Abstract
Free full text

VISUALIZATION OF MOLECULAR INTERACTIONS BY FLUORESCENCE COMPLEMENTATION
Abstract
The visualization of protein complexes in living cells enables validation of protein interactions in their normal environment and determination of their subcellular localization. The bimolecular fluorescence complementation (BiFC) assay has been used to visualize interactions among multiple proteins in many cell types and organisms. This assay is based on the association between two fluorescent-protein fragments when they are brought together by an interaction between proteins fused to the fragments. Modified forms of this assay have been used to visualize the competition between alternative interaction partners and the covalent modification of proteins by ubiquitin family peptides.
Interactions with different partners enable proteins to perform different functions in different cells. The multitude of potential interaction partners identified for many proteins suggests that the number of protein complexes in eukaryotes is much greater than the number of proteins encoded by their genomes. Combinatorial protein interactions are central to the developmental complexity and evolutionary adaptability of these organisms.
Protein interactions have been investigated using many different approaches. Many of the experimental approaches that enable direct detection of interactions such as co-precipitation or -purification require removal of the proteins from their normal environment. Many approaches that enable studies of protein interactions in their normal environments such as genetic analysis of compensatory mutations depend on indirect consequences of the interactions. The combined use of genetic and biochemical approaches has identified thousands of potential protein interactions, but the cell specificity and the subcellular localization of the great majority of these interactions remains unknown.
Direct visualization of protein complexes in living cells enables investigation of interactions in their normal environment. Two principal methods have been used to visualize protein complexes interactions in living cells. Fluorescence resonance energy transfer (FRET) analysis is based on changes in the fluorescence intensities or lifetimes of two fluorophores that are brought sufficiently close together1-3. Bimolecular fluorescence complementation (BiFC) analysis is based on the formation of a fluorescent complex by fragments of fluorescent proteins whose association is facilitated by an interaction between proteins that are fused to the fragments4,5. Other methods such as fluorescence cross-correlation spectroscopy have also been used 6. Studies of protein interactions using the FRET assay have been described in several recent reviews 1-3. This article focuses on interactions that have been visualized using the BiFC assay and on critical principles and assumptions underlying this approach. A fundamental difference between the FRET and BiFC approaches is that FRET analysis is based on measurement of the difference in fluorescence intensity or lifetime of one fluorophore in the presence and absence of a second fluorophore. In contrast BiFC analysis is based on the formation of a fluorescent complex from non-fluorescent constituents. This makes BiFC analysis potentially more sensitive and avoids interference from changes in fluorescence intensity or lifetime caused by cellular conditions unrelated to protein interactions. As discussed in detail below, this also prevents real-time analysis of complex formation using the BiFC assay.
One obstacle to many approaches for the visualization of protein interactions is that most proteins have multiple partners in the cell. Thus, under native conditions, only a subpopulation interacts with any particular partner. Interactions between the proteins under study with other cellular proteins can interfere with detection of the complex under investigation. This problem is often solved by overexpression of the proteins to outcompete endogenous interaction partners and to drive complex formation. This strategy carries the risk that protein overexpression produces non-native complexes or alters the properties of the complexes that are formed. One advantage of BiFC analysis is that complexes that are formed with other partners are invisible, enabling selective visualization of the complex under investigation.
The Bimolecular Fluorescence Compolementation assay
Many proteins can be divided into fragments that can associate to produce a functional complex. However, no general strategy for the design of such fragments has been articulated. Experimental applications of protein fragment complementation are therefore critically dependent on the identification of fragments that can associate with each other under relevant conditions (see Table 1 for a representative subset). Complementation assays using fragments of different proteins have different properties. BiFC has the advantage that the complex can be directly visualized in living cells without the need for staining with exogenous molecules that could affect detection of the interaction.
Table 1
Proteins that can be divided into two fragments that can form a functional complex
| Protein | Assay | Environment | Interaction Partners | Ref. |
|---|---|---|---|---|
| Ribonuclease | Enxyme activity | In vitro | Spontaneous | 37 |
| β-galactosidase | Growth, enzyme assay | E. coli, cells | Spontaneous, FKBP-FRB | 38,39 |
| Ubiquitin | UBP cleavage | Yeast | GCN4 zipper | 40 |
| DHFR | Growth, staining | E. coli, cells | GCN4, FKBP-FRAP | 41 |
| GFP | Intrinsic fluorescence | E. coli, in vitro | Designed zipper | 42 |
| YFP | Intrinsic fluorescence | Mammalian cells | bZIP, Rel family | 4 |
BiFC analysis enables visualization of protein interactions in living cells and organisms with minimal perturbation of the normal cellular environment (Fig. 1). Conditional fluorescence complementation was first demonstrated by expression of fragments of the enhanced yellow fluorescent protein (YFP) fused to either bZIP or Rel family proteins in mammalian cells4. Mutations that prevented the interactions reduced the fluorescence, demonstrating that the native protein interactions facilitated fluorescence complementation. The same fragments produce fluorescent complexes when they are fused to a variety of structurally dissimilar interaction partners (see below). The BiFC approach is therefore generally applicable for the visualization of protein interactions in living cells.

The association of the fluorescent-protein fragments can be enhanced by their molecular proximity such as when they are tethered in the same macromolecular complex. Fusion proteins that produce optimal signal must generally be empirically determined. For true in vivo interaction partners, it is virtually always possible to find fusion proteins that produces a detectable signal. The fluorescence intensity produced by bimolecular fluorescence complementation in living cells is generally less than 10% of that produced by intact fluorescent proteins. It is likely that only a subset of the fragments associate with each other since the fluorescence intensity of BiFC complexes produced in vitro is comparable to that of intact fluorescent proteins4. The image on the right shows BiFC analysis of the recruitment of a protein to subnuclear foci by dimerization with a mutated version. Image acquired by Nirmala Rajaram reproduced with permissions from the Americal Society for Microbiology © 2004 11.
BiFC analysis does not require structural information about the interaction partners. The association between the fluorescent-protein fragments does not require that the interaction partners position the fragments in a specific orientation or within a fixed distance from each other. Nevertheless, steric constrains can prevent association of the fragments within a complex. Peptide linkers between the fragments and the interaction partners may therefore facilitate fragment association. The interaction partners also do not need to form a complex with a long half-life, as potentially transient interactions can be trapped by the association of the fluorescent-protein fragments (see Box 1 on BiFC complex dynamics). It is not necessary for a large proportion of the interaction partners to associate with each other because cells have low background fluorescence and unassociated fragments do not interfere with detection of the complex.
An essential criterion for fusion proteins to be used for BiFC analysis is that the fluorescent-protein fragments do not associate with each other efficiently in the absence of an interaction between the proteins fused to the fragments. Since spontaneous association between the fragments can be affected by the nature of the fusion proteins, this must be tested for each case independently. Expression of some fluorescent-protein fragments that are not fused to interaction partners produces strong fluorescence7,8. The fluorescence produced by spontaneous association of the fragments used for BiFC analysis is frequently reduced when the fragments are fused to proteins that do not interact with each other. In many cases, high-level expression of fusion proteins results in fluorescent complex formation in the absence of specific interactions between proteins fused to the fragments. It is therefore essential to express the fusion proteins at levels comparable to their endogenous counterparts and to test the effects of mutations that eliminate the interaction on fluorescence complementation.
As in the case of all approaches that make use of fusion proteins, it is necessary to consider the possibility that the fusions affect the functions of the interaction partners. Ideally, the fusion proteins should be tested by substituting them for their endogenous counterparts. However, this approach is practical only in prokaryotes and yeast, so alternative assays to test the functions of the fusion proteins must be used in the other eukaryotes. It is also important to examine potential consequences of the stabilization of the interaction between the fusion proteins by bimolecular fluorescent complex formation (see Box 1). Trapping of the interaction partners by association of the fluorescent-protein fragments can alter the properties of the complex.
Visualization of protein interactions using the BiFC assay
BiFC analysis has been used to study interactions among a wide range of proteins in many different cell types and organisms (Table 2). The results have validated interactions between many putative interaction partners and have identified several new interactors. Discovery of the subcellular locations of many protein complexes has provided new insights into their functions.
Table 2
Examples of protein interactions that have been visualized using the BiFC assay
| Category | Class | Organism | References |
|---|---|---|---|
| Peptides | Coiled coil, heat shock | Escherichia coli, in vitro | 36, 42 |
| Nuclear proteins | bZIP, Rel, bHLHZIP, Bromodomain, SMAD, Interferon Regulatory Factor, Winged helix | Mammalian cells | 4,9,10,11-17,22,43 |
| Ubiquitination | E3 ligase-substrate, Peptide conjugates | Mammalian cells, Saccharomyces cerevisiae, Sinapsis alba, Petroselinum crispum) | 18,20,23,35 |
| Signaling | Mitogen activated protein kinase network, PKB and PDK kinases, Heterotrimeric G proteins, Phospholipases, Apoptosis, Photosensitivity | Mammalian cells, Dictyostelium discoideum, Aspergillus nidulans | 24,25,28,29,33,48,49 |
| Enzyme complexes | ACCS, P450 | E. coli, Mammalian cells | 50 |
| Membrane proteins | Integrin signaling, Arf GTPases, Lectins, Cytokine receptors | Mammalian cells | 19,21,27,30 |
| Nucleic acid binding | RNA binding, DNA binding | Mammalian cells, in vitro | 31,32,51,58 |
| Plant proteins | Transcription factors, protein modification, Flowering, Plastid division | N. benthamiana, N. tabacum, Arabidopsis thaliana, Allium sp. | 15,52-57 |
| Plant pathogens | Type IV secretion, Host interactions | Agrobacterium tumefaciens, Nicotiana tabacum | 44-47 |
Subnuclear localization
The BiFC assay has been used to study interactions among many different structural classes of transcription factors4,5,9-17. These studies have provided a more refined understanding of protein interactions in the nucleus and their effects on subnuclear localization (Fig. 1). In many cases, the localization of a protein complex differs from those of the interaction partners, demonstrating that complex formation can regulate localization4,10,11. Further studies of the mechanisms regulating subnuclear localization may contribute toward understanding of the roles of compartmentalization in nuclear functions.
Enxyme-substrate complexes
The BiFC assay has also been applied for the identification of several enzyme-substrate interactions, including those of ubiquitin ligases, kinases and guanine nucleotide exchange factors 18-23. Determination of the substrate specificities and sites of action of these enzymes in living cells has yielded new hypotheses for their functions. In Saccharomyces cerevisiae, the ubiquitin E3 ligase Grr1 interacts with the Hof1 regulator of cytokinesis within the bud neck during the M phase of the cell cycle20. Degradation of Hof1 by Grr1 is an important step for actomyosin contraction during cytokinesis in yeast20.
Signal transduction cascades
Using the BiFC assay, interactions among many signaling proteins in signal transduction networks have been visualized. The Smad family of transcription factors mediates responses to TGFβ signaling. BiFC analysis demonstrated that Smad3 complexes with Smad4 are localized to the nucleus whereas Smad3 association with Protein Kinase B (PKB)/Akt prevents its nuclear translocation22. BiFC analysis has therefore been particularly valuable for the visualization of membrane protein interactions because of the potential role of the membrane environment in controlling complex formation19,24-27. Contrary to expectation, the constrained mobilities of membrane proteins apparently neither prevent the association of the fluorescent-protein fragments nor eliminate the requirement for specific protein interactions.
Regulation of complex localization
The BiFC assay is ideally suited for visualization of the subcellular localization of protein complexes. Heterodimers formed by Jun and ATF2 transcription factors are cytoplasmic in unstimulated cells, and are translocated into the nucleus upon stimulation of stress-activated protein kinases4. Complexes formed by Max with different Myc and Mad family transcription factors are localized to different subnuclear locations10. Upon isopreterenol stimulation, the β and γ subunits of the heterotrimeric G-protein internalize as a complex that is separate from the β-adrenergic receptor24,28. A complex formed by the guanine nucleotide exchange factors GBF1 and the small GTPase Arf1 is recruited to the golgi in cells stimulated with brefeldin A21. Similarly, a complex formed by the Bcl-2 family proteins Bif-1 and Bax is relocated to mitochondria in cells induced to undergo apoptosis29. The BiFC assay therefore enables visualization of the recruitment of protein complexes to different subcellular locations.
Interactions involving post-translational modifications
Many interactions require specific post-translational modifications and the requirement for the modification can be tested in living cells using the BiFC assay. The bromodomain protein Brd2 binds selectively to acetylated histones, and complementation between these proteins required both the bromodomain of Brd2 and the H4 tail that contains the acetylation site13. Fluorescence complementation by the ERGIC53 receptor and cathepsins catZ and catC requires the lectin-binding domain of ERGIC53, suggesting that these interactions require ligand glycosylation30. BiFC can therefore be used to monitor changes in post-translational modifications that alter protein interactions in cells.
Macromolecular complexes and molecular scaffolds
The interaction that brings together the fluorescent-protein fragments need not be direct. Fusion proteins that are brought together by assembly in a macromolecular complex can produce bimolecular fluorescent complexes in the absence of direct contact between the proteins that are fused to the fragments. Likewise, fusion proteins that bind to the same molecular scaffold may support fluorescence complementation even if the fusion partners do not contact each other directly. Using this principle, RNA binding by the human zipcode-binding protein 1 ortholog (IMP1), the iron regulatory protein (IRP1), the fragile X mental retardation protein (FMRP), and the human Staufen homologue (hStau1) have been visualized in living cells31. Likewise, co-occupancy by zinc-finger DNA binding proteins on oligonucleotides has been detected in vitro32. Therefore, although the association of fluorescent-protein fragments in the BiFC assay is bimolecular, this assay is not limited to the visualization of binary interactions.
BiFC studies in different organisms
The BiFC assay has been used to study interactions in a variety of species from many different phyla (see column 4 of Table 2). The BiFC assay is therefore likely to be generally applicable for visualization of protein interactions in virtually every aerobically grown cell type and organism that can be genetically modified to express proteins that are fused to the fluorescent-protein fragments. All of the studies in higher eukaryotes or cultured cells from these organisms have utilized transient expression of the fusion proteins. Under these conditions, the levels of protein expression in individual cells are difficult to establish. Future studies using transgenic organisms with stable expression of the fusion proteins in specific tissues are likely to have even greater impact on the understanding of protein interactions in the physiological context.
Interaction screens
Complementation assays, and in particular the yeast two-hybrid transcription activation assay, have been used to identify new interaction partners for many proteins. The Ft1 protein was identified as an interaction partner of PKB/Akt using a BiFC-based library screening approach33. The advantage of BiFC-based screens is that the interactions can be detected within the cell, and the effects of stimuli on the interaction can be directly tested. One limitation of BiFC-based screens is that the intrinsic differences in protein expression levels probably influence which partners can be identified. Nevertheless, BiFC analysis has the potential to identify partners that interact with a protein of interest under specific cellular conditions. BiFC analysis could also be used to identify synthetic molecules or cellular factors that can modulate protein interactions.
Simultaneous visualization of multiple protein complexes
The transformation of cell biology that was caused by the green fluorescent protein has been accelerated by the identification of numerous variants with altered spectral and photophysical characteristics1. Bimolecular fluorescent complexes that are formed through the association of fragments from different fluorescent proteins have spectra that differ from those of complexes formed through the association of fragments from the same fluorescent protein5,34. The spectral differences between these complexes enable simultaneous visualization of multiple protein complexes in the same cell (Fig. 2). Direct comparison of the distributions of multiple complexes in the same cell eliminates the need to find markers for subcellular compartments that co-localize with the complex. It also allows determination whether differences in complex localization are a direct consequence of differences between their localization signals or are an indirect consequence of differences between the effects of these complexes on cellular functions. The latter possibility is difficult to exclude when comparing the distributions of the complexes in different cells.

Multicolor BiFC analysis is based on the formation of bimolecular fluorescent complexes with different spectra through interactions between proteins that are fused to different fluorescent-protein fragments. These complexes can be independently visualized by using different excitation and emission wavelengths. The panels at the bottom show multicolor BiFC analysis of two different complexes in the same cells. The images on the left and right were acquired using different excitation and emissions wavelengths (shown in false color) and the image in the center shows an overlay of these images. The images were acquired by Dr. Changdeng Hu5.
The multicolor BiFC assay can also be used to investigate the competition between mutually exclusive interaction partners for complex formation with a common partner in cells5,10. When two alternative interaction partners that are fused to fragments of different fluorescent proteins are expressed with a limiting amount of a common partner that is fused to a dually complementary fragment, different quantities of complexes are formed with each partner. The proportion of complexes that is formed with each interaction partner reflects the relative efficiencies of complex formation with each interaction partner in the cell. As bimolecular fluorescent complex formation is not reversible under all conditions, the relative efficiencies of complex formation do not necessarily reflect the equilibrium binding affinities of the interaction partners in the cell. Nevertheless, for complexes with dissociation rates that are faster than the rate of association between the fluorescent-protein fragments (t½ ≈ 1 min), it is likely that the relative amounts of bimolecular fluorescent complexes formed with each interaction partner reflect the relative efficiencies of complex formation between the unmodified proteins. Comparison of the efficiencies of complex formation between alternative interaction partners requires normalization for differences between the efficiencies of association of different fluorescent-protein fragments and the resulting fluorescence intensities5,10. The influence of steric constraints on the association between fragments fused to different interaction partners can be tested by using linker sequences of different lengths.
Applications of multicolor BiFC analysis
The multicolor BiFC assay has been used to determine the relative efficiencies of dimerization among the basic region – leucine zipper (bZIP) domains of the Fos, Jun and ATF2 transcription factors. In living cells, bFos-bJun heterodimers form more efficiently than either bFos-bATF2 or bJun-bATF2 heterodimers5. Jun-ATF2 heterodimers are thought to regulate the expression of many genes in response to cell stress. It is therefore plausible that heterodimer formation is regulated in response to cell stress or that regions outside the bZIP domains influence the dimerization specificities of the proteins.
Multicolor BiFC analysis has also been used to compare the relative efficiencies of Max interactions with the basic helix-loop-helix-zipper (bHLHZip) domain of Myc versus Mad family transcription factors10. Heterodimers formed by Max with Myc promote cell proliferation whereas heterodimers formed with Mad family proteins promote quiescence or differentiation in many cell types. As predicted based on in vitro studies, Max favored heterodimer formation with bMyc over homodimerization, and this preference was reversed by point mutations in the leucine zipper of Max. Surprisingly, Max formed heterodimers with Mad3 less efficiently than with bMyc whereas it formed heterodimers with Mad4 more efficiently than with bMyc. Therefore, the relative amounts of Max heterodimers with Myc and the Mad-family proteins depend on the specific Mad family proteins that are present in the cell.
Visualization of ubiquitin-family-peptide conjugates in cells
Conjugation of ubiquitin-family peptides to a wide range of protein substrates serves as a mechanism that modulates the stability and function of many proteins. A ubiquitin-mediated fluorescence complementation (UbFC) assay has been developed for the visualization of ubiquitin conjugates in living cells35. In the UbFC approach, the fluorescent-protein fragments are fused to ubiquitin and to a putative substrate protein. The covalent attachment of ubiquitin to a substrate can facilitate association between the fragments, enabling selective visualization of the ubiquitin conjugate (Fig. 3). The UbFC assay shares many of the characteristics of the BiFC assay. Of particular significance is its capacity to visualize a small subpopulation of modified proteins when high levels of unmodified proteins are also present. This is critical for studies of ubiquitin-family peptide modifications because the proportion of most proteins that are conjugated by a ubiquitin family peptide at any one time is small.
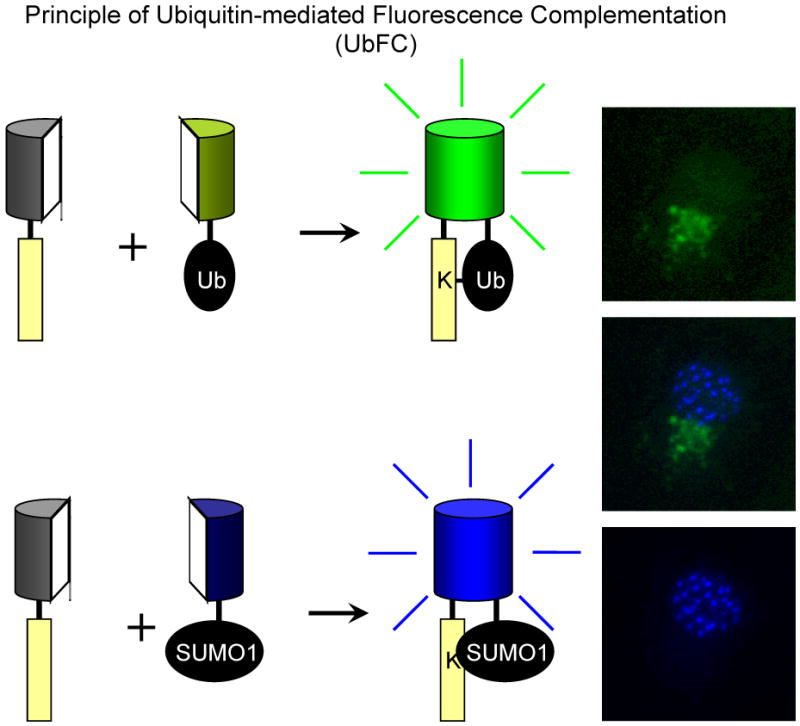
The ubiquitin-mediated fluorescence complementation (UbFC) assay is based on the association between fluorescent-protein fragments that are brought together by covalent conjugation of ubiquitin that is fused to one fragment to a substrate that is fused to the complementary fragment. The fluorescent-protein fragment must be fused to the N-terminal ends of ubiquitin-family peptides since their C-terminals ends are essential for conjugation to substrate proteins. Fusions to the N termini of ubiquitin and small ubiquitin-like modifier-1 (SUMO-1) do not interfere with the activities of the enzyme complexes that conjugate these peptides to protein substrates. It is essential to determine if the conjugates that contain the fluorescent-protein fragments retain the biological functions of the unmodified conjugates. Because ubiquitin can be conjugated in different monomeric and polymeric configurations to substrates, it is important to establish that the stoichiometry and configuration of the ubiquitin conjugate are not altered by the fusions. The images on the right show multicolor UbFC analysis of ubiquitin (green) and SUMO1 (blue) conjugation in the same cell. The images were acquired by Dr. Deyu Fang and are reproduced with permission from the National Academy of Sciences © 2004 35.
As in the case of the BiFC assay, it is essential to establish if mutations that prevent ubiquitination of the substrate affect the complementation signal. It is possible that steric constraints allow association of the fluorescent-protein fragments only when specific lysine residues are modified. Such effects of the steric arrangement of fluorescent-protein fragments on fluorescence complementation can be detected by comparing ubiquitin-mediated fluorescence complementation using substrates in which the fluorescent-protein fragments are fused to different ends of the protein.
The UbFC assay has been used to visualize ubiquitin-family peptide conjugates of the Jun protein35. Surprisingly, ubiquitinated Jun is exported from the nucleus, and is translocated to lysosomes for degradation. The UbFC assay can also be used to detect modifications by other ubiquitin family peptides such as the small ubiquitin-like modifier-1 (SUMO-1). UbFC analysis demonstrated that SUMO-1 modification of Jun induced translocation into subnuclear foci35. Multicolor UbFC analysis demonstrated that Jun modification by ubiquitin versus SUMO-1 induced translocation to different locations within the same cell. Recent studies have revealed a broad range of functions for ubiquitin family peptide modifications, and more will undoubtedly be discovered. The direct visualization of these conjugates using the UbFC assay is a valuable tool for the investigation of these modifications in living cells.
Future prospects and challenges
Studies of protein interactions and modifications have produced great advances in our understanding of biological regulatory mechanisms. The ability to visualize interactions and modifications in living cells and organisms is a critical requirement for further progress in this area. The BiFC and UbFC assays provide valuable tools for the visualization of non-covalent interactions and ubiquitin family peptide conjugation in living cells and organisms. Studies to date using these assays have established that they are generally applicable for the study of interactions among a variety of structurally and functionally dissimilar proteins as well as modifications by different ubiquitin family peptides. These assays have been used in a variety of organisms and cell types, and there do not appear to be any barriers to their use in an ever expanding array of experimental systems.
The present generation of fluorescence complementation assays are powerful tools, but have some characteristics that limit their utility. Chief among these limitations is the irreversible association of the fluorescent-protein fragments at least under some conditions. A better understanding of the dynamics of bimolecular fluorescent complex formation in living cells and the development of complementing fragments that minimally perturb the dynamic exchange of interaction partners would be valuable improvements. A second limitation is the intrinsic ability of fluorescent-protein fragments to associate with each other independent of an interaction between proteins fused to the fragments. This main source of background signal in the BiFC assay varies depending on the fusion proteins and their levels of expression. Development of fluorescent-protein fragments with a reduced tendency for intrinsic association, but without a reduced ability to associate when brought together by a protein interaction would make the assay less sensitive to the levels of protein expression. Finally, fluorescent protein engineering has produced an abundance of variants with useful characteristics1, but has not markedly increased the rates of the chemical reactions required for fluorophore formation. Given the importance of this transformation for all experiments using fluorescent proteins, acceleration of the chemistry of fluorophore formation would be useful. Bimolecular fluorescent complexes with novel properties could be identified through the screening of combinatorial libraries of fluorescent-protein fragments.
The specificity of protein interactions in cells is determined to a great extent by the competition between mutually exclusive interaction partners. A better understanding of the factors that influence the relative efficiencies of complex formation including localization, scaffolds, modifications, networks of alternative partners as well as intrinsic binding affinity is essential for understanding of the regulation of protein interactions. The multicolor BiFC assay provides a tool that enables investigation of many of these factors in the normal cellular environment. However, the efficiency of bimolecular fluorescent complex formation depends on both the efficiency of the interaction between the proteins under investigation as well as on the efficiency of association between the fluorescent-protein fragments. Determination of the relative efficiencies of fluorescent-protein fragment association when they are present in different complexes remains an important challenge. Differences between these efficiencies can be evaluated on a case by case basis, but a general method for the qualtitative comparison of the efficiencies of of complex formation would make using the multicolor BiFC approach more straightforward.
Many biological processes occur asynchronously in different cells in a population. Studies of such processes at a population level can miss relationships between the processes because these relationships are averaged over the entire population. The multicolor BiFC approach enables the simultaneous imaging of multiple protein interactions in the same cell. Comparison of several molecular interactions in the same cell can identify relationships between the interactions in an asynchronous cell population.
Fluorescence complementation assays will undoubtedly continue to find new applications in the future. Studies of protein interactions in transgenic plants and animals will enable investigation of the developmental and tissue-specific regulation of complex formation. The use of regulated vector systems to control fusion protein expression in cultured cells will enable control of the levels of protein expression at more uniform levels. The use of native transcription regulatory regions and “knock-in” expression constructs when possible will enable studies of the timing and the cell-type specificity of complex formation.
The BiFC approach could be used for purposes other than the study of protein interactions. The wide range of spectral variants that are produced by different combinations of fluorescent-protein fragments could be used for combinatorial tagging of cells in studies of cell lineages and migration. The signal produced by association of fluorescent-protein fragments could be used to study cell or organelle fusion and the resulting mixing of their contents. Many posttranslational modifications could be imaged through the use of protein domains that specifically recognize the modifications. Ultimately, strategies for the simultaneous control and monitoring of protein interactions and modifications may be realized.
References
Full text links
Read article at publisher's site: https://doi.org/10.1038/nrm1929
Read article for free, from open access legal sources, via Unpaywall:
https://europepmc.org/articles/pmc2512262?pdf=render
Citations & impact
Impact metrics
Article citations
Functional analysis of protein interactions using coupled bi-fluorescence complementation/GFP nanobody techniques.
Nucleic Acids Res, 52(14):e66, 01 Aug 2024
Cited by: 1 article | PMID: 38932691 | PMCID: PMC11317145
Real-time single-molecule imaging of transcriptional regulatory networks in living cells.
Nat Rev Genet, 25(4):272-285, 09 Jan 2024
Cited by: 4 articles | PMID: 38195868
Review
Protein-Protein Interactions: Bimolecular Fluorescence Complementation and Cytology Two Hybrid.
Methods Mol Biol, 2715:247-257, 01 Jan 2024
Cited by: 1 article | PMID: 37930533
Ubiquitination detection techniques.
Exp Biol Med (Maywood), 248(15):1333-1346, 01 Aug 2023
Cited by: 4 articles | PMID: 37787047 | PMCID: PMC10625345
Review Free full text in Europe PMC
Visualization of trans-interactions of a protocadherin-α between processes originating from single neurons.
iScience, 26(7):107238, 17 Jul 2023
Cited by: 1 article | PMID: 37534169 | PMCID: PMC10392085
Go to all (266) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation.
Chem Soc Rev, 38(10):2876-2886, 04 Sep 2009
Cited by: 129 articles | PMID: 19771334 | PMCID: PMC2980501
Review Free full text in Europe PMC
Bimolecular fluorescence complementation: visualization of molecular interactions in living cells.
Methods Cell Biol, 85:431-470, 01 Jan 2008
Cited by: 89 articles | PMID: 18155474 | PMCID: PMC2829325
Multicolor bimolecular fluorescence complementation (BiFC) analysis of protein interactions with alternative partners.
Cold Spring Harb Protoc, 2013(9):798-803, 01 Sep 2013
Cited by: 6 articles | PMID: 24003202
Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis.
Curr Protoc Protein Sci, Chapter 19:19.10.1-19.10.21, 01 Sep 2005
Cited by: 17 articles | PMID: 18429278
Funding
Funders who supported this work.






