Abstract
Free full text

Outer Membrane Vesicle Production by Escherichia coli Is Independent of Membrane Instability
Abstract
It has been long noted that gram-negative bacteria produce outer membrane vesicles, and recent data demonstrate that vesicles released by pathogenic strains can transmit virulence factors to host cells. However, the mechanism of vesicle release has remained undetermined. This genetic study addresses whether these structures are merely a result of membrane instability or are formed by a more directed process. To elucidate the regulatory mechanisms and physiological basis of vesiculation, we conducted a screen in Escherichia coli to identify gene disruptions that caused vesicle over- or underproduction. Only a few low-vesiculation mutants and no null mutants were recovered, suggesting that vesiculation may be a fundamental characteristic of gram-negative bacterial growth. Gene disruptions were identified that caused differences in vesicle production ranging from a 5-fold decrease to a 200-fold increase relative to wild-type levels. These disruptions included loci governing outer membrane components and peptidoglycan synthesis as well as the σE cell envelope stress response. Mutations causing vesicle overproduction did not result in upregulation of the ompC gene encoding a major outer membrane protein. Detergent sensitivity, leakiness, and growth characteristics of the novel vesiculation mutant strains did not correlate with vesiculation levels, demonstrating that vesicle production is not predictive of envelope instability.
Release of outer membrane (OM) vesicles has been observed for all gram-negative bacteria studied to date (reviewed in references 5, 15, and 16). Native vesicles are rounded structures with lumenal periplasmic components bounded by an outer layer of outer membrane proteins (Omps) and lipids (16). For Escherichia coli, our laboratory has reported that strain DH5α releases 0.23% of Omps F and C and 0.14% of OmpA into vesicles (12); other investigators have found vesicles to account for 0.2 to 0.5% of bacterial culture material (9, 20). Electron microscopy studies reveal bulging of the OM and subsequent fission of vesicles containing electron-dense material (16). These biochemical and microscopic observations suggest that OM vesicles are formed from protrusions that are pinched off from the OM in a manner that leads to the inclusion of periplasmic material.
The wide variety of strains and diversity of environments for which vesiculation has been observed suggest an important role for vesicle production in gram-negative bacterial growth and survival (5, 15, 16). Vesicle production varies with growth phase and nutrient availability, and vesicle-associated enzymes may aid in nutrient scavenging. Vesicle-mediated transfer of toxic components to other bacteria can eliminate competing species. In addition, interactions between eukaryotic cells and vesicles from pathogenic bacteria suggest a role for vesicles in pathogenesis (13).
We have conducted a screen to generate and identify mutants in E. coli that exhibit altered vesiculation levels. Both vesicle-overproducing and -underproducing mutants are of interest, as they offer insight into mechanisms involved in the physical formation and regulation of vesicle production. Vesiculation phenotypes did not correlate with defects in cell envelope stability or growth characteristics of mutant strains. Gene disruptions that cause vesiculation phenotypes were identified in genes involved in Omp expression, peptidoglycan synthesis, and the σE envelope stress response.
MATERIALS AND METHODS
Growth conditions and reagents.
E. coli strains were grown at 37°C in Luria-Bertani (LB) broth (EM Science) or on LB plates (LB with 15 g/liter agar). For Tn5 insertion mutants, kanamycin (Sigma) was added at 50 μg/ml. Unless otherwise noted, reagents were purchased from VWR.
Transposon-based gene disruption.
The Tn5-based EZ::TN <R6Kγori/KAN-2> Tnp Transposome (Epicentre) was introduced by electroporation according to the manufacturer's protocol.
Low-volume supernatant dot blot screen.
Kanamycin resistant mutants were grown overnight in 150 μl of LB broth in duplicate 96-well V-bottom plates. Mutants that did not grow were not considered further. Each plate contained a positive control of wild-type cells and a negative control of wild-type cells that had been washed; for the negative control, the washed cells were resuspended in fresh LB immediately prior to processing. Cells were pelleted (2,000 × g for 25 min at 4°C), and 5 μl of supernatant was transferred to a vacuum manifold system. Cellular debris was filtered by a Supor membrane (0.45-μm-pore-size polysulfone; Pall Gelman Sciences), and vesicles were collected by a second membrane (high protein binding, 0.45-μm-pore-size Protran nitrocellulose; Schleicher and Schuell). The nitrocellulose membrane was immunoblotted using rabbit anti-lipopolysaccharide (LPS) (Accurate; discontinued) or rabbit anti-E. coli OM as the primary antibody.
Large-scale protein and lipid-based screens.
Broth cultures (250 ml) were grown overnight. Cells were pelleted (10,000 × g for 10 min at 4°C), and culture supernatants were filtered (0.45-μm-pore size polyvinylidene difluoride) before centrifugation (38,400 × g for 1 h at 4°C). Pelleted vesicles were resuspended in Dulbecco's phosphate-buffered saline with added salt (0.2 M NaCl) and filter sterilized through 0.45-μm-pore-size Ultra-free spin filters (Millipore). A portion of the sterile resuspended vesicles was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Coomassie-stained OmpF, C, and A were quantitated by densitometry (NIH Image J software). A second portion was incubated with FM4-64 (Molecular Probes) (3.3 μg/ml in phosphate-buffered saline for 10 min at 37°C). Vesicles alone and FM4-64 probe alone were negative controls. After excitation at 506 nm, the emission at 750 nm was measured with a Molecular Devices SpectraMAX GeminiXS fluorometer. CFU values were determined by dilution plating. Vesicle production was calculated by dividing Omp densitometry units or lipid fluorescence units by CFU. Relative vesicle production was determined by dividing vesicle production of the mutants by that of the wild-type strain in each experimental group.
Identification of disrupted genes.
For the majority of the mutants, genomic DNA was prepared using the MasterPure DNA Purification Kit (Epicentre), and the insertion sites were sequenced using primers to the transposon (Tn). The remaining mutants were identified with a nested PCR technique using a set of random and fixed primers (24). BLAST homology searches were done at www.ncbi.nlm.nih.gov. Southern blotting to confirm single Tn insertions was performed as previously described (2) and visualized using a North2South Direct HRP labeling and detection kit (Pierce) with a probe complementary to a portion of the kanamycin insertion cassette.
Characterization assays.
Detergent sensitivity was assessed on LB plates supplemented with 0.5% deoxycholate. To assay release of periplasmic RNase, strains grown on LB plates overnight were overlaid with 0.6% agar containing 30 mg/ml type VI Torula yeast RNA (Sigma) and incubated 4 h before the addition of 1 N HCl to precipitate undigested RNA. Zones of clearing indicating leakage of periplasmic RNase I into the agar were measured. To assay release of periplasmic maltose binding protein (MBP) in shaking liquid cultures, strains were grown overnight at 37°C at 200 rpm. Cells were pelleted (10,000 × g for 10 min), and cell-free supernatants were generated using 0.45-μm-pore-size spin filters (Millipore). Vesicles were removed with 100-kDa spin filters (Millipore), and the remaining MBP was detected by SDS-PAGE and immunoblotting. MutL in the cell-free supernatant was quantitated by densitometry of the MutL band detected by SDS-PAGE and immunoblotting. For growth assays, LB was inoculated 1:100 with a saturated culture and incubated (200 rpm and 37°C), and turbidity (optical density at 600 nm [OD600]) was measured. Measurements of cyan fluorescent protein (CFP) production from an ompC operon fusion were performed as previously described (3). Cultures were inoculated at an OD600 of ~0.02 and grown to an OD600 of ~0.4. Chloramphenicol was added to a concentration of 75 μg/ml, and the cultures were placed on ice until CFP fluorescence was measured by fluorometry using excitation and emission wavelengths of 434 and 475 nm, respectively.
Electron microscopy.
Vesicles were applied to a 400-mesh copper electron microscopy grid (Electron Microscopy Sciences) and negatively stained with 2% uranyl acetate.
Vesicle flotation assay.
Periplasm was prepared as previously described (12). Pelleted vesicles or periplasm were adjusted to 50% (vol/vol) Optiprep (Greiner) in HS buffer (10 mM HEPES, pH 7.4, 150 mM NaCl) in a total of 3 ml, loaded to a 12.5-ml ultracentrifuge tube (Beckman), and layered with Optiprep/HS (2 ml of a 50% concentration, 2 ml 45%, 2 ml 40%, 2 ml 35%, and 1 ml 30%). After centrifugation (100,000 × g for 16 h at 4°C), fractions were removed sequentially from the top of the gradient and separated by SDS-PAGE. Proteins were detected using Coomassie blue stain or transferred to polyvinylidene difluoride membranes and immunoblotted using mouse-anti-MBP (Sigma), anti-mouse horseradish peroxidase (Sigma), and chemiluminescence reagents (Amersham Pharmacia Biotech).
OM antibody.
OM prepared from strain ATCC 35401 as previously described (10) was concentrated with a Centricon Plus-20 filter device (Millipore) and used in a standard rabbit protocol (Strategic Biosolutions) to generate a polyclonal antibody that was determined to be highly sensitive to vesicles produced by a variety of E. coli strains.
RESULTS AND DISCUSSION
Generation of transposon insertion mutants.
Laboratory E. coli strain DH5α was selected for mutagenesis due to its basal level of vesiculation. Since we wished to use a Tn5-based transposome to generate a library of mutants containing irreversible random chromosomal insertions of a kanamycin resistance cassette, we first needed to determine whether growth in kanamycin affected vesicle production. We transformed DH5α cells with pWSK130, a low-copy-number kanamycin resistance plasmid, and quantitated vesicle production. The effect of growth in the presence of 50 μg/ml kanamycin on vesicle production was within the average standard error of the mean (SEM; 0.09) for untreated wild-type bacteria. Therefore, vesiculation phenotypes described in this screen are not due to kanamycin treatment.
Identification of vesiculation mutants.
We designed a sensitive, high-throughput dot blot screen to detect OM vesicles in low-volume cultures. Approximately 4,000 kanamycin-resistant transformants were assayed in duplicate as described in Materials and Methods. Individual cell-free supernatants were adsorbed to a nitrocellulose filter, and vesicles were detected by immunoblotting using antibodies to OM components (Fig. (Fig.1A).1A). Culture supernatants with markedly increased or decreased signal intensity on blots from duplicate cultures indicated mutants with potentially altered vesiculation levels.
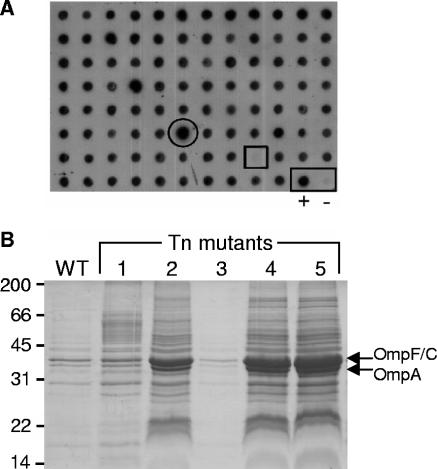
Representative data from the screen for vesiculation mutants. (A) Immunoblot with anti-LPS antibody of cell-free supernatants from cultures of insertion mutants. Representative high-vesiculation (circle) and low-vesiculation (square) mutant phenotypes are shown. Control wild-type (+) and washed wild-type (−) samples are at bottom right (rectangle). (B) Coomassie blue-stained SDS-PAGE of pelleted vesicles from cultures of wild-type DH5α (WT) and five Tn mutants demonstrates high vesiculation (mutants 2, 4, and 5) and low vesiculation (mutant 3) phenotypes. Molecular weight standard proteins (kDa) and the positions of Omps F, C, and A are labeled.
The amount of pelletable Omps and lipids in the cell-free culture supernatant of each potential vesiculation mutant culture was then compared with the amount produced by a wild-type control culture. These secondary screens eliminated selection due to variable antibody reactivity. For the protein-based screen, vesicle proteins were visualized by SDS-PAGE and Coomassie staining, and the intensities of the major vesicle protein bands OmpF, C, and A were quantitated by densitometry (Fig. (Fig.1B).1B). For the lipid reagent-based assessment, fluorescence was measured after incubation with the lipid probe FM4-64. Fluorescence of FM4-64 requires incorporation into membranes, and we determined that the response correlated linearly with an assay concentration of 0.04 to 7.6 μg/ml purified DH5α vesicle protein (data not shown). Protein- and lipid-based measurements were normalized for the amount of bacteria in the culture and divided by vesicle production of a wild-type culture included in each assay to determine relative vesicle production. These screens were performed at least twice for each mutant to establish reproducibility.
For most of the mutants, the lipid- and protein-based methods of determining relative vesiculation returned similar values (Table (Table1,1, columns 6 and 7). Since vesicles closely reflect the composition of the OM, the correlation between the two measurements suggests that the OM lipid-to-protein ratio for the majority of the mutant strains was not substantially different from the parent. Thus, the observed phenotypes reflected vesicle quantities rather than altered OM content. Differences observed for MK5B7, MK10F34, and MK10H29 suggest that these strains have OM compositions that differ from wild-type.
TABLE 1.
Summary of Tn mutant vesiculation and membrane integrity phenotypes
| Class and mutant no. | Gene disruption (insertion after codon no./total no. of codons) | Downstream effect | Location within operona | Description | Relative vesicle production (protein)b | Relative vesicle production (lipid)b | Detergent sensitivityc | RNase leakaged | MBP in soluble supernatant fractione | MutL leakagef | Doubling time (min)g |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type, DH5α | N/Ah | N/A | N/A | WT | WT | − | − | − | − | 75 | |
| Envelope structure | |||||||||||
    MK7E17 MK7E17 | ponB (363/844) | None (predicted) | Peptidoglycan synthesis | ++ | +++ | − | ++ | ++ | ++ | 225 | |
    MK10E29 MK10E29 | tolB (185/430) | Yes | tolB pal ybgF | Periplasmic protein; uptake of group A colicins | ++ | ++ | − | ++ | ++ | + | 120 |
    MK8F18 MK8F18 | pal (11/173) | Yes | tolB pal ybgF | Peptidoglycan-associated lipoprotein | ++ | +++ | + | ++ | ++ | − | 100 |
    MK5A43 MK5A43 | tolA (79/421) | None | ybgC tolQRA | Outer membrane integrity; phage infection | ++ | ++ | ++ | ++ | ++ | ++ | 120 |
    MK6F12 MK6F12 | tolB (269/430) | Yes | tolB pal ybgF | Periplasmic protein; uptake of group A colicins | ++ | +++ | ++ | ++ | ++ | − | 120 |
    MK4E44 MK4E44 | ompR (44/239) | Yes | ompR envZ | Response regulator for ompC and ompF | +i | + | − | + | + | − | 130 |
    MK10F34 MK10F34 | ompR (96/239) | Yes | ompR envZ | Response regulator for ompC and ompF | +i | ++ | + | ++ | + | − | 135 |
    MK6A33 MK6A33 | ompC (180/367) | None (predicted) | Outer membrane porin | +i | + | − | − | − | − | 95 | |
    MK12E45 MK12E45 | ompC (295/367) | None (predicted) | Outer membrane porin | +i | + | − | + | − | + | 105 | |
    MK1F40 MK1F40 | wzxE (253/416) | Yes (predicted) | wecDE wzxE wecF | Inner membrane translocase for a component of ECA | + | + | − | + | + | − | 105 |
    MK10G32 MK10G32 | waaG/rfaG (293/374) | Yes | waaQGPSBIJYZK | LPS core biosynthesis; glucosyl transferase | + | + | ++ | − | + | − | 90 |
    MK10H29 MK10H29 | waaG/rfaG (21/374) | Yes | waaQGPSBIJYZK | LPS core biosynthesis; glucosyl transferase | − | + | ++ | − | + | − | 120 |
| Envelope stress | |||||||||||
    MK6D31 MK6D31 | degS (347/355) | None (predicted) | degQ degS | IM serine protease for RseA; σE pathway | +++ | +++ | + | + | − | − | 90 |
    MK11F26 MK11F26 | degP (19/474) | None (predicted) | Periplasmic serine protease; σE- and Cpx-induced | +++ | +++ | + | − | − | − | 120 | |
    MK5B7 MK5B7 | rseA (168/216) | Yes | rpoE rseABC | Inhibitor of σE | ++i | +++ | + | + | + | − | 75 |
    MK7A41 MK7A41 | degP (51/474) | None (predicted) | Periplasmic serine protease; σE- and Cpx-induced | ++ | ++ | + | + | + | − | 95 | |
| Global effects | |||||||||||
    MK7C1 MK7C1 | tatC (73/258) | Yes (predicted) | tatBCD | IM secretion apparatus | ++ | +++ | ++ | ++ | ++ | − | 95 |
    MK9D4 MK9D4 | pnp (315/711) | None | rpsO pnp | Polynucleotide phosphorylase | + | + | − | − | + | − | 90 |
    MK9G12 MK9G12 | glnA (467/469) | Some | glnA + glnALG | Glutamine synthetase | − | − | + | − | ++ | − | 135 |
    MK6F18 MK6F18 | lysS/herC (429/505) | None (predicted) | prfB lysS | Lysyl tRNA synthetase | − | − | − | − | − | − | 75 |
    MK11A9 MK11A9 | pepP (16/441) | Yes (predicted) | ygfB pepP ubiH visC | Proline aminopeptidase | − | − | − | − | + | − | 180 |
| Unclassified contribution | |||||||||||
    MK8A44 MK8A44 | nlpI (114/294) | None (predicted) | OM lipoprotein | +++ | +++ | + | − | − | − | 120 | |
    MK5G18 MK5G18 | nlpI (205/294) | None (predicted) | OM lipoprotein | +++ | +++ | − | ++ | ++ | − | 100 | |
    MK7B29 MK7B29 | yieM (424/483) | None (predicted) | Uncharacterized open reading frame | + | + | − | − | − | − | 105 | |
    MK4A31 MK4A31 | ypjA (161/1569) | None (predicted) | Uncharacterized open reading frame; homology with autotransporter domain | − | − | − | + | ++ | − | 105 | |
    MK5A31 MK5A31 | nlpA (87/272) | None (predicted) | Lipoprotein 28 (inner membrane) | − | − | − | − | − | − | 110 |
Mutants were retained that reproducibly displayed greater than 3-fold or less than 0.7-fold vesicle production relative to wild type as measured by either the Omp- or lipid-based vesicle assays (Fig. (Fig.2;2; Table Table1,1, columns 6 and 7). Mutants were analyzed for filamentation which would distort CFU determinations. With the exception of slight chaining for MK7C1, Gram staining of wild-type and mutant cultures grown to mid-log or stationary phase revealed no significant morphological differences (data not shown). Growth curves were measured for all of the mutants, and their log-phase doubling times were calculated (Table (Table1,1, column 12). In general, more extreme vesiculation phenotypes corresponded to longer doubling times; however, all mutant cultures achieved an OD600 of >1.5 by 27 h at 37°C.
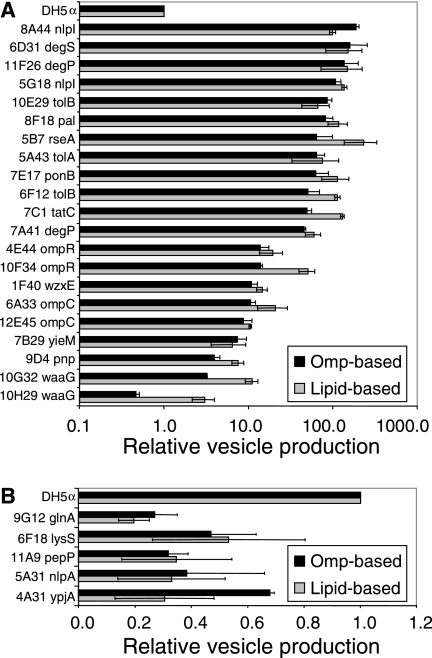
Relative increase (n-fold) in vesicle production of Tn mutants. Vesicle-overproducing mutants (A) and vesicle-underproducing mutants (B) are identified by protein- and lipid-based methods. Relative vesicle production/CFU compared to wild-type strain DH5α was calculated using Omp bands (black bars) or incorporation of the lipid probe FM4-64 (gray bars). Values are the average ± SEM (n ≥ 2).
Of 4,000 transformants screened, 26 unique insertion mutants were confirmed to have reproducible protein- and/or lipid-based vesiculation phenotypes. Only five of the disruptions resulted in decreased vesiculation (undervesiculation) as calculated by both protein and lipid measurements, and no null vesiculation mutants were found. While the screen was not genome saturating and did not allow us to assay mutations that would cause severe growth defects, the relatively low number of vesicle-underproducing mutants suggests that vesicle production may be an important process for growth of gram-negative bacteria.
Disrupted genes were identified by sequencing (Table (Table1,1, column 2). Potential operon disruptions are indicated in Table Table11 (columns 3 and 4). We noted that several of the vesiculation mutant insertions are in genes that are in close proximity. However, our insertions did not follow the periodicity or loose Tn5 consensus site reported for nonrandom Tn5 target insertion (8), suggesting that the clustering of insertions reflects a genuine relationship of these genes to vesiculation. Genetic complementation will be conducted in further studies to definitively establish the relationship of the observed phenotypes to the disrupted loci.
Characterization of membrane integrity.
To determine whether vesiculation is directly linked to cell envelope instability, we extensively characterized each of the mutants for deoxycholate sensitivity, periplasmic leakage, and cytoplasmic leakage (Table (Table1,1, columns 8 to 11). Leakage of periplasmic protein was assayed under both solid and shaking liquid growth conditions since shear forces in a shaking culture could reveal weakened membranes. Sensitivity to detergent did not correlate with leakage of periplasmic components, as demonstrated by MK7E17 and MK11F26, suggesting that the chemical barrier and containment functions of the OM are separable. Mutants were also tested for compromised inner membrane integrity by assessing leakage of cytoplasmic MutL, which indicates defects of the inner membrane and/or elevated levels of cell lysis.
The mutants displayed various levels of detergent sensitivity and periplasmic or cytosolic leakage, but there was no specific correlation between vesicle production and membrane instability (Table (Table1,1, columns 6 to 11). For example, both MK11F26 and MK7E17 produced highly elevated quantities of vesicles. However, MK11F26 had little to no integrity defects, while the compromised nature of the MK7E17 envelope was severe. Likewise, while both MK4A31 and MK6F18 showed decreased vesicle production, MK4A31 displayed a loss of envelope integrity, whereas MK6F18 did not. The precise contribution of membrane fragments from compromised cells to the apparent vesiculation phenotypes of these mutants cannot be distinguished by our assays. However, MutL was not detected in the cell-free supernatants of mutants exhibiting the highest levels of vesiculation (MK8A44, MK6D31, MK5B7, and MK11F26), demonstrating that these high vesiculation phenotypes occur without cell lysis. Thus, we conclude that vesiculation levels are not predictive of compromised cell envelopes or vice versa.
From these data, we have determined that there is no distinct correlation between detergent sensitivity, outer and inner membrane leakiness, filamentation, doubling time, and vesicle production. Therefore, genes identified in this mutant screen are useful in revealing particular bacterial factors that influence vesiculation.
Heightened vesiculation does not require increased OM component synthesis.
Releasing increased quantities of vesicle material could conceivably require a heightened rate of Omp synthesis. We tested this possibility by introducing Tn5 disruptions causing vesicle overproduction into MDG131, an MC4100-based K-12 E. coli strain containing an operon fusion of cfp to ompC (3). The activity of the reporter was measured by a fluorescence assay to compare the behavior of the wild-type and vesicle-overproducing strains. Decreased Omp expression upon activation of the σE pathway has been previously reported (26) and was confirmed in our assay for the vesicle-overproducing rseA::Tn5 mutant (Fig. (Fig.3).3). The levels of the CFP reporter in the other mutants with vesicle overproduction did not vary substantially from wild type. Therefore, vesicle overproduction does not appear to require increased OM component synthesis. These strains do, however, tend to grow more slowly than wild type (Table (Table1,1, column 12), which could be a compensating effect due to a need to maintain the OM.
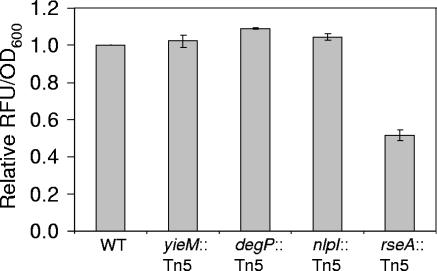
Vesicle overproduction does not require increased Omp expression. CFP fluorescence measurements corresponding to ompC expression (relative fluorescence units [RFU]) for transductants containing vesicle overproduction mutations from MK7B29 (yieM::Tn5), MK11F26 (degP::Tn5), MK8A44 (nlpI::Tn5), and MK5B7 (rseA::Tn5). Values were corrected for culture optical density at an OD600 of 0.4 and normalized to wild-type MDG131. Values are the average ± SEM (n ≥ 2).
Mutations that alter envelope structure affect vesiculation.
A majority of the genes identified in this screen influence envelope structure, which includes the OM, the peptidoglycan, and the biochemical ties between them (Table (Table1).1). The phenotypes resulting from these disruptions are likely to result from the loss of a single gene product in some mutants and from more wide-ranging global effects caused by specific deletions in others. All of the mutants with disruptions in genes known to be important for envelope structure exhibited increased vesiculation.
Identification of the tolA, tolB, and pal loci was anticipated, as E. coli strains with mutations in these genes have been previously reported to display increased vesiculation phenotypes (4). These gene products form bridges between the OM, peptidoglycan, and inner membrane, and mutations in these genes also cause severe periplasmic leakage and detergent intolerance (4). In addition to these previously reported phenotypes, we observed leakage of the cytoplasmic protein MutL for MK5A43 (tolA) and MK10E29 (tolB), suggesting compromised inner membrane integrity.
MK7E17 contains a disruption in ponB, which encodes the bifunctional transglycosylase transpeptidase PBP1b. Transpeptidases and transglycosylases are critical in growth and cross-linking of the peptidoglycan. This mutant is likely to produce an inactive PBP1b truncate, and, in fact, MK7E17 has integrity defects that resemble those observed in E. coli upon overexpression of inactive versions of PBP1b (18). Our SDS-PAGE analysis of MK7E17 supernatant material suggests that the bulges observed previously are probably composed primarily of OM (data not shown). We hypothesize that a ponB truncate expressed in MK7E17 interferes with peptidoglycan remodeling (but not enough of the truncate is expressed so as to be fatal, as was the case for the overexpressed inactive constructs) and that continued OM growth results in bulging and release as OM vesicles.
Porins contribute to the uniquely porous characteristic of the gram-negative bacterial OM. We anticipated recovering mutants in genes encoding these major OM proteins since our protein-based screen relied on Omp quantitation. However, they also influence the structure of the cell envelope, as porins OmpF and OmpC interact with peptidoglycan and TolA of the Tol/Pal complex (7). Two mutants with disruptions in ompR (MK4E44 and MK10F34) and two with disruptions in ompC (MK6A33 and MK12E45) were identified as having moderately increased vesiculation levels. OmpR is an osmotic response regulator in a two-component system that controls expression of OmpC and OmpF (25). Lipid-based measurements of the MK10F34 ompR::Tn5 mutant may be higher than the protein-based calculations due to reduced expression of one or both of the OmpF/C proteins (Fig. (Fig.22).
LPS is a major structural element of the bacterial OM and has three constituent components: lipid A, core oligosaccharide, and, in strains that have maintained the ability to produce it, O-antigen polysaccharide. Previous studies have reported that the type and/or length of LPS O antigen affects OM vesicle formation (23). However, since the strain we mutagenized does not produce O antigen, the vesiculation mutant phenotypes identified by this screen cannot be the result of differences in that component.
We found that mutations in waaG, a gene in the operon responsible for LPS core biosynthesis, also caused altered vesiculation phenotypes. MK10G32 has a disruption in the C-terminal region of the gene, whereas the Tn5 insertion in MK10H29 is in the N-terminal region. From these insertion locations, we hypothesize that MK10G32 may maintain some WaaG activity but that the allele in MK10H29 is completely nonfunctional. Polar mutations in the waa operon have pleiotropic effects including detergent sensitivity and a reduction in the quantity of Omps in the OM (28). The consequences of waaG mutations on the composition of the OM make it difficult to draw specific conclusions as to the role of LPS structure in vesiculation. For example, decreased OM Omp content is the likely explanation for the lower than wild-type appearance of the protein-based vesicle determination for MK10H29 (Fig. (Fig.2).2). Nevertheless, the combination of protein- and lipid-based phenotypes presented here supports a role in vesicle production for the LPS core oligosaccharide component.
MK1F40 has a disruption in wzxE, which encodes a flippase for membrane translocation of the lipid III substrate for enterobacterial common antigen biosynthesis (27). This disruption likely also affects the downstream wecF gene. WecF is required for the conversion of lipid II to lipid III, and it has been proposed that lipid II accumulation in wecF mutants perturbs the OM (6). Thus, the vesicle overproduction phenotype of MK1F40 may be linked to cell surface changes resulting from an enterobacterial common antigen biosynthesis defect and/or OM perturbation caused by lipid II accumulation.
Vesiculation in response to global changes in bacteria.
Many of the mutants with vesicle underproduction phenotypes have gene disruptions that are likely to incur global changes in the bacteria (Table (Table1).1). For example, MK9G12 contains a disruption at the penultimate codon of glnA. Rather than this minor truncation of glutamine synthetase, the low-vesiculation phenotype may be due to altered expression of downstream genes glnL and glnG encoding nitrogen regulators II and I (22). MK6F18 contains an insertion in the constitutively expressed lysyl-tRNA synthetase gene lysS (21). Vesicle production by this mutant may be reduced due to decreased protein synthesis; however, a simple reduction in the number of proteins per vesicle is not sufficient to explain both the lipid- and protein-based vesiculation phenotypes. The MK11A9 insertion is in the pepP proline aminopeptidase gene, but the disruption could also affect the downstream genes for oxygenase UbiH or oxidoreductase VisC; thus, the genetic basis for the phenotype of this mutant is not yet known.
In the case of MK7C1, the vesiculation phenotype may be due to global or structural effects. This mutant has a disruption in the gene for TatC, an integral inner membrane component of the Tat (twin arginine translocation) secretion machinery. Tat substrates include the cell wall hydrolyzing amidases AmiA and AmiC (11), whose mislocalization could contribute to vesicle overproduction in the mutant. However, the many pleiotropic effects of tat disruptions, as evidenced by the substantial loss of membrane integrity, make it difficult to pinpoint the specific basis of the phenotype.
Vesiculation in response to envelope stress.
Notably, our discovery of four vesiculation mutants with transposon disruptions in three different genes of the σE pathway strongly suggests a link between this pathway and vesiculation (Table (Table1).1). The σE pathway is an envelope stress response system that is activated by protein misfolding signals in the bacterial extracytoplasmic space to drive transcription of various stress-responsive genes (reviewed in reference 1). Two of the genes identified in our screen, degS and rseA, code for stress signal transmitters in the σE stress response pathway, and the other, degP, codes for a downstream effector.
The relatively mild membrane perturbation of the σE pathway mutants indicates that their vesiculation phenotypes are not related to structural defects, and this was confirmed by additional analyses. Vesicles purified from the MK6D31 degS, MK5B7 rseA, and MK11F26 degP mutant strains were observed by electron microscopy to be similar in morphology to wild type (Fig. (Fig.4A).4A). We also assayed periplasmic MBP content of the vesicles to confirm intactness. Pelleted vesicle material was subjected to equilibrium density centrifugation. Vesicles migrate upward due to their high lipid content, while non-lipid-associated proteins remain at the bottom. In vesicle gradients prepared from the wild-type and mutant strains, MBP peaked with OMP proteins in the lightest density fractions (Fig. (Fig.4B).4B). In contrast, free MBP from a periplasmic control remained in the highest density fractions (Fig. (Fig.4B,4B, bottom panel).
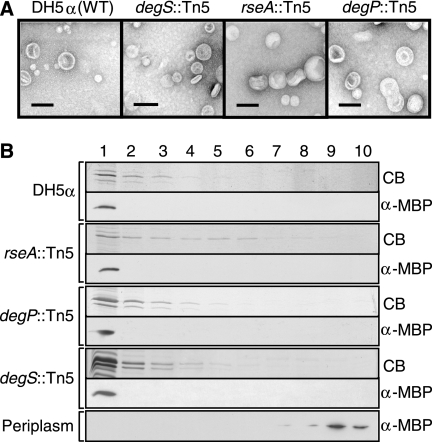
Vesicles produced by strains with mutations in σE response pathway genes are released intact. (A) Electron micrographs of vesicles from the wild type and mutant strains MK6D31, MK5B7, and MK11F26. Size bar, 100 nm. (B) Equilibrium density gradients of vesicles from the wild type and mutant strains (listed in the panel A legend). Periplasm from DH5α was applied to the same gradient (bottom panel). Fraction 1 is the least dense; fraction 10 is the most dense. Prominent vesicle protein bands are visible in light fractions by Coomassie blue (CB)-stained SDS-PAGE; MBP was visualized by immunoblotting (α-MBP). rseA::Tn5 samples were concentrated by precipitation with 20% trichloroacetic acid.
Disruption of genes in the σE pathway can affect the activity of the pathway in a variety of ways: certain disruptions result in low σE activity, while others cause no change or even hyperactivation of the σE response (1). Thus, it is interesting that all of the σE pathway mutants we obtained caused increased vesiculation. A detailed analysis of the σE pathway mutants supports the role of vesiculation as a general envelope stress response (A. J. McBroom and M. J. Kuehn, submitted for publication). Both impairment and hyperactivation of the σE pathway are likely to result in accumulation of material in the cell envelope, which we propose induces heightened vesiculation. When σE activation is prevented, accumulation results from a failure to process or degrade misfolded polypeptides, and in cases where the σE pathway is constitutively hyperactive, accumulation results from a large influx of σE-transcribed proteins to the envelope. It is possible that vesicle formation under stressing conditions occurs via a mechanism that differs from the typical vesiculation process.
Other vesiculation mutants discovered in this screen may also affect the σE pathway. Multicopy plasmid expression of a sequence mapping to the mtrA deaD nlpI pnp genomic region increases σE activity (17). In addition, LPS alterations such as those in the waaG mutants, or the loss of wecF function, which may be the case for the MK1F40 wzxE::Tn5 mutant, result in activation the σE envelope stress pathways (6, 19).
Disruptions in unclassified genes affecting vesiculation.
The remaining mutants encode uncharacterized or hypothetical components that have no known structural role or relationship to envelope stress (Table (Table1),1), and thus it is interesting to consider that their role in vesiculation may be mechanistically direct. For example, NlpI is an OM-anchored lipoprotein containing tetratricopeptide repeat motifs thought to contribute to protein complex formation (29). The cell envelope integrity of the two nlpI::Tn5 mutants is dissimilar, likely reflecting differences in the activities or stabilities of the two truncated products. YpjA, disrupted in MK4A31, is a hypothetical Omp (14) that could directly impact vesiculation. NlpA, a nonessential periplasmic lipoprotein tethered to the inner membrane (30), is also a potential candidate for direct involvement. Our observation of vesicle underproduction is the first phenotype reported for an nlpA mutant. Examining the effects of combining these mutations with mutations in other categories will yield further insight regarding the contribution of specific factors to vesicle production.
Summary.
We have identified 20 genes whose disruption causes altered vesiculation levels. Since our analysis was not genome-saturating, we presume that vesicle biogenesis is also influenced by additional loci that are currently undetermined. Our characterization of vesiculation mutants reveals that vesiculation is not a consequence of bacterial lysis or disintegration of the bacterial envelope and cannot be simply correlated with membrane instability. Few low-vesiculation mutants and no null-vesiculation mutants were identified in our screen, suggesting that vesiculation may be a process important in the growth of gram-negative bacteria. In addition to mutations that affected protein synthesis, localization, and envelope structure, we also discovered that vesiculation levels were altered by mutation of an envelope stress response pathway. Furthermore, we identified a number of envelope components that may affect vesiculation in a direct manner. Study of vesiculation phenotypes for corresponding mutations in other gram-negative bacteria will reveal whether vesicle production is based on a common mechanism.
Acknowledgments
This work was supported by a National Science Foundation Fellowship (to A.P.J.), NIGMS, NIAID, a Duke University Undergraduate Research Fellowship (to S.V.), an NIH graduate training grant, and a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease Award (to M.J.K.).
The authors thank S. Bauman, C. Lucaveche, and M. Reedy for electron microscopy assistance, P. Modrich for MutL antibody, M. Goulian for MDG131, S. Kushner for pWSK130, and B. Webster for technical advice.
REFERENCES
Articles from Journal of Bacteriology are provided here courtesy of American Society for Microbiology (ASM)
Full text links
Read article at publisher's site: https://doi.org/10.1128/jb.00498-06
Read article for free, from open access legal sources, via Unpaywall:
https://jb.asm.org/content/188/15/5385.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/101856103
Article citations
Released bacterial ATP shapes local and systemic inflammation during abdominal sepsis.
Elife, 13:RP96678, 20 Aug 2024
Cited by: 2 articles | PMID: 39163101 | PMCID: PMC11335348
Marine Delivery Vehicles: Molecular Components and Applications of Bacterial Extracellular Vesicles.
Mar Drugs, 22(8):363, 09 Aug 2024
Cited by: 0 articles | PMID: 39195479 | PMCID: PMC11355966
Review Free full text in Europe PMC
Budding and explosive membrane vesicle production by hypervesiculating Escherichia coli strain ΔrodZ.
Front Microbiol, 15:1400434, 20 Jun 2024
Cited by: 0 articles | PMID: 38966389 | PMCID: PMC11222570
Bacterial extracellular vesicles: biotechnological perspective for enhanced productivity.
World J Microbiol Biotechnol, 40(6):174, 20 Apr 2024
Cited by: 1 article | PMID: 38642254 | PMCID: PMC11032300
Review Free full text in Europe PMC
Extracellular vesicles in mycobacteria: new findings in biogenesis, host-pathogen interactions, and diagnostics.
mBio, 15(5):e0255223, 03 Apr 2024
Cited by: 0 articles | PMID: 38567992 | PMCID: PMC11077946
Review Free full text in Europe PMC
Go to all (225) article citations
Data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
NlpI-mediated modulation of outer membrane vesicle production through peptidoglycan dynamics in Escherichia coli.
Microbiologyopen, 4(3):375-389, 08 Mar 2015
Cited by: 70 articles | PMID: 25755088 | PMCID: PMC4475382
Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response.
Mol Microbiol, 63(2):545-558, 05 Dec 2006
Cited by: 419 articles | PMID: 17163978 | PMCID: PMC1868505
Modulation of bacterial outer membrane vesicle production by envelope structure and content.
BMC Microbiol, 14:324, 21 Dec 2014
Cited by: 97 articles | PMID: 25528573 | PMCID: PMC4302634
Regulated proteolysis: control of the Escherichia coli σ(E)-dependent cell envelope stress response.
Subcell Biochem, 66:129-160, 01 Jan 2013
Cited by: 37 articles | PMID: 23479440
Review
Funding
Funders who supported this work.
NIAID NIH HHS (1)
Grant ID: R21 AI063239
NIGMS NIH HHS (1)
Grant ID: R01 GM066140




