Abstract
Free full text

From the Cover
Structure of a heparin-dependent complex of Hedgehog and Ihog
Abstract
Hedgehog (Hh) signaling molecules mediate key tissue-patterning events during animal development, and inappropriate activation of Hh signaling in adults has been associated with human cancers. Recently, a conserved family of type I integral membrane proteins required for normal response to the Hh signal was discovered. One member of this family, Ihog (interference hedgehog), functions upstream or at the level of Patched (Ptc), but how Ihog participates in Hh signaling remains unclear. Here, we show that heparin binding induces Ihog dimerization and is required to mediate high-affinity interactions between Ihog and Hh. We also present crystal structures of a Hh-binding fragment of Ihog, both alone and complexed with Hh. Heparin is not well ordered in these structures, but a basic cleft in the first FNIII domain of Ihog (IhogFn1) is shown by mutagenesis to mediate heparin binding. These results establish that Hh directly binds Ihog and provide the first demonstration of a specific role for heparin in Hh responsiveness.
Hedgehog (Hh) is a secreted signaling molecule that mediates key tissue-patterning events during both vertebrate and invertebrate development (1–3). Hh also plays an important role in the maintenance and regulation of stem cells in adult organisms (4, 5). Abnormal activation of the Hh signaling pathway has been implicated in the initiation and growth of many human tumors (6), and drugs targeting the Hh pathway are under development (7).
Hh is secreted but undergoes two lipid modifications that restrict its free diffusion and facilitate transport to appropriate target sites (8, 9). Hh binding to Ptc, a 12-pass integral membrane protein with homology to bacterial resistance–nodulation–division (RND) transporters (10) is a central event in Hh signaling and blocks the ability of Ptc to inhibit the seven-pass integral membrane protein Smoothened (Smo), a positive regulator of Hh responses (10). Recent genetic and RNAi experiments implicate the membrane-associated proteins Ihog and dally-like protein (Dlp) in Hh responsiveness (11–14). Dlp, a member of the glypican family of heparan sulfate proteoglycans (HSPGs) (15), has dual roles in mediating Hh responsiveness and in transport of the Hh signal to distant cells (12–14). Heparan sulfate glycosaminoglycan chains, in particular, have also been implicated in Hh movement by the observation that tout velu, which encodes a heparan sulfate copolymerase (16, 17) that acts on Dlp (12), is required for normal Hh transport.
Ihog, a Drosophila protein previously known as CG9211, is a type I transmembrane protein with four Ig domains followed by two FNIII domains, a membrane-spanning region, and a cytoplasmic region of no known function (11). Ihog is homologous to another Drosophila protein, CG32796 or brother of Ihog (BOI), and two mammalian proteins, CDO and BOC (18, 19), that are also components of the Hh signaling pathway (11, 20, 21). Several observations indicate that Ihog may function as a coreceptor for Hh: (i) reduction of Ihog expression results in diminished Hh binding and responsiveness, (ii) epistasis experiments place Ihog function upstream or at the level of Ptc, (iii) the Ihog extracellular region is able to pull down HhN from conditioned medium, and (iv) coexpression of Ptc and Ihog results in a synergistic increase in Hh binding to the cell surface (11).
The first FNIII domain of Ihog (IhogFn1) is necessary and sufficient to pull down HhN, but both FNIII domains are needed to synergize with Ptc and reconstitute Ihog function in cell-based signaling assays (11). Complicating interpretation of Ihog function is the observation that purified HhN and Ihog extracellular regions do not appreciably interact (see Fig. 2A), indicating that a binary interaction between Ihog and Hh cannot explain Ihog function. To investigate the role of Ihog in Hh signaling, we initiated structural and biophysical studies of functional fragments of Drosophila melanogaster Ihog and Hh.
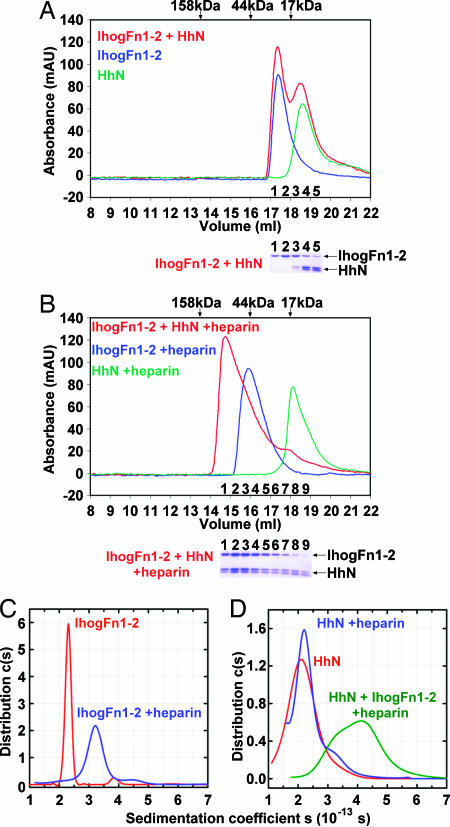
IhogFn1–2 forms a complex with HhN in the presence of heparin. (A) Size-exclusion chromatography elution profiles of HhN (green), IhogFn1–2 (blue), and IhogFn1–2 combined with HhN (red). Elution volumes of molecular weight standards are indicated with arrows above the chromatograms. Below is a Coomassie blue-stained SDS/PAGE analysis of IhogFn1–2+HhN fractions. HhN appears as a doublet, owing to partial proteolysis. (B) Same as A, except that each solution now contains heparin decasaccharide. Below the chromatograms is a Coomassie blue-stained SDS/PAGE analysis of fractions from the IhogFn1–2+HhN+heparin run. (C) Sedimentation coefficient distributions c(s20,w) calculated for IhogFn1–2 alone at 75 μM (red) and at 80 μM in the presence of 1 equivalent of heparin decasaccharide (blue). (D) Sedimentation coefficient distributions c(s20,w) for 29 μM HhN (red), 29 μM HhN in the presence of 1 equivalent of heparin decasaccharide (blue), and a mixture of 10 μM HhN, 10 μM IhogFn1–2, and 50 μM heparin decasaccharide (green).
Results and Discussion
Structure of Ihog FNIII Domains.
The crystal structure of IhogFn1 was determined by multiwavelength anomalous diffraction and refined to 1.35-Å resolution (Table 2, which is published as supporting information on the PNAS web site). The crystal structure of a tandem repeat of both Ihog FNIII domains (IhogFn1–2) was then determined by molecular replacement and refined to 2.4-Å resolution (Table 1; and see Table 3, which is published as supporting information on the PNAS web site). Because the IhogFn1 structure is essentially identical in both crystals (rmsd of 0.63 Å for 108 shared Cαs), subsequent discussion will focus on the IhogFn1–2 structure. Both FNIII repeats in IhogFn1–2 adopt the canonical β-sandwich topology of FNIII domains (22) but fold back on one another in an unusual “horseshoe-like” arrangement (Fig. 1A). This domain arrangement has not been observed in previous multiFNIII repeat structures (23, 24), but a similar arrangement has been observed for repeated Ig-like β-sandwich domains in axonin-1 (25) and hemolin (26).
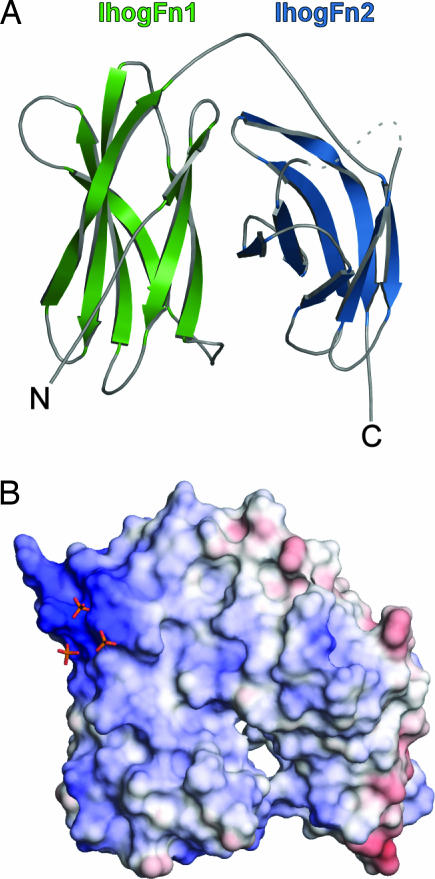
IhogFn1–2 adopts a horseshoe-like structure. (A) Ribbon diagram of IhogFn1–2. IhogFn1 is colored green, and IhogFn2 is colored light blue. A disordered loop is indicated by the dashed line. (B) An electrostatic potential surface of IhogFn1–2, shown in the same orientation as in A. The scale is calibrated to −12 kT/e and +12 kT/e for red and blue, respectively. Three sulfate ions are shown as orange and red sticks. All structure images were generated with PyMOL (http://pymol.sourceforge.net).
Table 1.
Crystallographic refinement statistics
| IhogFn1–2 | IhogFn1–2/HhN | |
|---|---|---|
| Resolution, Å | 50–2.4 | 50–2.2 |
| Rwork/Rfree, % | 24.4/27.5 | 19.8/24.6 |
| Average protein B-factor | 38.4 | 39.7 |
| rmsd bond lengths, Å | 0.008 | 0.015 |
| rmsd bond angles,° | 1.66 | 1.46 |
The interdomain interaction at the core of the IhogFn1–2 structure buries 1,110 Å2 of surface area and exhibits a high degree of shape complementarity (0.74), values consistent with a stable interface (27, 28). Additionally, when two threonines, T534 and T630, with side chains that form a hydrogen bond across the interdomain interface, are substituted with cysteine, a disulfide bond is formed, as judged by altered mobility of the variant protein in SDS/PAGE experiments (data not shown). Introduction of this disulfide bond, which fixes the protein in the horseshoe conformation, has no effect on Ihog function in cell-based signaling assays, indicating that this conformation represents a biologically active form of the protein (Fig. 4A).
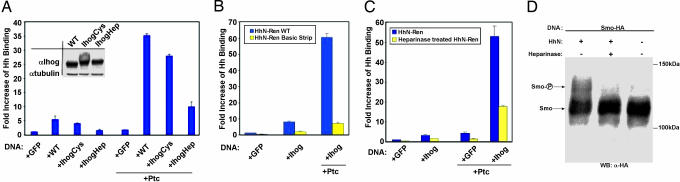
Heparin is required for Ihog function and Hh responsiveness. (A) Effects of Ihog mutations on HhN binding to Ihog-expressing Drosophila cultured cells. (Inset) Western blot of Ihog protein from transfected cells used in the binding assay. (B) Effects of HhN basic strip mutations on HhN binding to Ihog-expressing Drosophila cultured cells. (C) The ability of HhN to bind to cells expressing Ihog is diminished after treatment of the HhN-containing conditioned medium with heparinase. (D) Western blot of HA-tagged Smo from Ihog-expressing cells incubated with HhN-containing conditioned medium. Smo phosphorylation is decreased after heparinase treatment of the medium.
IhogFn1 and IhogFn1–2 Bind Heparin.
A second striking feature of the IhogFn1–2 structure is a localized region of positive charge that encompasses seven basic residues on IhogFn1 (R503, K507, R508, K509, R547, R549, and K567). This region is conserved in invertebrate and vertebrate Ihog homologs (Fig. 6, which is published as supporting information on the PNAS web site), and multiple sulfate ions are bound to this site in both the IhogFn1 and IhogFn1–2 crystals (Fig. 1B). The conservation of this basic region and the presence of bound sulfate ions led us to test whether IhogFn1–2 binds heparin, a highly sulfated polysaccharide similar to the heparan chains of HSPGs, which are known to play a role in Hh signaling (17). Pull-down experiments and heparin-column chromatography show that both IhogFn1 and IhogFn1–2 bind heparin-agarose and elute at >800 mM NaCl (Fig. 7, which is published as supporting information on the PNAS web site, and data not shown).
Heparin Promotes Interactions Between IhogFn1–2 and HhN.
The observation that IhogFn1–2, like HhN, binds heparin suggested that a requirement for heparin might explain the failure of purified HhN and IhogFn1–2 to interact, as judged by size-exclusion chromatography (Fig. 2A). Pull-down experiments, which previously detected an interaction between HhN and IhogFn1, were carried out in heparin-containing medium (11). When size-exclusion chromatography experiments with HhN alone, IhogFn1–2 alone, and a mixture of HhN and IhogFn1–2 were repeated in the presence of a molar excess of heparin decasaccharide, two new interactions were observed. Heparin decasaccharide produced a shift in the elution time of IhogFn1–2 alone that is indicative of dimer formation (Fig. 2B), and mixtures of HhN and IhogFn1–2 shifted to faster eluting fractions, consistent with formation of complexes having HhN/IhogFn1–2 stoichiometries of up to 2:2 (Fig. 2B). Heparin is thus not only essential for high-affinity interactions between purified HhN and IhogFn1–2 but also promotes IhogFn1–2 dimerization.
Analytical ultracentrifugation (AUC) experiments with HhN, IhogFn1–2, and heparin were carried out to investigate the strength, stoichiometry, and potential cooperativity of IhogFn1–2 and HhN complexes. Sedimentation velocity and sedimentation equilibrium experiments show that IhogFn1–2 alone is monomeric in solution but exists in a monomer–dimer equilibrium, with dimerization constants of 60 ± 5 μM and 430 ± 10 μM in the presence of heparin decasaccharide and hexasaccharide, respectively (Fig. 2C; and see Fig. 8, which is published as supporting information on the PNAS web site). Similar experiments with HhN in the presence of heparin oligosaccharides show no evidence for HhN oligomerization (Fig. 2D). The ubiquitous presence of heparan sulfate and high effective concentrations of membrane proteins at the cell surface suggest that Ihog and its homologs are likely to be constitutively dimerized on the cell surface, consistent with an earlier observation that vertebrate Ihog homologs coimmunoprecipitate from cell lysates (18).
AUC experiments with mixtures of HhN and IhogFn1–2 at 20 μM total protein and in the presence of excess heparin decasaccharide also demonstrate the formation of HhN/IhogFn1–2 complexes with a maximal 2:2 stoichiometry (Fig. 2D; and see Fig. 9 and Table 4, which are published as supporting information on the PNAS web site). By modeling the expected AUC results for given distributions of HhN/IhogFn1–2 complexes, equilibrium constants that best fit the data were obtained. Slight cooperativity between various subcomplexes is required to achieve good fits to the experimental data, but all well fit models yield values of dissociation constants for HhN/IhogFn1–2 interactions in the 0.4–8.0 μM range (see Materials and Methods).
Hh Binds in a Cleft on IhogFn1.
A complex of IhogFn1–2 and HhN formed in the presence of heparin decasaccharide crystallized in 2.0 M Na,K-phosphate, and these crystals diffracted x-rays to 2.2-Å resolution. The structure of this complex was determined by molecular replacement and refined to final values of 0.198 and 0.246 for Rcryst and Rfree, respectively (Fig. 3A and Tables 1 and 3). Two essentially identical 2:2 complexes of HhN/IhogFn1–2 are present in the asymmetric unit. The structures of IhogFn1–2 and HhN within this complex are fundamentally unchanged relative to their unbound conformations; superposition of complexed and uncomplexed forms of IhogFn1–2 results in an rmsd of 1.24 Å for 208 shared Cαs, and superposition of the complexed form of HhN with an uncomplexed form of murine Sonic HhN (66% amino acid sequence identity) (29) results in an rmsd of 0.68 Å for 142 shared Cαs. HhN domains are homologous to prokaryotic zinc hydrolases (29), but, unlike Sonic HhN, no evidence for a bound metal ion is found in the HhN electron density. This result is not unexpected, because two key metal-coordinating residues are not conserved in HhN, and the signaling ability of HhN does not appear to rely on an intrinsic catalytic activity (30).
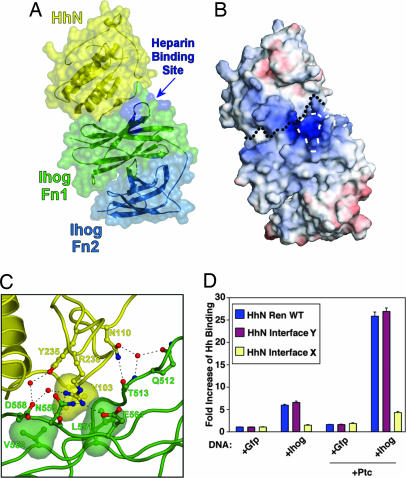
HhN binds to a cleft on IhogFn1. (A) Semitransparent molecular surface of the HhN/IhogFn1–2 complex superimposed on a ribbon diagram of the molecules. HhN is colored yellow, IhogFn1 is green, and IhogFn2 is light blue. The four residues that when mutated lead to loss of heparin binding are colored dark blue. (B) Electrostatic potential surface of the HhN/IhogFn1–2 complex, shown in the same orientation as in A. The black dotted line marks the boundary between HhN and IhogFn1–2. The four residues that when mutated lead to loss of heparin binding are outlined with white dashes. The color scale is calibrated to −12 kT/e and +12 kT/e for red and blue, respectively. (C) HhN/IhogFn1–2 interface. The four Hh residues mutated in the HhN/Ihog-interface 1 mutant are represented as balls and sticks, as are nearby IhogFn1–2 residues. Bridging waters are represented by red spheres and hydrogen bonds are shown as dashed lines. (D) Effects of HhN-interface mutations on HhN binding to Ihog-expressing Drosophila cultured cells.
Two different contacts between HhN and IhogFn1–2 are observed in the crystal lattice, one mediated by IhogFn1, and another mediated primarily by IhogFn2, but mutagenesis experiments clearly identify the interface restricted to IhogFn1 as physiologically relevant (Fig. 3 A and D). Restriction of the Ihog/Hh interface to IhogFn1 accounts for experiments showing that IhogFn1 alone is sufficient both to pull down HhN from conditioned media (11) and to bind HhN in the presence of heparin, as judged by gel filtration (data not shown). The IhogFn1 and HhN interface buries ≈1,180 Å2 and has a shape complementarity parameter of 0.70, values comparable to those of known biological interfaces, and consists of a core of three hydrophobic residues surrounded by predominantly polar interactions (Fig. 3C). The central residue of the hydrophobic contact is V103 of HhN, which nestles between L551 and V553 of Ihog. Another key residue is R238 of HhN, which makes four hydrogen bond or salt-bridge contacts to three Ihog residues, one through a water molecule. A majority of the Ihog residues at the HhN/IhogFn1–2 interface, including R238 and V103 of HhN and V553 of Ihog, are conserved among invertebrate homologs but not among vertebrate homologs (Fig. 10, which is published as supporting information on the PNAS web site). This conservation pattern is consistent with pull-down assays, which indicate that the FNIII domain primarily responsible for mediating interactions with Hh has shifted from IhogFn1 in Drosophila (invertebrate) to the domain homologous to IhogFn2 in CDO and BOC (vertebrate) (11, 20).
Heparin Is Required for Ihog Function and Hh Responsiveness.
Although present in the crystallization buffer, no evidence for well ordered heparin is apparent at or near the HhN/IhogFn1–2 interface. This absence may stem from the presence of 2.0 M phosphate and 0.2 M sulfate ions in the crystallization buffer, which are likely to compete for heparin binding. An electrostatic potential map of the complex does, however, reveal that the conserved region of positive charge on IhogFn1 noted earlier is adjacent to the HhN/IhogFn1–2 interface and is contiguous with a strip of positive charge on HhN (Fig. 3B). This observation, coupled with the absence of conformational changes in HhN or IhogFn1–2 that could arise from allosteric effects, suggests that heparin promotes HhN/IhogFn1–2 interactions by binding both proteins and bridging this interface.
To verify that this positively charged strip is responsible for mediating heparin binding and to assess the role of heparin binding in Ihog function, basic amino acids in this strip in both Ihog (R503, K507, K509, and R547) and Hh (K105, R147, R213, and R239) were mutated to glutamate. The IhogFn1–2 variant (IhogFn1–2Hep) failed to bind heparin Sepharose (data not shown), and full-length Ihog with the four IhogFn1–2Hep substitutions failed to support normal Hh binding in cell-based assays (Fig. 4A). Similarly, the Hh variant (HhN-Ren Basic Strip) showed a dramatic reduction in its ability to bind to cells overexpressing Ihog (Fig. 4B). These data demonstrate that the positively charged region of IhogFn1 is important for both heparin and HhN binding. Although the IhogFn1–2Hep substitutions are adjacent to the HhN/IhogFn1–2 interface, they do not form any part of the contact surface, and their influence on HhN binding is almost certainly indirect through interference with heparin binding. The fact that mutating basic residues on HhN that are adjacent to but not within the HhN/IhogFn1–2 interface also greatly decreases HhN binding to the cell surface supports the likelihood that heparin promotes HhN/Ihog interactions by directly binding and spanning the HhN/Ihog complex.
The importance of heparin for mediating interactions between HhN and cell-surface components was further verified by performing cell-binding experiments after heparinase treatment (Fig. 4C). Incubation of HhN-conditioned medium with heparinase reduced HhN binding to Ihog- and/or Ptc-overexpressing cells by ≈3-fold, indicating that heparin contributes significantly to high-affinity interactions between Hh and its receptor complex in this assay. To investigate whether the heparin-dependence of HhN binding to cells translates into a loss of Hh responsiveness, cells transfected with HA-tagged Smo were incubated with HhN-conditioned medium that was pretreated with heparinase (Fig. 4D). HhN-dependent Smo phosphorylation is largely absent after heparinase treatment, indicating that heparin is required for normal pathway activation. A role for heparin in vertebrate Hh responsiveness appears to be conserved, with HSPGs having been shown to exert both positive and negative effects on Shh activity (31).
IhogFn1–2 Forms a Symmetric Dimer.
Because both gel-filtration and AUC experiments indicated the formation of IhogFn1–2 dimers, the HhN/IhogFn1–2 crystal lattice was inspected for IhogFn1–2 contacts. Two possible Ihog/Ihog dimers were observed. One dimer is mediated primarily by regions on IhogFn1 (Fig. 5) and another primarily by IhogFn2. An IhogFn1–2 variant bearing amino acid substitutions designed to distinguish between these potential dimer interfaces proved insoluble and unsuitable for further studies, but two observations suggest that the interface mediated primarily by IhogFn1 is responsible for the Ihog dimerization observed in the presence of heparin: (i) IhogFn1 alone dimerizes in the presence of heparin (Fig. 11, which is published as supporting information on the PNAS web site), and (ii) the IhogFn1-mediated interface results in a self-limited dimer in contrast to the predominantly IhogFn2-mediated interface, which would allow extended filaments. No evidence for IhogFn1–2 complexes larger than dimers is observed in either AUC or size-exclusion chromatography experiments (Fig. 2).
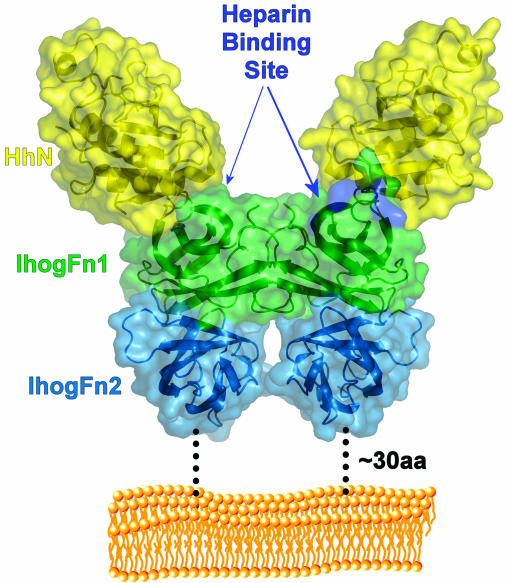
The 2:2 HhN/IhogFn1–2 complex. Model of the HhN/IhogFn1–2 complex relative to the cell surface. The molecules are shown as ribbons inside semitransparent molecular surfaces. Dotted lines represent the ≈30-aa linker that exists between the end of IhogFn2 and the transmembrane domain. A phospholipid bilayer is shown in orange.
Electron density for a sulfated monosaccharide is present in a crevice formed between subunits in the IhogFn1-mediated dimer (Fig. 12, which is published as supporting information on the PNAS web site), but the precise nature of this monosaccharide and whether it is part of a larger disordered polysaccharide are unclear. No atomic model has thus been fit to this density, but its position between IhogFn1–2 subunits may explain the formation of IhogFn1–2 dimers in the presence of heparin. This site is distinct from the heparin-binding site identified by charge and mutagenesis, and modeling suggests that it is unlikely that a single heparin decasaccharide could contact both sites simultaneously.
Concluding Remarks.
The HhN/IhogFn1–2 crystal structure reveals a 2:2 complex that supporting experiments indicate is likely to represent a physiological complex (Fig. 5). In this complex, each Hh molecule contacts only a single Ihog molecule, and a pair of 1:1 Hh/Ihog complexes forms an entirely Ihog-mediated dimer to produce the 2:2 complex. This arrangement is consistent with the minimal cooperativity observed between Hh/Ihog interactions and Ihog dimerization. Ihog dimers are heparin-dependent and probably constitutive on the cell surface, which may facilitate binding to multimeric forms of Hh through an increase in avidity (32, 33). The observed 2:2 complex is able to accommodate four Ig domains at the IhogFn1–2 N terminus without introducing clashes, and ≈30 aa intervene between this complex and the cell membrane, rendering its orientation relative to the membrane uncertain.
Heparin is also required to mediate IhogFn1–2/HhN interactions and appears to do so by binding to basic regions on Hh and Ihog that are adjacent in the Hh/Ihog complex. The dependence of both this interaction and Ihog dimerization on heparin provides a direct mechanism by which Hh responsiveness could be modulated by changes in the availability or nature of heparin or HSPGs. Heparan sulfate and HSPGs have been indirectly implicated in many signaling pathways, but this result demonstrates a direct role for heparin in mediating a high-affinity receptor–ligand interaction, reminiscent of the role of heparin in mediating interactions between fibroblast growth factor and its receptors (34).
The crystal structures presented here reveal that no changes in IhogFn1–2 conformation or oligomerization state occur after HhN binding, and it has been demonstrated that the cytoplasmic domain of Ihog is dispensable for Hh signaling (11). Taken together, these data suggest that a primary role of Ihog is to bind HhN and that signaling must be coordinated with other molecules. Several observations indicate that Ihog and Ptc behave synergistically with respect to HhN binding and signaling (11), and the simplest model explaining all of these data is that HhN forms a ternary complex with Ihog and Ptc, and this complex has a greater affinity for HhN than either Ihog or Ptc alone. The HhN/IhogFn1–2 complex characterized here provides a concrete basis for testing this and other models of the role of Ihog in Hh signaling.
Materials and Methods
More detailed descriptions may be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Cloning, Expression, and Purification of Hh and Ihog Fragments.
DNA fragments encoding D. melanogaster Ihog residues 466–577 (IhogFn1) and 466–679 (IhogFn1–2) were amplified by PCR and cloned into pT7HMT (35). Proteins were expressed in Escherichia coli strain B834(DE3) and purified by immobilized metal ion chromatography (IMAC), cation-exchange chromatography, and gel filtration.
A fragment of the D. melanogaster Hh gene encoding residues 85–248 (HhN) was subcloned into the pT7HMT vector. Expression and purification were carried out similarly to IhogFn1–2, except that solutions contained β-mercaptoethanol to avoid aggregation.
The IhogFn1–2 heparin-binding mutant (IhogFn1–2Hep) was created by using megaprimer PCR mutagenesis. Briefly, the substitutions R503E, K507E, K509E, and R547E were introduced during three rounds of PCR, and the mutated gene was subcloned back into pT7HMT as described above. A similar approach was used to create the IhogFn1–2Cys mutant, which contains the T534C and T630C substitutions.
For binding assays, protein expression in Drosophila cell culture was driven by the Actin 5C promoter. Expression vectors directing expression of Ptc, Dlp, and Renilla luciferase-tagged HhN were generated as described (13, 36). The HhN/Ihog interface mutants were created by using megaprimer PCR mutagenesis. The interface 1 mutant contained four HhN substitutions (V103A, N110A, Y235F, and R238A), and the interface 2 mutant contained three HhN substitutions (D191A, H193A, and Q196A).
Crystallization.
Native IhogFn1–2 was crystallized by vapor diffusion in hanging drops. Crystals were grown by mixing 1 μl of protein at 6 mg/ml in double-distilled (dd)H2O with 1 μl of reservoir buffer (50 mM Tris·HCl, pH 7.9, 23% wt/vol PEG 3350, and 200 mM (NH4)2SO4) diluted 1:1 with ddH2O. The protein crystallized in space group P212121 with unit cell dimensions a = 39.38, b = 65.24, and c = 84.21 Å.
Purified IhogFn1–2 and HhN in 20 mM Tris, pH 8.0, and 200 mM NaCl were mixed at a 2:1 molar ratio, respectively, in the presence of β-mercaptoethanol and Tris(2-carboxyethyl)phosphine (TCEP). The mixture was concentrated to ≈6 mg/ml total protein to which an amount of heparin decasaccharide equimolar with IhogFn1–2 was added. Before each tray was set up, the complex solution was diluted to ≈3 mg/ml with fresh 20 mM Tris, pH 8.0, 200 mM NaCl, and 2 mM TCEP. Initial crystals were grown by the vapor-diffusion method in hanging drops at 20°C, by mixing 1 μl of diluted protein complex solution with 1 μl of reservoir buffer (2.0 M Na,K-phosphate pH 6.4, 200 mM LiSO4, and 50 mM CAPS, pH 10.6). The complex crystallized in space group P21 with unit cell dimensions a = 75.35, b = 70.00, and c = 155.70 Å, with β = 90.18°.
Data Collection and Structure Determination.
Diffraction data from native IhogFn1–2 crystals were collected at beamline 14-BM-C of the Advanced Photon Source at Argonne National Laboratory, Argonne, IL. Diffraction data from native IhogFn1–2/HhN complex crystals were collected at beamline X4A of the National Synchrotron Light Source at Brookhaven National Laboratory, Upton, NY. Details of the structure determinations can be found in Supporting Materials and Methods. Data collection and refinement statistics are presented in Table 3, and a complete description of the asymmetric unit for each structure is provided in Table 5, which is published as supporting information on the PNAS web site.
Heparin Pull-Down Assay.
Purified protein (5 μg) was added to 1 ml of PBS with Tween 20 (PBS-T) and 20 μl of heparin-agarose slurry (Sigma) and incubated for 1 h at room temperature. The resin was pelleted by centrifugation at 5,000 × g for 30 s, and the supernatant was discarded. The resin was washed three times with PBS-T and heated to 100°C for 5 min in Laemmli loading buffer. Bound proteins were resolved by SDS/PAGE and stained with Coomassie blue.
In Vivo Binding Assay.
Drosophila S2-R+ cells in 6-well plates were transfected by using Fugene 6 (Roche, Indianapolis, IN). Forty-eight hours after transfection, cells were washed once with cold culture medium (or twice with serum-free culture medium for heparinase-treatment experiments) and incubated at 4°C for 1 h with conditioned medium containing HhN-Ren proteins. After extensive washing, cells were lysed, and Renilla luciferase activity was measured. Protein expression levels for different Ihog variants were determined for identically transfected cells by Western blot using a rat polyclonal antibody as described (11).
Heparinase Treatment.
Conditioned medium containing HhN-Ren protein was incubated with heparinase (Seikagaku Corporation, Tokyo, Japan) at 37°C for 2 h. Heparinase-treated conditioned medium was cooled down to 4°C before addition to S2-R+ cells.
HhN-Induced Smo Phosphorylation.
Drosophila S2-R+ cells, which produce endogenous Ihog, were plated in 24-well plates and transfected for expression of Smo-HA. Forty-eight hours after transfection, cells were washed twice with serum-free culture medium and incubated at 25°C for 4 h with conditioned medium containing HhN protein (either heparinase-treated medium or untreated medium) or control medium without HhN protein. After washing with PBS, cells were lysed, and Smo phosphorylation was measured by Western blot using rat polyclonal antibody for HA (Roche).
Analytical Ultracentrifugation.
All samples were prepared in PBS (pH 7.4) by using protein stocks having an A280 of ≈3.0 and were handled on ice or at 4.0°C. Sedimentation-equilibrium studies measuring the interactions of IhogFn1–2 and IhogFn1 with heparin were conducted at 4.0°C on an Optima XL-A analytical ultracentrifuge (Beckman, Fullerton, CA). Samples were studied at rotor speeds of 10, 14, 18, 22, and 26 thousand rpm, and data were acquired at a wavelength of 280 nm or 294 nm. Data collected in the presence of one equivalent of sucrose octasulfate (SOS), heparin hexasaccharide, or decasaccharide, were analyzed globally in terms of a reversible monomer–dimer equilibrium of the 1:1 IhogFn1–2/heparin complex.
Sedimentation equilibrium studies measuring the interaction between HhN and IhogFn1–2 were conducted at 4.0°C and 280 nm at rotor speeds of 10, 12, 14, 16, and 18 thousand rpm. HhN data, with and without one equivalent of heparin decasaccharide, were collected at 10, 20, and 30 μM. HhN/IhogFn1–2 mixtures were studied at three different loading ratios (9.3:9.7 μM, 13.1:7.0 μM and 5.8:12.1 μM) in the presence of 50 μM heparin decasaccharide.
Sedimentation velocity experiments were conducted in duplicate at 4.0°C on a Beckman Optima XL-A analytical ultracentrifuge. Samples (loading volume of 300–350 μl) were studied at a loading absorbance (λ = 280 or 294 nm) of ≈1.0. One hundred to 120 scans were acquired at 58,000 rpm by using the shortest time delay possible. Molecular modeling used structural data for IhogFn1–2 to calculate the sedimentation coefficient based on a horseshoe model with the program HYDROPRO Version 5A (37). An extended form of IhogFn1–2 was modeled in a similar fashion, as were various IhogFn1–2 and HhN complexes.
Size-Exclusion Chromatography.
Mixtures of IhogFn1–2 (63 μM), D. melanogaster Hedgehog N-terminal domain (HhN) (62 μM), or heparin decasaccharide (190 μM) were adjusted to 260 μl with PBS and incubated for 45 min at room temperature. Approximately 200 μl of the solution were then passed through a Superdex 200 10/300 column (GE Healthcare, Buckinghamshire, U.K.) equilibrated in PBS at 4°C. Heparin oligosaccharides were purchased from Neoparin (Alameda, CA).
Acknowledgments
We thank the beamline staffs of X4A at the National Synchrotron Light Source and 14-BM-C at the Advanced Photon Source for assistance with x-ray data collection and Wei Yang and Samuel Bouyain for a critical reading of the manuscript. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (to R.G.). D.J.L is supported by grants from NIH and the Department of Defense, and P.A.B. is an investigator of the Howard Hughes Medical Institute. J.S.M. is supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2IBB, 2IBG, and 2IC2).
See Commentary on page 17069.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.0606738103
Read article for free, from open access legal sources, via Unpaywall:
http://www.pnas.org/content/103/46/17208.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics

Discover the attention surrounding your research
https://www.altmetric.com/details/102447727
Article citations
Identification of small molecule antagonists of sonic hedgehog/heparin binding with activity in hedgehog functional assays.
Biochim Biophys Acta Gen Subj, 1868(11):130692, 14 Aug 2024
Cited by: 0 articles | PMID: 39151833
Hedgehog on the Move: Glypican-Regulated Transport and Gradient Formation in <i>Drosophila</i>.
Cells, 13(5):418, 27 Feb 2024
Cited by: 1 article | PMID: 38474382 | PMCID: PMC10930589
Review Free full text in Europe PMC
The role of glycosaminoglycan modification in Hedgehog regulated tissue morphogenesis.
Biochem Soc Trans, 51(3):983-993, 01 Jun 2023
Cited by: 5 articles | PMID: 37222258 | PMCID: PMC10317163
Review Free full text in Europe PMC
Cellular and molecular mechanisms of Hedgehog signalling.
Nat Rev Mol Cell Biol, 24(9):668-687, 17 Mar 2023
Cited by: 42 articles | PMID: 36932157
Review
Hedgehog is relayed through dynamic heparan sulfate interactions to shape its gradient.
Nat Commun, 14(1):758, 10 Feb 2023
Cited by: 4 articles | PMID: 36765094 | PMCID: PMC9918555
Go to all (62) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Protein structures in PDBe (3)
-
(1 citation)
PDBe - 2IBBView structure
-
(1 citation)
PDBe - 2IC2View structure
-
(1 citation)
PDBe - 2IBGView structure
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla.
Nature, 455(7215):979-983, 14 Sep 2008
Cited by: 111 articles | PMID: 18794898 | PMCID: PMC2679680
The ihog cell-surface proteins bind Hedgehog and mediate pathway activation.
Cell, 125(2):343-357, 01 Apr 2006
Cited by: 152 articles | PMID: 16630821
Competitive coordination of the dual roles of the Hedgehog co-receptor in homophilic adhesion and signal reception.
Elife, 10:e65770, 18 May 2021
Cited by: 6 articles | PMID: 34003115 | PMCID: PMC8131103
Interactions between Hedgehog proteins and their binding partners come into view.
Genes Dev, 24(18):2001-2012, 01 Sep 2010
Cited by: 136 articles | PMID: 20844013 | PMCID: PMC2939362
Review Free full text in Europe PMC
Funding
Funders who supported this work.




