's instructions before mounting. Essentially the same protocol was used for the detection of P-ERK1/2 in Hep2 cells with the following exception. A mouse anti-P-ERK antibody (Sigma) was used at a 1:100 dilution, and the fluorescently labeled secondary antibodies described above were replaced with a tyramide signal amplification kit (Molecular Probes), which was used according to the manufacturer's instructions. To quantify the extent of agonist-induced ruffling in M1MR- and AT1AR-transfected Hep2 cells, 100 appropriately transfected cells were counted and scored as ruffling or nonruffling. Cells on two coverslips were counted per transfection, and the scoring was repeated on at least three separate transfections.
Confocal images were taken at room temperature with a Bio-Rad MRC 1024 laser scanning confocal system with a Nikon Plan Apo 60× oil immersion lens and an Optiphot 2 microscope equipped with Bio-Rad Lasersharp 2000 software to acquire the images. Images were optimized for contrast in Adobe Photoshop, but no further manipulations were made.
siRNA transfection.
M2 and A7 cells (approximately 4 × 106 cells/dish) and Hep2 cells (approximately 7 × 106 cells/dish) were transfected with siRNAs directed against βarr, FLNA, or control siRNA (20 μg per 100-mm dish) by use of Genesilencer as previously described (1) or by electroporation (for Hep2 cells). The siRNAs targeting βarr or control siRNA have been described previously (1), and the siRNA sequence targeting FLNA is 5′-GGGCTGACAACAGTGTGGTGC-3′, corresponding to base pairs 3074 to 3094 relative to the start codon as previously described for use in an shRNA vector (36). All assays were performed 72 h after siRNA transfection.
Purification of TAT-HA-FLNA(22-24) peptide.
An overnight culture of BL21(DE3)pLysS (Novagen) transformed with His6-TAT-HA-FLNA(22-24) plasmid was diluted 1/20 and grown for 5 h at 37°C. Bacteria were resuspended and lysed by sonication in 8 M urea, 20 mM HEPES, 100 mM NaCl, pH 8.0 (lysis buffer). Insoluble material was eliminated by centrifugation at 17,000 × g for 20 min, and the filtered supernatant was applied to a HiTrap chelating high-performance column (Amersham Biosciences) loaded with 100 mM NiSO4. The column was washed with lysis buffer, and bound protein was eluted with a 10 to 250 mM imidazole gradient in lysis buffer. Elution was monitored at a wavelength of 254 nm. Fractions containing protein were collected, diluted with 2 volumes of 25% glycerol in PBS, and applied to a Sephadex G-25 column preequilibrated with PBS-20% glycerol. The excluded fraction was characterized by SDS-PAGE and Coomassie staining.
Preparation of GST-βarr and in vitro binding assays.
GST-βarr1 or -βarr2 fusion proteins were expressed in BL21(DE3)pLysS (Novagen) and purified on a GSTrap fast-flow column (Amersham Biosciences) according to the manufacturer's instructions. Products eluted with 10 mM glutathione were desalted on a HiTrap desalting column (Amersham Biosciences) in PBS and characterized by SDS-PAGE and Coomassie staining.
For in vitro binding assays, 25-μg portions of GST fusion proteins were immobilized on 20 μl glutathione-Sepharose beads for 1 h at 4°C in PBS. Beads were washed twice in 1 ml PBS and twice in binding buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, supplemented with protease inhibitors). Fifty μg of TAT-HA-FLNA(22-24) was then added in a final volume of 500 μl of binding buffer. After 1 h at 4°C, beads were washed four times with 1 ml of binding buffer. Complexes were applied on 10% SDS-PAGE gels, and proteins were revealed by Ponceau red staining and Western blotting with anti-HA 3F10 monoclonal antibody and anti-GST polyclonal antibody (generous gift of N. Varin, Institut Cochin, Paris, France).
RESULTS
Identification of FLNA as a βarr-interacting protein.
To identify proteins interacting with βarr, we made use of a cytoplasmic yeast two-hybrid system based on the membrane translocation of Sos called the SRS (4). The SRS uses cdc25H, a strain of yeast which grows normally at 25°C but which harbors a thermosensitive mutation in the yeast Ras GEF Cdc25, abolishing growth at 37°C. Membrane localization of hSos bypasses the requirement for a functional yeast Ras GEF, Cdc25, permitting growth at the restrictive temperature. Interaction between two-hybrid proteins can result in hSos membrane localization. For this method, the bait is fused to hSos, whereas the cDNA-derived prey proteins are fused to a myristylation signal targeting them to the membrane. Protein-protein interaction results in the membrane translocation of hSos, allowing the yeast to grow at 37°C in the presence of galactose.
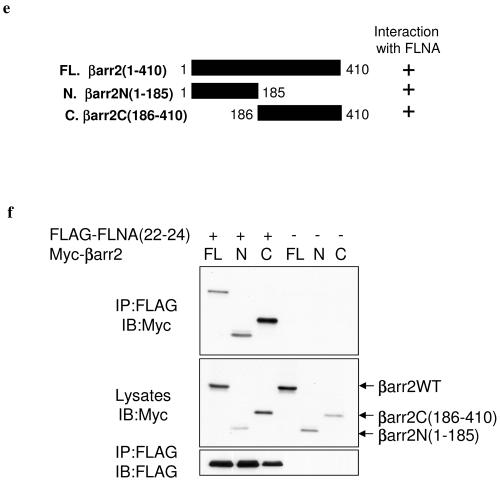
As full-length βarr2 self-activated the SRS (Fig. (Fig.1a)1a) we used a C-terminally truncated form of βarr2 (pSosGlyβarr2ΔC1) containing amino acid residues 1 to 337 to screen a human thymus cDNA library (Fig. (Fig.1b).1b). Approximately 1.5 × 106 library colonies were screened, and 10 of the 135 positive clones identified were found to encode the actin-binding protein FLNA (Fig. (Fig.1c).1c). The smallest partial clone obtained in the SRS screen [pMyr-FLNA(22-24)] contained amino acids 2363 to 2647 of FLNA, corresponding to the majority of repeat 22 through to its carboxy terminus (Fig. (Fig.1c).1c). The specificity of the interaction was confirmed by growth on Leu−/Ura−/Trp− plates containing galactose at 37°C following retransformation of pMyr-FLNA(22-24) into pSosGlyβarr2ΔC1-expressing cdc25H yeast (Fig. (Fig.1d1d).
Identification of FLNA as a βarr-binding protein. (a) An adaptor coding for five glycine residues was inserted between the cDNAs for hSos and full-length βarr2 to reduce potential structural interference between the two proteins upon expression. When the pSosGlyβarr2 chimera (containing amino acids 1 to 410 of βarr2) was introduced into cdc25H yeast in the absence of prey proteins, it promoted growth at 37°C, while a control plasmid containing the SosGly fusion alone did not. This indicates that full-length βarr2 on its own is capable of targeting Sos to the membrane and is therefore not suitable as a bait. (b) To identify potential viable baits, a series of βarr2 C-terminal truncations (containing amino acids 1 to 316, 1 to 337, 1 to 359, 1 to 380, or 1 to 392) was cloned in frame to and downstream of the hSosGly coding sequence. Cdc25H yeast cells were transformed with the truncations, incubated for 3 days, and subsequently replica plated and assessed for growth at 37°C. Growth at 37°C of the truncations is indicated. βarr2ΔC1, a bait containing amino acids 1 to 337 and giving no background in yeast, was subsequently used to screen the library. (c) Schematic representation of FLNA indicating the actin-binding domain (ABD), 24 repeat regions, and two hinge domains (H1 and H2). The 10 partial FLNA clones identified in the SRS screen of a human thymus cDNA library are depicted as black bars. (d) Retransformation of the smallest prey plasmid, pMyr-FLNA(22-24), containing amino acids 2363 to 2647 of FLNA, into pSosGlyβarr2ΔC1-containing cdc25H yeast. Colonies that grew at 25°C following transformation were patched onto both galactose (prey expression-induced) and control glucose (prey expression-repressed) Trp−/Leu−/Ura− dropout plates. Growth at 37°C was assessed 2 or 3 days later. Yeast patches grew on galactose-containing plates only at 37°C, confirming the interaction between βarr2 and FLNA. (e) The L40 yeast reporter strain containing the indicated plasmids was analyzed for histidine auxotrophy. Transformants were patched on selective medium with histidine (+His) and then replica plated onto medium lacking histidine (−His) and incubated for 2 or 3 days. Growth in the absence of histidine indicates interaction between full-length βarr1 and βarr2 with FLNA(22-24). The Ras-Raf interaction was used as a positive control.
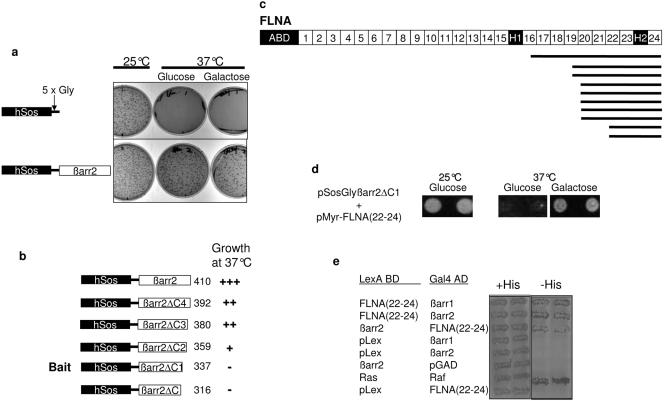
To ascertain if FLNA could interact with full-length βarr, we used a classical two-hybrid system in L40 reporter yeast. Cotransfection of FLNA(22-24) with βarr1 or βarr2 constructs conferred the ability of yeast to grow on Leu−/Trp−/His− plates, whereas cotransfection of FLNA(22-24) or βarr with control vectors did not (Fig. (Fig.1e),1e), indicating that both full-length βarr1 and βarr2 are able to interact with FLNA. Confirming the findings obtained by the yeast two-hybrid system, βarr2 containing a FLAG epitope tag coimmunoprecipitated with Myc-tagged FLNA(22-24) from COS (Fig. (Fig.2a)2a) and HeLa cells (data not shown). Similarly, FLAG-epitope-tagged FLNA(22-24) coimmunoprecipitated with βarr1 (Fig. (Fig.2b)2b) and βarr2 (data not shown) from COS cells. Endogenous full-length FLNA also coimmunoprecipitated with FLAG-epitope-tagged βarr2 (Fig. (Fig.2c),2c), and, finally, endogenous FLNA could be coimmunoprecipitated with endogenous βarr1 from Hep2 cells (Fig. (Fig.2d2d).
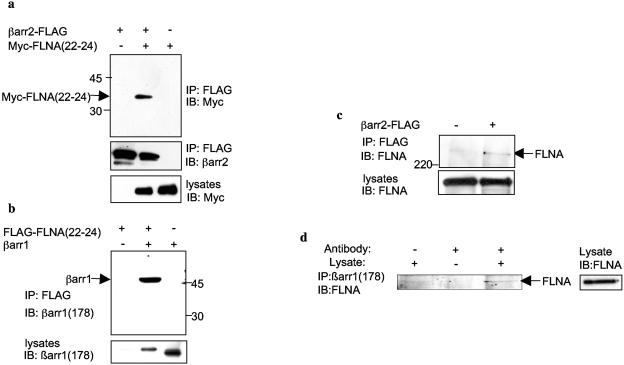
βarr and FLNA associate in mammalian cells. (a) COS cells growing in 100-mm dishes were transfected with 1 μg of C-terminally FLAG-tagged βarr2 (βarr2-FLAG) or empty control vector with 1 μg Myc-FLNA(22-24) containing amino acids 2363 to 2647 of FLNA or empty control vector. Immunoprecipitations (IP) were performed as described in Materials and Methods. Immunoprecipitates were analyzed by SDS-PAGE and probed with polyclonal anti-Myc antibody at a 1:1,000 dilution. Lysates of Myc-FLNA(22-24) represent 10% of the input used in the coimmunoprecipitation experiment. IB, Western blotting. (b) COS cells were transfected with 1 μg of FLAG-tagged FLNA(22-24) or empty control vector with or without 1 μg of βarr1 cDNA in pCMV5. Immunoprecipitates were analyzed by SDS-PAGE and probed with anti-βarr1-specific antiserum at a 1:1,000 dilution. Lysates of βarr1 represent 5% of the input used in the coimmunoprecipitation experiment. (c) COS cells were transfected with 2 μg of βarr2-FLAG or empty control vector. Immunoprecipitates were analyzed by SDS-PAGE and probed with an anti-FLNA antibody at a 1:1,000 dilution to detect endogenous full-length FLNA. Lysates of FLNA represent 5% of the input used in the coimmunoprecipitation experiment. (d) Endogenous FLNA from Hep2 cell lysates was coimmunoprecipitated with endogenous βarr using anti-βarr1 antibodies [βarr(1178)]. Immunoprecipitates were analyzed as described above.
As coimmunoprecipitation experiments cannot rule out the possibility that the interaction between FLNA and βarr occurs indirectly via other proteins, we performed pulldown experiments using purified recombinant proteins. GST fusion proteins of βarr1 and βarr2 were incubated with purified His6-TAT-HA-FLNA(22-24). Complexes were isolated with glutathione-Sepharose, subjected to SDS-PAGE, and transferred to nitrocellulose. Ponceau red staining revealed that purified FLNA(22-24) can interact directly with both βarr1 and βarr2 (Fig. (Fig.3).3). Binding was specific, as no FLNA(22-24) was detected in the presence of GST alone. Taken together, these data indicate that FLNA is a βarr-binding protein and that the interaction takes place via amino acids 2363 to 2647 of FLNA, corresponding to the distal two-thirds of repeat 22 through to its carboxy terminus.
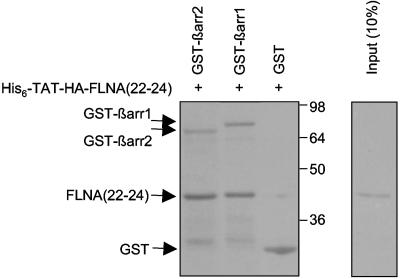
Direct binding of βarr to FLNA(22-24) in vitro. GST alone, GST-βarr1, or GST-βarr2 bound to glutathione-Sepharose was incubated with purified His6-TAT-HA-FLNA(22-24). Complexes were isolated and washed as described in Materials and Methods. Ten percent of the His6-TAT-HA-FLNA(22-24) peptide used in the pulldown experiments was loaded as a control. After samples were resolved by SDS-PAGE, the gel was transferred to nitrocellulose membrane and stained with Ponceau red solution. Positions of purified proteins are depicted. The identities of these proteins were confirmed by immunoblot experiments using anti-HA and anti-GST antibodies (not shown). GST-βarr1 retained 21% ± 5% of the input, and GST-βarr2 retained 17% ± 2% (n = 4).
Mapping the binding domains of FLNA and βarr.
To further refine the βarr-binding region of FLNA within repeat regions 22 to 24, we created a series of FLNA(22-24) truncations. Deletion either of repeat 24 or of repeat 24 and the second hinge domain (H2) together had no effect on the binding of βarr to FLNA in coimmunoprecipitation experiments (Fig. 4a and b). Similarly, removal of amino acids 2363 to 2385 from repeat 22 of FLNA had no effect on βarr-FLNA interaction (Fig. 4a and b). In contrast, the removal of all of repeat 22 resulted in the complete loss of the ability of βarr to bind FLNA (Fig. 4a and b). This indicates that amino acids 2386 to 2427 contained within repeat 22 of FLNA are essential for βarr binding. Similar results were obtained in yeast two-hybrid experiments (data not shown). This region is located near the regions of FLNA which interact with most FLNA-binding GPCRs, encompassing repeat domains 14 to 21 or repeat 24 (5, 13, 20, 31, 39). Of particular interest with regard to MAPK signaling is the fact that this region (residues 2386 to 2427 of FLNA) is contained within previously identified binding sites for the two FLNA-interacting MAP2K proteins, MEK1 (amino acids 2282 to 2647) and MKK4 (amino acids 2282 to 2454) (34). In an attempt to identify the specific residues contained within amino acids 2386 to 2427 of FLNA that are responsible for the binding of βarr, a series of quintuple alanine-scanning substitutions in this region were created in FLNA(22-24) and tested for their abilities to bind βarr in coimmunoprecipitation experiments (Fig. 4c and d). The data demonstrate that several successive substitutions severely impair or abolish the ability of βarr to interact with FLNA(22-24). These results therefore indicate that multiple amino acid residues contained within the 2386 to 2420 FLNA region likely participate in βarr binding.
To determine which region of βarr is important for FLNA(22-24) binding, coimmunoprecipitation experiments were carried out using either the N-terminal part (amino acids 1 to 185, corresponding to the short N-terminal extremity and to the amino-terminal globular domain) or the C terminal part (186 to 410, corresponding to the carboxy-terminal globular domain and C-terminal tail) of βarr2 (Fig. 4e and f). FLNA(22-24) was actually found to bind both domains of βarr2, indicating the existence of a complex interaction which involves the presence of multiple binding sites on βarr for FLNA. Similar data were obtained in yeast two-hybrid experiments (data not shown).
FLNA(22-24) enhances GPCR-induced βarr-mediated activation of ERK in COS cells.
βarr have recently been shown to play an important role in MAPK signaling, functioning as scaffolds for GPCR-stimulated MAPK activation by binding component kinases of the cascades, such as Raf-1 and ERK2. Interestingly, FLNA has also been implicated in the regulation of MAPK signaling with regard to the GPCR CaR (5, 20, 34) and, as mentioned above, interacts with the MAP2Ks MKK4 and MEK1. The MEK1-binding site on FLNA overlaps the βarr-binding site identified here. Since the βarr-MEK1 complex may occur, at least in part, through indirect interaction (33), we investigated whether the interaction between βarr and FLNA contributes to agonist-evoked GPCR activation of ERK, looking at the effect of FLNA(22-24) overexpression on βarr-mediated ERK activation in COS cells. Overexpression of βarr1 (Fig. 5a and b) or βarr2 (data not shown) moderately enhanced M1MR- and AT1AR-induced GFP-ERK2 phosphorylation, as assessed by immunoblotting of COS cell lysates with anti-phospho-ERK antibodies, in agreement with previous findings (46). GFP-ERK2, which has been shown to behave like wild-type ERK1/2 (46), was used in these experiments to control for ERK phosphorylation in the transfected pool of cells. When βarr1 was overexpressed in combination with FLNA(22-24), M1MR-induced ERK activation was significantly potentiated (enhancement of 48% ± 13%; P < 0.05 [n = 4]) compared to stimulated control cells overexpressing βarr1 alone (Fig. (Fig.5a),5a), indicating a role for FLNA in GPCR-induced ERK activation. A similar potentiation was seen with the AT1AR when βarr1 was overexpressed with FLNA(22-24) (Fig. (Fig.5b).5b). As activation of ERK can occur via both G protein-dependent and G protein-independent (i.e., βarr-dependent) mechanisms, we made use of a mutant AT1AR, which is mutated in its DRY motif (DRY/AAY) and thereby rendered severely impaired in Gαq/11 coupling in COS cells (24), to determine if the observed potentiation of ERK activation by FLNA(22-24) mentioned above does indeed involve a βarr-dependent pathway. A number of recent studies indicate that activation of ERK via this mutant receptor is completely independent of G proteins and is exclusively mediated by βarr (48, 2). AngII-induced activation of ERK via the DRY/AAY receptor was increased fourfold by βarr1 and further enhanced approximately twofold when βarr1 and FLNA(22-24) were both overexpressed (Fig. (Fig.5c).5c). This finding confirms that the increase in active ERK observed when FLNA(22-24) and βarr1 are both overexpressed is via a βarr-dependent pathway and not via a Gαq/11-dependent pathway. Since FLNA(23-24) did not coimmunoprecipitate with βarr (Fig. (Fig.4),4), we anticipated that this fragment would not potentiate βarr-mediated activation of ERK as observed with FLNA(22-24). As expected, AngII-induced activation of ERK via the AT1AR was potentiated when βarr1 was overexpressed with FLNA(22-24) but not with FLNA(23-24) (Fig. (Fig.5d).5d). This finding is consistent with the hypothesis that the βarr-binding site contained within FLNA(22-24) contributes to the increased activation of ERK when both proteins are overexpressed.
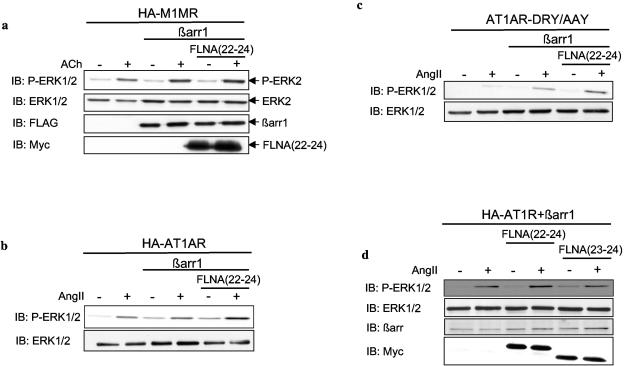
GPCR-induced βarr-mediated activation of ERK is enhanced by FLNA(22-24). (a) Serum-starved COS cells transfected with plasmids encoding HA-M1MR, GFP-ERK2, FLAG-βarr1, or empty control vector and Myc-FLNA(22-24) or empty control vector were left untreated or were stimulated with 100 μM ACh for 5 min at 37°C. Cell lysates were resolved by SDS-PAGE, and phosphorylation of ERK2 was assessed by immunoblotting (IB) using an anti-phospho-ERK1/2 antibody at a 1:1,000 dilution. Total GFP-ERK2, FLAG-βarr1, and Myc-FLNA(22-24) expression levels were confirmed by probing the immunoblots with anti-ERK1/2 (1:20,000 dilution), anti-FLAG (1:1,000 dilution), and anti-Myc (1:1,000 dilution) antibodies, respectively. Gels were scanned, and phospho-ERK2 and total ERK2 levels were determined using NIH ImageJ. The phospho-ERK2:total ERK2 ratio was calculated for receptor-stimulated cells expressing M1MR, βarr1, and ERK2 (lane 4), and this value was normalized to 100% ERK activation. The phospho-ERK2:total ERK2 ratio was subsequently calculated for ACh-stimulated cells expressing M1MR, βarr1, ERK2, and FLNA(22-24) (lane 6), which, when compared to cells expressing M1MR, βarr1, and ERK2 (100%), gave a result of 148% ± 13%. The data represent the mean ± SEM of four independent experiments (P < 0.05). (b and c) Serum-starved COS cells transfected with plasmids encoding HA-AT1AR (b) or AT1AR-DRY/AAY (c), GFP-ERK2, FLAG-βarr1, or empty control vector and Myc-FLNA(22-24) or empty control vector were left untreated or were stimulated with 1 μM AngII for 5 min at 37°C. Cell lysates were resolved by SDS-PAGE, and phospho-ERK2 and total ERK2 levels were assessed by immunoblotting as described for panel a. Expressions of FLAG-βarr1 and Myc-FLNA(22-24) were confirmed by immunoblotting as described for panel a (not shown). (d) Serum-starved COS cells transfected with plasmids encoding HA-AT1AR, GFP-ERK2, FLAG-βarr1, and either Myc-FLNA(22-24), Myc-FLNA(23-24), or empty control vector were left untreated or were stimulated with 1 μM AngII for 5 min at 37°C. Cell lysates were resolved by SDS-PAGE, and expressions of phospho-ERK2, total ERK2, FLAG-βarr1, Myc-FLNA(22-24), and Myc-FLNA(23-24) were assessed by immunoblotting as described for panel a.
Taken together, the above results indicate that the interaction of βarr with FLNA may control the extent of GPCR-induced βarr-mediated ERK activation. As the carboxy terminus of FLNA has been implicated in binding to MAPK cascade components (see above), we tested whether FLNA(22-24) expression could increase the previously reported interaction between βarr and ERK2. The amount of GFP-ERK2 detected in FLAG coimmunoprecipitates was markedly increased upon coexpression of FLAG-βarr1 with FLNA(22-24) (Fig. (Fig.6),6), which is consistent with the idea that FLNA can facilitate the formation of βarr-ERK complexes.
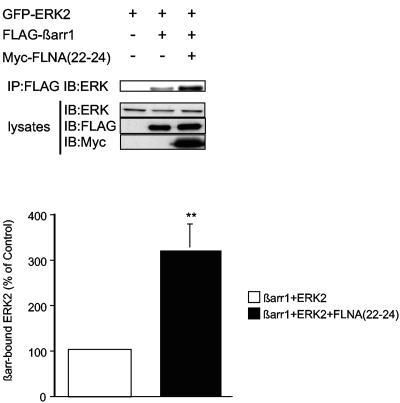
FLNA(22-24) enhances βarr-ERK2 association. COS cells were transiently transfected with plasmids encoding GFP-ERK2, FLAG-βarr1, and Myc-FLNA(22-24) as indicated. FLAG immunoprecipitates were probed for coprecipitated ERK2. Equivalent expression of constructs was confirmed by immunoblotting of cell lysates. The histogram shows the amount of GFP-ERK2 detected in FLAG-βarr1 immunoprecipitates. Data are normalized to 100% for cells expressing βarr and GFP-ERK2 and represent the mean ± SEM of five independent experiments (**, P < 0.02).
FLNA is essential for full βarr-mediated ERK activation following M1MR activation.
Since FLNA(22-24) potentiates GPCR-mediated βarr-dependent ERK activation, we investigated whether this functional activity could be recapitulated by the full-length protein. The specific effect of FLNA expression on βarr-mediated ERK activation was investigated by use of FLNA-deficient M2 cells and their FLNA-replete counterpart, A7 cells (Fig. (Fig.7),7), as a model. Both cell types contained equivalent amounts of the MAPK signaling proteins βarr, MEK1, and ERK (Fig. (Fig.7a,7a, left panels), and the distributions of βarr in both cell types were similar (Fig. (Fig.7a,7a, right panels). This model was used to confirm that maximal ERK activation requires cooperation between βarr and full-length FLNA. siRNA against βarr, recently employed to investigate the role of βarr in GPCR-induced ERK activation (1, 2, 48), was used to inhibit βarr expression in both M2 and A7 cells. The siRNA directed against βarr led to an approximately 50% decrease in endogenous βarr levels in both M2 and A7 cells, as assessed by Western blotting (Fig. (Fig.7b).7b). In A7 cells transfected with the M1MR and control siRNA oligonucleotides, a robust activation of ERK was observed following receptor activation (Fig. (Fig.7c,7c, lanes 1 and 2). The siRNA-mediated reduction of βarr in A7 cells impaired M1MR-induced ERK activation by approximately 30% (Fig. (Fig.7c,7c, lanes 3 and 4). M2 cells transfected with M1MR and control siRNA oligonucleotides demonstrated reduced receptor-activated levels of active ERK compared to A7 cells (Fig. (Fig.7c,7c, lanes 6 and 2, respectively). In M2 cells with reduced levels of βarr following transfection with specific siRNA, the induction of ERK was similar to that of cells transfected with control oligonucleotides (Fig. (Fig.7c,7c, lanes 8 and 6, respectively). Similar results were obtained using the AT1AR (data not shown).
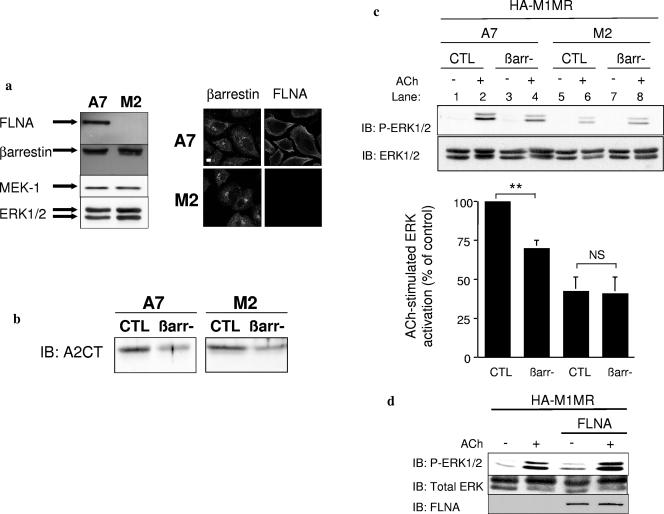
Requirement of FLNA for M1MR-induced βarr-mediated ERK activation. (a) M2 and A7 cells were lysed, and equivalent amounts of proteins were subjected to SDS-PAGE and immunoblotted with the indicated antibodies (left panel) as described in Materials and Methods. M2 and A7 cells were grown and fixed on coverslips as described in Materials and Methods. Cells were then processed for fluorescence microscopy using A2CT directed against βarr and an anti-FLNA antibody and revealed by Alexa Fluor 594- or 647-conjugated antibodies (right panel). Scale bar, 5 μm. (b) Serum-starved M2 and A7 cells transfected with control (CTL) or βarr siRNA oligonucleotides were lysed 72 h posttransfection, and equivalent amounts of protein were subjected to SDS-PAGE and immunoblotted (IB) with the anti-βarr antibody A2CT at a 1:5,000 dilution. (c) Serum-starved M2 and A7 cells transfected with a plasmid encoding HA-M1MR and either control or βarr1/2 siRNA oligonucleotides were left untreated or were stimulated with 100 μM ACh for 5 min at 37°C. Flow cytometry was performed with anti-HA antibodies on M2/A7 cells transfected with the HA-M1MR to determine receptor density at the cell surface. Mean fluorescence intensity in A7 cells (n = 3) was 3.76 ± 0.35 times higher than that in M2 cells. A7 cells are considerably larger than M2 cells, and the number of surface receptors/milligram protein was finally found to be equivalent between the cell types. Equal receptor amounts of cell lysates (corresponding to equivalent protein levels) were resolved by SDS-PAGE, and phosphorylation of ERK1/2 was assessed by immunoblotting using an anti-phospho-ERK1/2 antibody (1:1,000 dilution). Total ERK1/2 levels were controlled using an anti-ERK1/2 antibody (1:20,000 dilution). Data shown represent the mean ± SEM calculated from three independent experiments. The results in each experiment were normalized to the level of ERK phosphorylation in stimulated A7 cells transfected with control siRNA. Significance was evaluated using a t test. **, P < 0.02. NS, not significant. (d) Serum-starved M2 cells transfected with a plasmid encoding HA-M1MR and either pCMV-FLNA encoding full-length FLNA (32) or a control plasmid were left untreated or were stimulated with 100 μM for 5 min at 37°C. P-ERK1/2 and total ERK1/2 levels were assessed by immunoblotting as described for panel c. FLNA was detected using an anti-FLNA antibody (1:1,000 dilution).
Finally, to confirm that the different levels of ERK activation achieved in M1MR-transfected M2 and A7 cells were correlated with the presence or absence of FLNA, full-length FLNA cDNA was transiently transfected in M2 cells; this enhanced ERK activation following M1MR stimulation (Fig. (Fig.7d).7d). Taken together, these data indicate that to achieve full activation of ERK via the M1MR in this model system, the simultaneous expression of both FLNA and βarr is essential.
Colocalization of βarr, FLNA, and active ERK in membrane ruffles in Hep2 cells following M1MR activation.
FLNA is an actin cross-linker that is appreciated as an important player in actin remodeling. It is enriched in membrane ruffle structures, which contain bundles of actin filaments that are more densely packed than those of the underlying cell lamellae. βarr have recently been shown to be involved in GPCR-induced actin cytoskeletal changes, which require βarr-dependent MAPK activation (16, 17, 23). If βarr, FLNA, and active ERK do cooperate to orchestrate receptor-mediated cell shape changes, they would be expected to colocalize in areas of active cytoskeletal remodeling following receptor activation. We therefore extended our studies to Hep2 cells, which undergo substantial membrane ruffling in response to various stimuli (10). In Hep2 cells expressing M1MR, agonist treatment (acetylcholine) for 5 min resulted in extensive cell shape change with the rapid formation of membrane ruffles, a marker of cytoskeletal reorganization (Fig. (Fig.8A,8A, compare panels a [unstimulated control cells] and c). Actin was localized to these membrane structures following receptor stimulation, as assessed by phalloidin staining (Fig. (Fig.8A,8A, compare panels b and d). Under these conditions, 70% ± 0.8% (n = 3) of the M1MR-transfected cells formed ruffles (Fig. (Fig.9D).9D). Similar results were obtained with the AT1AR (Fig. (Fig.8B,8B, compare panels a and b [unstimulated control cells] with c and d), with 68% ± 3.6% (n = 3) of transfected cells demonstrating ruffling following angiotensin II stimulation (Fig. (Fig.9E).9E). Interestingly, receptor, endogenous βarr, and FLNA were colocalized in these membrane ruffles following agonist stimulation, as shown in Fig. Fig.8C,8C, panels a through d. As anticipated, active phosphorylated ERK was also found in ruffles following receptor activation (Fig. (Fig.8C,8C, panels e through h). As FLNA(22-24), which lacks the actin-binding domain, was found to potentiate ERK activation (Fig. (Fig.5)5) like full-length FLNA, one would expect to observe a colocalization of FLNA(22-24) with phosphorylated ERK in membrane ruffles following receptor stimulation. As shown in Fig. Fig.8C,8C, panels i through l, FLNA(22-24) was indeed colocalized with activated receptor in the membrane ruffles of Hep2 cells. To rule out potential artifactual colocalization of proteins due to variations of cell thickness in the ruffling cells, Hep2 cells were cotransfected with M1MR and YFP-tubulin as a negative control. Activated M1MR, but not tubulin, was concentrated in actin-rich ruffles (Fig. (Fig.8C,8C, panels m through p). This suggests that the observed enrichment of M1MR, βarr, and FLNA immunofluorescence in the membrane ruffles of agonist-treated cells does indeed reflect true colocalization. In conclusion, the GPCR-stimulated colocalization of βarr, FLNA, and P-ERK at membrane ruffles is consistent with a cooperative role for βarr- and FLNA-mediated ERK activation in this GPCR-dependent process.
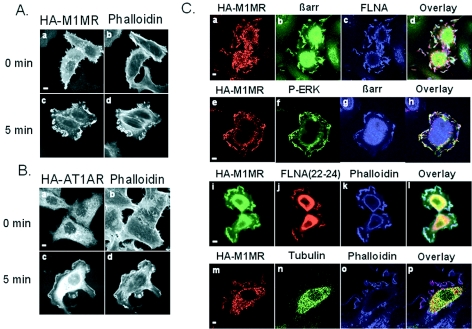
M1MR, βarr, FLNA, and active ERK colocalize in membrane ruffles in Hep2 cells following agonist stimulation. (A) Hep2 cells transfected with HA-M1MR were left untreated or were stimulated with 100 μM ACh for 5 min, fixed, processed for immunofluorescence with an anti-HA (3F10) antibody, and revealed using an Alexa Fluor 488 antibody (a and c). Alexa Fluor 594 phalloidin was used to stain for actin (b and d). (B) Hep2 cells transfected with HA-AT1AR were left untreated or were stimulated with 1 μM AngII for 5 min, fixed, and processed for immunofluorescence as described for panel A. (C) Hep2 cells transfected with HA-M1MR were stimulated with 100 μM ACh for 5 min, fixed, and processed for immunofluorescence using anti-HA (panels a and e), anti-βarr (panels b and g), anti-FLNA (panel c), and anti-P-ERK1/2 (panel f) antibodies. Hep2 cells cotransfected with HA-M1MR and Myc-FLNA(22-24) (panels i through l) or HA-M1MR and YFP-tubulin (panels m through p) were stimulated with 100 μM ACh for 5 min, fixed, and processed for immunofluorescence as described for panel A. Panel n shows fluorescence emitted by YFP-tubulin. Scale bars, 5 μm.
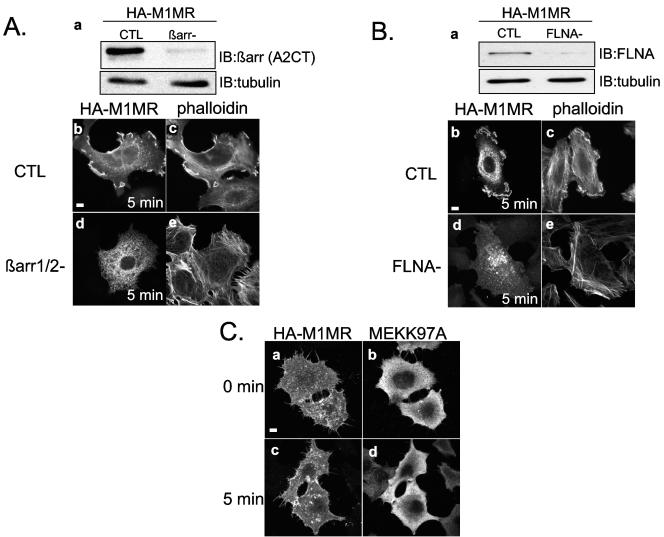
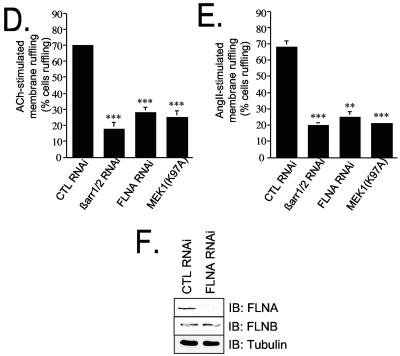
βarr, FLNA and active ERK are all required to achieve full M1MR- and AT1AR-induced ruffle formation in Hep2 cells. (A) (a) Representative blot showing Hep2 cells transfected with either control (CTL) or βarr RNAi oligonucleotides. IB, immunoblotting. (b through e) Hep2 cells transfected with HA-M1MR and either control or βarr siRNA oligonucleotides were stimulated with 100 μM ACh for 5 min, fixed, and processed for immunofluorescence using an anti-HA (3F10) antibody (b and d) and Alexa Fluor 594 phalloidin to stain for actin (c and e). (B) (a) Representative blot showing Hep2 cells transfected with either control or FLNA RNAi oligonucleotides. (b through e) Hep2 cells transfected with HA-M1MR and either control or FLNA siRNA oligonucleotides were stimulated with 100 μM ACh for 5 min, fixed, and processed for immunofluorescence as described for panel A. (C) Hep2 cells cotransfected with HA-M1MR and MEK(K97A) were left untreated or were stimulated with 100 μM ACh for 5 min, fixed, and processed for immunofluorescence using anti-HA (3F10) and anti-MEK antibodies. (D) Histogram showing the quantification from three independent experiments of ACh-induced membrane ruffling in Hep2 cells transfected with the HA-M1MR and either control, βarr1/2, or FLNA RNAi oligonucleotides or MEK(K97A) expression vector. Phalloidin staining was routinely used for quantification, and approximately 100 appropriately transfected cells were scored as ruffling or nonruffling per experiment. The results shown represent the means ± standard errors of the means from three independent transfections. ***, P < 0.001. Significance was evaluated using a t test. (E) Histogram showing the quantification from three independent experiments of AngII-induced membrane ruffling in Hep2 cells transfected with the HA-AT1AR and either CTL, βarr1/2, or FLNA RNAi oligonucleotides or MEK(K97A) expression vector. Approximately 100 appropriately transfected cells were scored as ruffling or nonruffling per experiment. The results shown represent the mean ± standard error of the mean from three independent transfections. **, P < 0.02; ***, P < 0.001. Significance was evaluated using a t test. (F) Representative blot showing Hep2 cells transfected with either control or FLNA RNAi oligonucleotides. Immunoblotting was performed with anti-FLNA or anti-FLNB isoform-specific antibodies (1:1,000 dilution; Chemicon). The blot demonstrates that the knockdown is specific for FLNA and does not affect FLNB. Scale bars, 5 μm.
Essential requirement for βarr, FLNA, and active ERK for M1MR- and AT1AR-induced membrane ruffling in Hep2 cells.
To investigate the requirement for βarr and FLNA in M1MR- and AT1AR-induced ruffling in Hep2 cells, we employed siRNAs to reduce the expression of endogenous levels of these proteins. M1MR-transfected Hep2 cells showed a significant reduction in endogenous βarr levels following siRNA cotransfection (Fig. (Fig.9A,9A, panel a) and exhibited a severely impaired ruffling response. Only 18% ± 3.8% (n = 3) of the M1MR- and βarr RNAi-cotransfected cells ruffled compared to cells cotransfected with M1MR and control siRNA (Fig. (Fig.9A,9A, compare panels b and c with d and e and Fig. Fig.9D).9D). Similar results were obtained by use of Hep2 cells cotransfected with the AT1AR and βarr RNAi, where, compared to their control RNAi-transfected counterparts, only 20% ± 1.59% (n = 3) of the ruffling response was observed (Fig. (Fig.9E9E and data not shown). Similarly, a reduction in FLNA levels by RNAi (Fig. (Fig.9B,9B, panel a) inhibited M1MR-induced membrane ruffling compared to that of control RNAi-transfected cells (Fig. (Fig.9B,9B, compare panels d and e with b and c and Fig. Fig.9D);9D); the reduction was comparable to that observed in βarr-depleted cells. Essentially identical results were obtained using the AT1AR receptor (Fig. (Fig.9E9E and data not shown). To further substantiate the functional connection between the cooperative effect of βarr and FLNA on cytoskeletal rearrangement, on one hand, and ERK activation, on the other hand, the direct involvement of active ERK in ruffle formation was investigated in the same model system. Transfection of Hep2 cells with a plasmid encoding MEK1(K97A), a dominant negative mutant of MEK1 which interacts with ERK and inhibits its activation, reduced M1MR-stimulated membrane ruffling to levels similar to those observed for cells transfected with βarr1/2 and FLNA siRNAs. This finding indicates that active ERK is also required for ruffle formation in these cells (Fig. (Fig.9C,9C, panels c and d, and D). Similar results were obtained with Hep2 cells cotransfected with the AT1AR and MEK1(K97A) (Fig. (Fig.9E9E and data not shown). Taken together, these results indicate an essential requirement for βarr, FLNA, and active ERK in M1MR- and AT1AR-mediated cell shape change in our system, with the selective inhibition of each component being sufficient to cause a profound (60% or more) impairment of ruffle formation.
DISCUSSION
The studies described here indicate that the scaffolding and actin-binding protein FLNA interacts with βarr and support a model where these two molecules cooperate to achieve full GPCR- and βarr-dependent activation of ERK. We also present evidence to suggest that the extent and the subcellular localization of the ERK activation, which is achieved through this double scaffold, is capable of controlling cytoskeletal rearrangements such as the formation of membrane ruffles in Hep2 cells.
The involvement of FLNA in ERK activation downstream of an activated GPCR was documented for the CaR (5, 20), and it was proposed that the functions of FLNA in this process would be to stabilize the receptor at the plasma membrane and to target signaling proteins to the receptor (50). Our data indicate that βarr, which interact with both FLNA- and ligand-activated GPCRs, likely represent a third critical component of the GPCR-FLNA signaling complex. In particular, genetic or siRNA-mediated reduction of either βarr or FLNA was found to significantly impair M1MR-induced ERK activation. FLNA has also been shown to have effects on ERK signaling for receptors of other families, such as the insulin receptor (19). In FLNA-deficient M2 cells, activation of ERK via the insulin receptor is enhanced compared to that seen for FLNA-replete A7 cells, and FLNA has thus been proposed to play a negative role in ERK signaling via this receptor (19). Our findings on M1MR-induced ERK activation in M2 and A7 cells are in clear contrast to this result, with the divergent effects observed upon FLNA depletion likely explicable by different mechanisms used to activate ERK by GPCRs and the insulin receptor in this cell model.
FLNA binds the MAP2Ks MEK1 (weak interaction) and MKK4 (strong interaction) (34), which are upstream activators of ERK and JNK, respectively. The minimal βarr-binding site on FLNA identified by deletion mapping and alanine-scanning mutagenesis appears to be located between amino acids 2386 and 2420, contained within tandem repeat 22 of FLNA. Interestingly, the βarr-binding site is contained within the previously identified binding sites for MEK1 (amino acids 2282 to 2647; repeats 21 to 24) and MKK4 (amino acids 2282 to 2454; repeats 22 to 23) (34). It is therefore tempting to speculate that FLNA plays a positive role in βarr-mediated ERK activation by acting as a platform for the efficient recruitment of signaling molecules involved in this signaling pathway in an example of macromolecular cooperation. Indeed, the formation of signaling complexes between MEK1, ERK, and βarr appears to depend, at least in part, on indirect interaction, as the overexpression of c-Raf1 increases the binding of both kinases to βarr (33). Accordingly, FLNA could perform a role similar to that of c-Raf1 by promoting the formation of a stabler or more-efficient βarr-MAPK scaffold. Evidence in favor of this model is provided by coimmunoprecipitation experiments that demonstrate an increase in βarr-associated ERK2 in the presence of FLNA(22-24) and by the observation that full-length FLNA or FLNA(22-24) can potentiate βarr-mediated activation of ERK. In addition, as full-length FLNA binds to cytoskeletal elements at the periphery of the cytoplasm, including subcortical actin, it would serve to localize and amplify the βarr-MAPK cascade within this cell compartment. Of interest is the fact that βarr2 can also bind to the MAP3K ASK1, to JNK3, and indirectly, via JNK3, to the FLNA-binding MAP2K MKK4 to assemble an efficient JNK3 cascade (35). The role of FLNA in the signaling of this βarr2-MAPK scaffold therefore certainly merits further investigation. Finally, the functional significance of the interaction between FLNA and βarr is not restricted to MAPK signaling only. During the preparation of this work, it was reported that the dopamine D3 receptor, FLNA, and βarr form a basal ternary signaling complex (26). Increases in GPCR kinase activity decreased the stability of the complex, resulting in a reduction of D3R G protein-signaling potential.
FLNA is enriched in membrane ruffles, which are subcellular compartments characterized by inhibited actin filament turnover, a high level of other actin cross-linkers, such as ezrin, and a high level of Rac activity (9). Membrane ruffles are thought to arise following inefficient lamellipodium adhesion and retraction towards the cell body. In Hep2 cells overexpressing the M1MR or AT1AR, short-term agonist stimulation causes rapid membrane ruffling. Activated receptor, FLNA, βarr, and active ERK are localized to these ruffles. In this study, we have demonstrated that knockdown of endogenous FLNA or βarr levels inhibits M1MR- and AT1AR-induced ruffling. Transfection of a dominant negative MEK [MEK1(K97A)], which cannot activate ERK, also inhibits M1MR- and AT1AR-induced ruffling. Taken together, these findings indicate that βarr, FLNA, and active ERK are functionally important in the formation of membrane ruffles in Hep2 cells following M1MR and AT1AR stimulation.
Among the other FLN isoforms, FLNC has been reported to be expressed predominantly in muscle, whereas FLNB was found to be broadly distributed (43). FLNA knockdown with specific siRNA did not change the level of FLNB expression in Hep2 cells, as indicated by immunoblot studies with specific antibodies (Fig. (Fig.9F).9F). This observation is consistent with the fact that the cooperation between βarr and FLN is mostly restricted to the FLNA isoform. Alternatively, the existence of a marked difference in FLNA and FLNB concentrations in Hep2 cells might explain why the specific reduction of FLNA levels caused submaximal inhibition of membrane ruffling in these cells.
Other GPCRs, such as protease-activated receptor-2 (16) and fMLPR (8), induce membrane ruffling upon agonist activation. In the case of protease-activated receptor-2, the ruffling response has been shown to be βarr and ERK dependent (16) whereas for the fMLPR, βarr regulates ruffling via a Ral GDP dissociation stimulator-RalA effector pathway (8). Lymphocytes derived from βarr2-deficient mice have altered chemotactic responses (14), and CXCR4- and AT1AR-mediated chemotaxis in HEK cells occurs via βarr2- and p38-dependent signaling pathways (23, 44). These data together with our results indicate that following GPCR stimulation, βarr play an important role in regulating the actin cytoskeleton and subsequent cell shape change in processes such as cell motility. Our results also suggest that FLNA could serve to localize βarr-mediated MAPK activation to specific cell compartments, providing a link between βarr-mediated signaling events and cytoskeletal reorganization.
In addition to binding MAPK cascade components, repeat regions 22 to 24 of FLNA are important in recruiting Rho family GTPases and some regulatory cofactors implicated in cytoskeletal regulation. The small GTPases Rho, Rac, and Cdc42 bind FLNA repeats 22 to 24 constitutively, and RalA binds repeat 24 in a GTP-dependent manner. Trio, a guanine nucleotide exchange factor for Rho GTPases, binds FLNA via repeats 23 and 24 (7), suggesting that switching Rho GTPases bound to FLNA on and off may serve to regulate the spatial positioning of actin assembly within the cell. This is of particular interest with regard to βarr, as it was recently shown that βarr1 is involved in the activation of the small GTPase RhoA, promoting stress fiber formation following AT1AR stimulation (6).
To conclude, we have identified the actin binding and scaffolding molecule FLNA as a βarr-binding partner, in agreement with recently published data (26). βarr and FLNA act cooperatively to activate the MAPK ERK following GPCR stimulation. In addition, βarr, FLNA and active ERK play obligate roles in cell shape change in Hep2 cells following GPCR stimulation. We propose that FLNA integrates βarr-mediated signaling events and actin cytoskeleton regulation.
Acknowledgments
We thank A. Aronheim, S. Benichou, J. L. Benovic, M. G. Caron, S. Dowdy, R. J. Lefkowitz, C. J. Loy, H. Lunsady, Y. Ohta, R. Seger, and T. Stossel for providing cDNAs and reagents as detailed in the text; L. Achour for her contribution to fluorescence-activated cell sorter experiments; J. Liotard for technical assistance; and A. Benmerah, H. Enslen, F. Porteu, and S. Shenoy for helpful discussion.
This work was funded by grants from the Wellcome Trust, the Agence Nationale pour la Recherche, Ligue Contre le Cancer (Comité de l'Oise), CNRS, and INSERM. During the performance of this work, M.G.H.S. was supported by postdoctoral fellowships from the Wellcome Trust, Fondation pour la Recherche Medicale, and SIDACTION.
REFERENCES
Articles from Molecular and Cellular Biology are provided here courtesy of Taylor & Francis
Full text links
Read article at publisher's site: https://doi.org/10.1128/mcb.26.9.3432-3445.2006
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc1447405?pdf=render
HAL Open Archive
http://www.hal.inserm.fr/inserm-00069767
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1128/mcb.26.9.3432-3445.2006
Article citations
Boolean Modeling of Biological Network Applied to Protein-Protein Interaction Network of Autism Patients.
Biology (Basel), 13(8):606, 10 Aug 2024
Cited by: 0 articles | PMID: 39194544 | PMCID: PMC11352122
Protein-Protein Interaction Network Extraction Using Text Mining Methods Adds Insight into Autism Spectrum Disorder.
Biology (Basel), 12(10):1344, 18 Oct 2023
Cited by: 1 article | PMID: 37887054 | PMCID: PMC10604135
Neighborhood socioeconomic deprivation and individual-level socioeconomic status are associated with dopamine-mediated changes to monocyte subset CCR2 expression via a cAMP-dependent pathway.
Brain Behav Immun Health, 30:100640, 14 May 2023
Cited by: 2 articles | PMID: 37251548 | PMCID: PMC10220312
Interactions between β-arrestin proteins and the cytoskeletal system, and their relevance to neurodegenerative disorders.
Front Endocrinol (Lausanne), 14:957981, 09 Feb 2023
Cited by: 2 articles | PMID: 36843600 | PMCID: PMC9947276
Review Free full text in Europe PMC
Mechanical Activation of the β2-Adrenergic Receptor by Meningococcus: A Historical and Future Perspective Analysis of How a Bacterial Probe Can Reveal Signalling Pathways in Endothelial Cells, and a Unique Mode of Receptor Activation Involving Its N-Terminal Glycan Chains.
Front Endocrinol (Lausanne), 13:883568, 02 May 2022
Cited by: 2 articles | PMID: 35586623 | PMCID: PMC9108228
Review Free full text in Europe PMC
Go to all (81) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1.
Nat Cell Biol, 4(9):681-690, 01 Sep 2002
Cited by: 222 articles | PMID: 12198493
Filamin a binds to CCR2B and regulates its internalization.
PLoS One, 5(8):e12212, 17 Aug 2010
Cited by: 16 articles | PMID: 20808917 | PMCID: PMC2923182
Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway.
J Biol Chem, 278(29):27096-27104, 06 May 2003
Cited by: 41 articles | PMID: 12734206
The filamins: organizers of cell structure and function.
Cell Adh Migr, 5(2):160-169, 01 Mar 2011
Cited by: 288 articles | PMID: 21169733 | PMCID: PMC3084982
Review Free full text in Europe PMC





